Abstract
The cytokines interleukin 7 (IL-7) and interleukin 4 (IL-4) regulate lymphoid differentiation and function and activate the transcription factor Stat5. Using mice deficient for the 2 highly related transcription factors, Stat5a and Stat5b (Stat5a/b−/−), we investigated the role of Stat5 for B-cell differentiation, expansion, and function. Peripheral blood B cells of Stat5-deficient mice are significantly reduced, but no proliferation defects in response to various mitogenic stimuli are found. Also, IgM and IgG1 antibody production and immunoglobulin class switching are not affected. Pre- and pro-B cells of Stat5-deficient animals were found to have reduced responses to IL-7. Pro- and pre-B cells are the target cells of the abloncogene and numerous studies have suggested that Stat5a/b is essential for transformation by derivatives of the Abelson(abl) gene. To assess the role of Stat5a/b in transformation, we have evaluated the ability of variousabl derivatives to transform cells from Stat5a/b-deficient mice in vitro or in vivo. We demonstrate that the absence of Stat5a/b is not essential for the induction of lymphoid or myeloid tumors in vivo or on the ability to transform bone marrow cells in vitro.
Introduction
The cytokines interleukin 7 (IL-7) and interleukin 4 (IL-4) regulate important aspects of lymphoid development and function and both activate the highly related transcription factors Stat5a and Stat5b in B and T cells.1,2 In addition, strong Stat5a/b activation was found in leukemias and has been hypothesized to be essential for the transformation process induced by derivatives of the Abelson (abl) oncogene.3-5 In these studies we addressed the importance of Stat5a/b activation by IL-7, IL-4, or transforming protein tyrosine kinases using Stat5-deficient mice.6
Disruption of the IL-7 gene and its high affinity receptor (IL-7R) results in a dramatic loss of T- and B-cell precursors in the bone marrow.7-9 Developing B cells undergo distinct IL-7–dependent stages of expansion characterized by the expression of specific surface markers (classified by Hardy et al10 and Osmond et al11). Disruption of the IL-4 gene has shown its importance in mature B-cell proliferation and immunoglobulin class switching toward IgG1 and IgE as well as T helper 2 cell differentiation and T-cell proliferation.12,13 IL-4 activates Stat5a, Stat5b, and Stat6 and we have previously shown that Stat5-deficient T cells cannot proliferate in response to IL-2 or IL-4.14-16 Stat6-deficient mice have a block in T helper 2 cell differentiation and immunoglobulin class switching toward IgE, but B- and T-cell proliferation in response to IL-4 was not drastically reduced and immunoglobulin class switching toward IgG1 was unaltered.17 Lastly, IL-2 has been implicated in IgM production through Stat5a/b.18 19 Therefore, we addressed the question of whether B-cell development, proliferation, or immunoglobulin production and class switching is impaired in Stat5a/b-deficient mice.
B-cell precursors are the target cells of the abl oncogene and IL-7 can reconstitute multiple aspects of v-abl–mediated signaling, among which the activation of Stat5a/b is considered essential.20-23 Theabl gene was initially identified as a transforming gene transduced into a Moloney leukemia virus (Ab-MuLV) and capable of producing B-cell tumors in mice.24 Subsequently, it was identified as a gene that fused with the break point cluster gene (bcr) in leukemias containing a (9;22) translocation termed the Philadelphia chromosome, which is associated with chronic myeloid leukemia (CML), although it occurs also in acute lymphocyte leukemia (ALL).25,26 In both cases, the Abl protein is truncated and has increased protein kinase activity that is required for its transforming activity. A variety of substrates are phosphorylated in v-abl or bcr-abltransformed cells. Among the substrates that have been described as strongly activated and potentially critical for abl-induced transformation are Stat5a/b.3-5,20-23 27-30 Hence, we tested this widely held hypothesis by assessing the ability of v-abl or bcr-abl to induce disease in Sta5a/b-deficient mice.
Materials and methods
In vitro transformation assays
To determine numbers of growth factor–independent colonies induced by MSCV-bcr-abl p210/IRES-GFP and MSCV-bcr-abl p185/IRES-GFP, bone marrow from Stat5a/b(+/−) and (+/+) versus Stat5a/b (−/−) mice was cocultivated for 48 hours with GP+E86 producer cell lines in the presence of 6 μg/mL polybrene, IL-6 (50 ng/mL), IL-3 (25 ng/mL), and stem cell factor (SCF) (50 ng/mL). Three days later cells were harvested, washed, and plated in methylcellulose at a density of 1 × 105 cells/mL. Cells that were cocultivated with GP+E86 producer cells infected with the empty vector were used as negative control. The rate of infection of control and Stat5-deficient bone marrow was found comparable (10% to 20% in individual experiments). For Ab-MuLV infections, bone marrow was harvested, infected for 30 minutes with viral supernatant derived from A010 cells (a generous gift from Dr N. Rosenberg), enriched with 5 ng/mL IL-7 and 7 μg/mL polybrene and β-mercaptoethanol. The transformed, as well as the control, mock infected cells were maintained for 36 hours on IL-7, producing feeder layers before cloning in methylcellulose. Mock infected cells were used as negative controls and did not result in growth factor–independent colonies. IL-7–dependent colony assays were performed using methylcellulose containing IL-7 (Stemcell Technology, Vancouver, British Columbia, Canada) as described in Teglund et al.6
Determination of absolute B-cell numbers in the peripheral blood
The number of white blood cells of whole blood was determined using a Coulter Counter. Blood smears were then analyzed using light microscopy to determine the percentage of lymphocytes. At least 100 white blood cells were counted. The percentage of lymphocytes was then multiplied by the total white blood cell count to determine the absolute number of lymphocytes per microliter (ALC). The percentage of B and T cells within the lymphocyte population was then determined by FACS, and these percentages were multiplied by the ALC to determine the absolute number of B cells per microliter.
[3H]-thymidine incorporation assays
To measure [3H]-thymidine incorporation into mature B cells, B220+ splenic B cells were purified by FACS sorting (all cells were Thy1.2 negative and CD19 positive; purity was greater than 99% for B220). The 1 × 105 cells per well were plated into 96-well round bottom plates and stimulated with either 100 ng/mL IL-4, 20 μg/mL α-IgM, 1 μg/mL α-CD40, 10 ng/mL LPS or combinations of IL-4 with α-IgM or α-CD40. Forty-eight hours after plating, [3H]-thymidine was added for the remaining 12 hours.
Immunizations and serum immunoglobulin detection
To measure the T-cell–dependent response, mice were immunized with 100 μg ovalbumin (Sigma Chemical, St Louis, MO) in phosphate-buffered saline (PBS) and complete Freund's Adjuvant (Gibco BRL, Gaithersburg, MD) injected intraperitoneally. Serum samples were obtained 14 days after immunization and ovalbumin-specific responses were determined by enzyme-linked immunosorbent assay (ELISA), as described below. To measure the T-cell–independent immune response, mice were immunized with 50 μg NP-Ficoll (Biosearch Technologies, Novato, CA) in PBS injected intraperitoneally. Plasma samples were obtained 10 days after immunization and NP-specific antibody titers were determined by ELISA, as described below. Antibody titer was calculated as the fold increase compared with the average preimmunization titers of the wild-type control mice.
For the ELISAs, mice were bled via the retro-orbital plexus, and serum was collected after a 15-minute microcentrifuge spin at 4°C. Serum was stored in aliquots at −80°C and total serum Ig antibody concentrations were determined by standard ELISA. The 96-well maxisorp plates (NUNC, Roskilde, Denmark) were coated overnight at 4°C with 50 μL of 10 μg/mL ovalbumin or NP-BSA (bovine serum albumin). Plates were washed with PBS supplemented with 0.05% Tween 20 and blocked with a 3% BSA solution in PBS for 2 hours at room temperature. After 3 washes, diluted plasma samples were added to the coated wells for overnight incubation at 4°C. Plasma samples were serially diluted in wash buffer supplemented with 0.5% BSA. After overnight incubation, plates were washed and polyclonal alkaline phosphatase conjugated anti-IgM or anti-IgG1 (Southern Biotechnology Associates, Birmingham, AL) was added to the wells at 1 μg/mL and left at room temperature for 2 hours. The phosphatase substrate, p-Nitrophenyl phosphate (Sigma Chemical), was dissolved in 100 mmol/L Tris, 100 mmol/L sodium chloride, 5 mmol/L magnesium chloride and 10% diethanolamine, pH 9.5, and added to the plates for 30 minutes, at which time the OD405 was measured on a Bio Rad microplate reader (Richmond, CA).
Bone marrow transduction and in vivo transformations
A bicistronic retroviral vector using the murine stem cell virus (MSCV) long terminal repeats was used for expression of eitherbcr-abl p185 or p210. These constructs were generously provided by Dr Owen Witte. Viral supernatant was collected after transient transfection of 293T cells and ecotropic producer cell lines generated by repeated infection of GP+E86 cells with viral supernatant.31 Freshly isolated bone marrow cells were preactivated for 48 hours in medium containing IL-3 (25 ng/mL), IL-6 (50 ng/mL), and SCF (50 ng/mL) and were consequently cocultured on irradiated (1500 rads) ecotopic producer cell lines for 48 hours in the presence of 6 μg/mL polybrene. Lethally irradiated wild-type mice were reconstituted with the transduced bone marrow by tail vein injection (3 × 106 cells per mouse) and checked daily for the onset of disease. Diseased mice were killed. Blood samples and histopathologic sections were analyzed and used to diagnose the mice.
Flow-cytometric analysis
Single cell suspensions were preincubated with αCD16/CD32 antibodies (Pharmingen) to prevent nonspecific Fc receptor–mediated binding. Thereafter, aliquots of 2 to 5 × 105 cells were stained with monoclonal antibodies conjugated with fluorescent markers (Pharmingen, San Diego, CA) and analyzed by FACSscan (Becton-Dickinson, Franklin Lakes, NJ).
Protein analysis
Analysis was performed as described previously.32Briefly, cells were lysed in buffer containing 50 mmol/L HEPES buffer (pH 7.5), 0.1% Tween-20, 150 mmol/L NaCl, 1 mmol/L EDTA, 20 mmol/L β-glycerophosphate, 0.1 mmol/L sodium vanadate, 1 mmol/L sodium fluoride, 10 μg/mL each aprotinin and leupeptin (both from Sigma Chemical), and 1 mmol/L PMSF. Protein concentrations were determined using a BCA-kit as recommended by the manufacturer (Pierce, Rockford, IL). To assess expression levels of the various forms of the Abl proteins, 100 μg total protein per sample was electrophoretically resolved on a 7.5% polyacrylamide gel containing SDS, and transferred onto Immobilon membranes. Membranes were probed with a rabbit polyclonal antiserum directed against Abl (Santa Cruz Biotechnology Inc, Santa Cruz, CA). Sites of antibody binding were detected using protein A–conjugated horseradish peroxidase (EY Laboratories, San Mateo, CA) with chemiluminescent detection (ECL detection kit, Amersham, Arlington Heights, IL). Stat proteins were immunoprecipitated out of 600 μg of cell lysate using polyclonal antisera, resolved on a 7.5% gel, blotted, and probed with a monoclonal antibody directed against phosphotyrosine (4G10, UBI) as described in detail in Moriggl et al.16 Thereafter, blots were stripped and reprobed with antisera directed against the individual Stat proteins (Transduction Laboratories, Santa Cruz, CA).
Northern blot analysis
RNA of cell lines was isolated by RNAzol and separated on 1% agarose gels (20 μg per lane). A murine 0.2-kilobase (kb)BamHI-HindIII complementary DNA (cDNA) fragment for CIS, a 0.24-kb EcoRI fragment forOSM and a 1.2-kb EcoRI fragment for GAPDH, as loading control, were labeled by α-[32P]dCTP using a random labeling kit (Amersham). The membrane was hybridized using Rapid hybridization solution (Amersham), followed by stringent washes (final wash: 0.2 × SSC/0.1% SDS at 65°C).
Results
Stat5a/b deficiency results in reduced numbers of pre- and pro- B cells
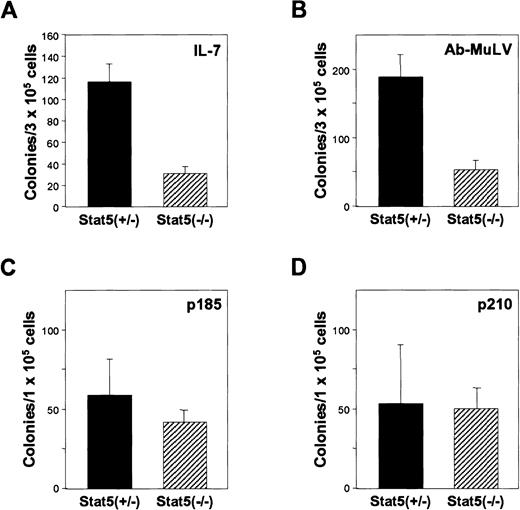
In the initial studies of Stat5a/b-deficient mice,6 a significant reduction in IL-7–induced bone marrow colonies was observed. Because this population contains the cells that are transformed by abl, we further characterized these changes. As illustrated, the frequency of IL-7 colony-forming cells was significantly reduced (Figure 1A), as was the size of the colonies, consistent with our previous results. FACS analysis of the bone marrow demonstrated that there was an overall reduction of B cells by 70% (13% ± 1% CD19-positive cells vs 4% ± 3% in the Stat5a/b-deficient mice). The reduction is found preferentially within the B220-positive, CD43-intermediate and -low populations (Figure 2A,B). Together, the results suggest that, in the absence of Stat5a/b, there is a preferential loss of pre- and pro-B cells in the bone marrow.
Stat5 contributes to IL-7–dependent B-cell precursor expansion, but
abl-induced transformation is independent of Stat5. (A) Ability to form IL-7–dependent colonies. Bone marrow was cloned in methylcellulose containing IL-7. (B) Ability to form Ab-MuLV–induced colonies. Bone marrow was infected with Ab-MuLV and cloned in methylcellulose under growth factor–free conditions. (C, D) Bone marrow from Stat5-deficient as well as control animals was cocultivated for 48 hours with GP+E86 producer cell lines for MSCV-bcr-abl p210/IRES-GFP and MSCV-bcr-ablp185/IRES-GFP. Three days after transduction, the cells were plated in methylcellulose.
Stat5 contributes to IL-7–dependent B-cell precursor expansion, but
abl-induced transformation is independent of Stat5. (A) Ability to form IL-7–dependent colonies. Bone marrow was cloned in methylcellulose containing IL-7. (B) Ability to form Ab-MuLV–induced colonies. Bone marrow was infected with Ab-MuLV and cloned in methylcellulose under growth factor–free conditions. (C, D) Bone marrow from Stat5-deficient as well as control animals was cocultivated for 48 hours with GP+E86 producer cell lines for MSCV-bcr-abl p210/IRES-GFP and MSCV-bcr-ablp185/IRES-GFP. Three days after transduction, the cells were plated in methylcellulose.
Decreased numbers of B-cell precursors in the bone marrow and mature B cells in the blood of Stat5-deficient mice.
(A, B) FACS analysis of fresh bone marrow isolated from Stat5 (+/−) and Stat5 (−/−) mice. Data represent percent of IgM-negative lymphoid populations. B220 and CD43 staining allows the distinction in B-cell precursor subsets. (C) Peripheral blood was withdrawn from 3-month-old Stat5-deficient mice and their heterozygous littermates. Absolute numbers of B220-positive cells per microliter are shown. (D) B220-positive, Thy1.2-negative cells were sorted by FACS, plated in 96-well round bottom dishes and stimulated as indicated. 48 hours after stimulation, [3H]-thymidine was added. Thymidine incorporation in heterozygous cells (■) and in Stat5-deficient cells (▨) is shown. (E) Determination of the antibody response to the T-cell–dependent antigen ovalbumin. Ovalbumin-specific serum IgM and IgG1 titration curves are shown. Each dilution curve represents the average of 5 mice. ■, heterozygous mice; ▪, Stat5a/b-deficient mice. (F) measurement of the serum IgM response against the T-cell–independent antigen NP-Ficoll. Results are the average of 5 mice. Preimmunization titers, ▪; postimmunization titers, ▨.
Decreased numbers of B-cell precursors in the bone marrow and mature B cells in the blood of Stat5-deficient mice.
(A, B) FACS analysis of fresh bone marrow isolated from Stat5 (+/−) and Stat5 (−/−) mice. Data represent percent of IgM-negative lymphoid populations. B220 and CD43 staining allows the distinction in B-cell precursor subsets. (C) Peripheral blood was withdrawn from 3-month-old Stat5-deficient mice and their heterozygous littermates. Absolute numbers of B220-positive cells per microliter are shown. (D) B220-positive, Thy1.2-negative cells were sorted by FACS, plated in 96-well round bottom dishes and stimulated as indicated. 48 hours after stimulation, [3H]-thymidine was added. Thymidine incorporation in heterozygous cells (■) and in Stat5-deficient cells (▨) is shown. (E) Determination of the antibody response to the T-cell–dependent antigen ovalbumin. Ovalbumin-specific serum IgM and IgG1 titration curves are shown. Each dilution curve represents the average of 5 mice. ■, heterozygous mice; ▪, Stat5a/b-deficient mice. (F) measurement of the serum IgM response against the T-cell–independent antigen NP-Ficoll. Results are the average of 5 mice. Preimmunization titers, ▪; postimmunization titers, ▨.
In the peripheral blood of Stat5a/b-deficient mice, there is also a reduction in B220-positive B cells (Figure 2C) to levels approximately 6% that of control mice. However, the peripheral B cells that are present responded normally to stimulation with IL-4 and B-cell receptor engagement (Figure 2D), as well as to stimulation with anti-CD40 in the presence of IL-4. Moreover, B-cell numbers in the spleen are not reduced in Stat5a/b-deficient animals nor did we find any alterations or abnormalities in the distribution of mature versus immature B cells in the spleen (data not shown). Immunization of Stat5a/b-deficient mice with T-cell–dependent or –independent antigens resulted in comparable levels and isotypes of antibody production (Figure 2E,F). Together the results indicate that Stat5a/b-deficient mice have a preferential deficiency in the early IL-7–dependent phase of pre- and pro-B-cell development.
Stat5a/b-deficient cells are susceptible to Ab-MuLV– andbcr-abl–induced transformation in vitro
The transforming activity of Ab-MuLV and the bcr-abloncogenes includes the ability to eliminate growth factor requirements of bone marrow progenitors. We therefore initially examined the response of bone marrow cells from Stat5a/b-deficient mice to be transformed to growth factor independence by Ab-MuLV andbcr-abl oncogenes. As illustrated in Figure 1B, the number, but not the size (data not shown) of growth factor–independent Ab-MuLV–induced bone marrow colony-forming cells was reduced in cells from Stat5a/b-deficient mice. As expected, immunocytochemical staining of the Ab-MuLV–transformed colonies revealed a B-lymphoid phenotype (data not shown). Therefore, we conclude that the number of Ab-MuLV targets are reduced consistent with the reduction in IL-7–responsive bone marrow cells. However, the cells that are present can be transformed in vitro comparable to wild-type cells.
To explore the requirement of Stat5 proteins for bcr-abltransformation, the ability to confer cytokine-independent growth of Stat5a/b-deficient bone marrow cells was examined. Bone marrow cells from Stat5a/b-deficient and wild-type mice were cocultured with cell lines producing MSCV-based retroviruses expressingbcr-abl-p210 or bcr-abl-p185 and GFP expressed from an internal ribosome entry site (IRES). Three days after infection, the cells were plated in methylcellulose and colony formation assessed. Analysis of the transformed cells by Wright-Giemsa staining and FACS analysis showed a myeloid phenotype for thebcr-abl p210 transformed cells (data not shown). The cells stained positive for the surface marker Mac1 and displayed myeloid features independent of whether Stat5a/b were present. Transduction with bcr-abl p185 resulted in myeloid as well as B lymphoid transformants. In Stat5 (+/−) bone marrow bcr-abl p185 transduction results in lymphoid and myeloid transformants, in Stat5 (−/−) bone marrow, the transfomed cells were mainly of myeloid phenotype consistent with a reduced availability of lymphoid precursor cells (data not shown). This finding may account for the slight, but not significant reduction of bcr-abl p185-induced colonies from Stat5a/b-deficient bone marrow. It should also be noted that the predominance of myeloid lineage colonies in these experiments may be influenced by the use of IL-3, IL-6, and SCF during the cocultivation for retroviral transduction.
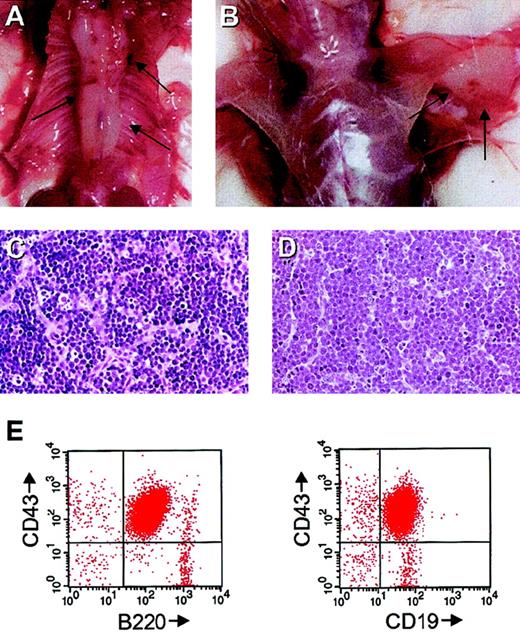
Stat5a/b-deficient mice are susceptible to Ab-MuLV–induced tumors in vivo
To assess the requirement for Stat5a/b for in vivo transformation, newborn mice were infected with replication-defective Ab-MuLV. Because the mice were obtained from crosses that use homozygous Stat5a/b-deficient males and heterozygous females, the heterozygous offspring served as controls. The majority (14 of 19; 74%) of the heterozygous animals developed B-cell lymphomas within 8 to 12 weeks, whereas the remainder survived longer than 6 months. Among the Stat5a/b-deficient animals, 7 of the 13 animals died from splenomegaly as a result of extramedullary hematopoiesis and anemia that is characteristically seen in nonmanipulated animals.16 Among the remaining 6 mice, 4 developed typical Ab-MuLV–induced pro-B-cell lymphomas at sites that were comparable to those seen with heterozygous control mice with a latency of 9 to 12 weeks (4 of 6; 67%). Histologically, the Stat5a/b-deficient tumors were identical to the tumors seen with control animals (Figure3C,D). Also comparable to the tumors from control animals, the cells expressed B220, CD43, and CD19 (Figure3E,F). In the 2 cases examined, 1 × 107Stat5a/b-deficient tumor cells induced solid tumors in secondary SCID recipients (data not shown). Therefore, we conclude that Stat5a/b-deficient mice are susceptible to classical Ab-MuLV in vivo transformation.
Ab-MuLV induces pro-B-cell tumors in vivo.
(A,B) Stat5-deficient mice with Ab-MuLV–induced tumors embedding the spine and in the axilla. Arrows indicate the tumors. (C) Hematoxylin-eosin stain of a section of a Stat5 heterozygous tumor. (D) Hematoxylin-eosin stain of a section of a Stat5-deficient tumor. (E) FACS analysis of a Stat5-deficient tumor displaying typical pro-B-cell markers.
Ab-MuLV induces pro-B-cell tumors in vivo.
(A,B) Stat5-deficient mice with Ab-MuLV–induced tumors embedding the spine and in the axilla. Arrows indicate the tumors. (C) Hematoxylin-eosin stain of a section of a Stat5 heterozygous tumor. (D) Hematoxylin-eosin stain of a section of a Stat5-deficient tumor. (E) FACS analysis of a Stat5-deficient tumor displaying typical pro-B-cell markers.
Stat5a/b-deficient cells are susceptible to transformation bybcr-abl in vivo
The in vivo transforming activities of bcr-abl-p210 andbcr-abl-p185 were examined by infecting bone marrow cells and reconstituting lethally irradiated mice.31 Infection of bone marrow cells from both wild-type and Stat5a/b-deficient mice resulted in the rapid onset of disease (18-24 days) with comparable pathologic alterations in all the recipients. The affected organs were lung (Figure 4A), spleen (Figure 4B), and liver (not shown). Histopathologic studies revealed extramedullary hematopoiesis in the spleen (Figure 4C), infiltration of tumor cells in the liver (Figure 4D), and lung (data not shown), resulting in the punctuated bleedings described by others.34 35 Analysis of peripheral blood from tumor-bearing mice was performed by blood smears and FACS (typical examples are illustrated in Figure 4E,F). Thebcr-abl-p210 infection of wild-type bone marrow cells induced 8 myeloid lineage leukemias of the 9 leukemias phenotyped as defined by the expression of the myeloid lineage markers Mac1 and Gr1. In contrast, among the tumors derived from Stat5a/b-deficient cells that were phenotyped, 4 were of B-cell lineage, 2 had myeloid lineage markers, and 5 tumors contained both myeloid and B-cell lineage cells. The cell lineages of the tumors derived from bcr-abl-p185 were comparable between infection of wild-type or Stat5a/b-deficient bone marrow (summarized in Table 1). In both cases, B-cell lineage and myeloid lineage tumors or mixed tumors were observed.
Typical alterations of lethally irradiated recipient mice reconstituted with Stat5a/b-deficient
bcr-abl-infected bone marrow. (A) Macroscopic appearance of the lungs. Note the punctuated bleedings associated with focal accumulation of myeloid cells. (B) Macroscopic appearance of the spleen. Note the loss of normal splenic architecture produced by the nodular sites of tumor. (C) Histologic section of a spleen stained with hematoxylin-eosin (HE). Extramedullary hematopoiesis and focal accumulations of tumor cells are evident. (D) Histologic section of a liver stained with HE demonstrates the perivascular accumulation of infiltrating myeloid cells. (E) Blood smear (top) and FACS analysis (bottom) of a myeloid lineage tumor. (F) Blood smear (top) and FACS analysis (bottom of a lymphoid lineage tumor).
Typical alterations of lethally irradiated recipient mice reconstituted with Stat5a/b-deficient
bcr-abl-infected bone marrow. (A) Macroscopic appearance of the lungs. Note the punctuated bleedings associated with focal accumulation of myeloid cells. (B) Macroscopic appearance of the spleen. Note the loss of normal splenic architecture produced by the nodular sites of tumor. (C) Histologic section of a spleen stained with hematoxylin-eosin (HE). Extramedullary hematopoiesis and focal accumulations of tumor cells are evident. (D) Histologic section of a liver stained with HE demonstrates the perivascular accumulation of infiltrating myeloid cells. (E) Blood smear (top) and FACS analysis (bottom) of a myeloid lineage tumor. (F) Blood smear (top) and FACS analysis (bottom of a lymphoid lineage tumor).
In vivo transformation of Stat5a/b-deficient cells bybcr-abl transforming genes
| BM cells . | Gene . | Myeloid B cell . | Mixed . | NT* . | |
|---|---|---|---|---|---|
| Wild-type | bcr-ablp210 | 8 | 0 | 1 | 6 |
| Stat5a/b(−/−) | bcr-ablp210 | 2 | 4 | 5 | 4 |
| Wild-type | bcr-ablp185 | 2 | 4 | 2 | 0 |
| Stat5a/b(−/−) | bcr-ablp185 | 2 | 4 | 0 | 2 |
| BM cells . | Gene . | Myeloid B cell . | Mixed . | NT* . | |
|---|---|---|---|---|---|
| Wild-type | bcr-ablp210 | 8 | 0 | 1 | 6 |
| Stat5a/b(−/−) | bcr-ablp210 | 2 | 4 | 5 | 4 |
| Wild-type | bcr-ablp185 | 2 | 4 | 2 | 0 |
| Stat5a/b(−/−) | bcr-ablp185 | 2 | 4 | 0 | 2 |
In vivo transformation: Lethally irradiated mice were reconstituted with bone marrow from Stat5-deficient and control heterozygous animals that has been cocultivated with GP + E86 producer cells for MSCV-bcr-abl p210/IRES-GFP and MSCV-bcr-abl p185/IRES-GFP for 48 hours. Sick animals were killed and the peripheral blood, as well as histologic sections, were analyzed for the presence of leukemic cells. The cell lineages involved in the tumors were characterized based on FACS analysis of peripheral blood samples.
BM indicates bone marrow.
Not determined.
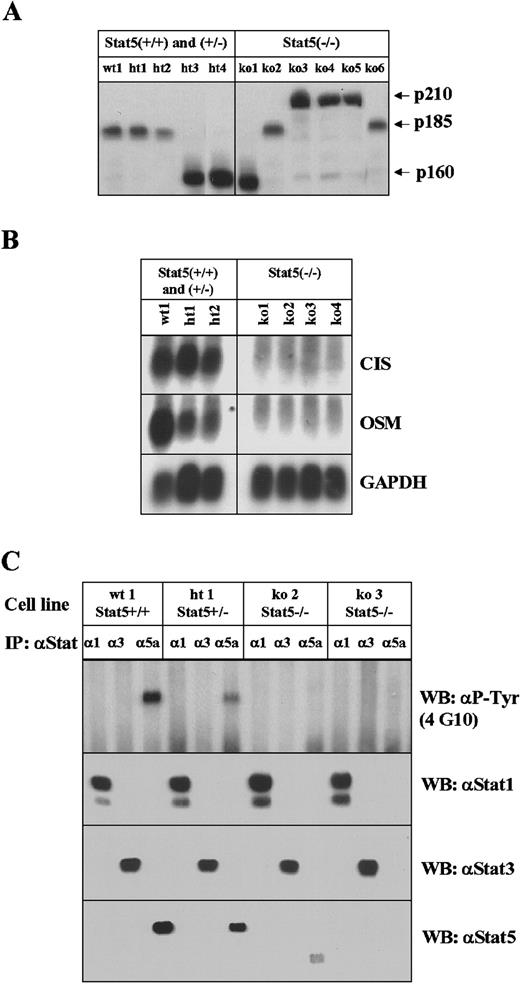
For further analysis, cell lines were derived from tumors of wild- type and Stat5a/b-deficient bone marrow (Table2). With the exception of one myeloid cell line from a Stat5a/b heterozygous animal expressing Mac1 and Gr1, the cell lines that were established were of B-cell lineage and expressed B220, CD43, and CD19. Cell lines were obtained from bothbcr-abl– and Ab-MuLV–infected wild-type and Stat5a/b-deficient blood. As illustrated in Figure5A, the various Abl proteins were highly expressed in both control and Stat5a/b-deficient tumor cell lines. As illustrated (Figure 5B), tumor cell lines from control bone marrow expressed both the Stat5a/b regulated genes, CIS andOSM, whereas the tumor cell lines from Stat5a/b-deficient animals or bone marrow did not express either gene. This result demonstrates that a redundant pathway, capable of replacing Stat5a/b function in the regulation of these 2 genes, was not activated. Lastly, tyrosine phosphorylation of various Stats is shown in Figure 5C. Consistent with previous observations, Stat5a/b is tyrosine phosphorylated in tumor cell lines from bcr-ablp185–transformed control bone marrow. This is not seen in tumor cell lines from Stat5a/b-deficient marrow cells. No significant activation of Stat1 or Stat3 was seen in any of the Stat5a/b-deficient tumor cell lines.
Properties of cell lines
| Cell line . | Stat5a/b . | Abl-protein . | Immunophenotype . |
|---|---|---|---|
| wt1 | +/+ | p185 | B220/CD19 |
| ht1 | +/− | p185 | B220/CD29 |
| ht2 | +/− | p185 | Mac1/Gr1 |
| ht3 | +/− | p160 | B220/CD19 |
| ht4 | +/− | p160 | B220/CD19 |
| ko1 | −/− | p160 | B220/CD19 |
| ko2 | −/− | p185 | B220/CD19 |
| ko3 | −/− | p210 | B220/CD19 |
| ko4 | −/− | p210 | B220/CD19 |
| ko5 | −/− | p210 | B220/CD19 |
| ko6 | −/− | p185 | B220/CD19 |
| Cell line . | Stat5a/b . | Abl-protein . | Immunophenotype . |
|---|---|---|---|
| wt1 | +/+ | p185 | B220/CD19 |
| ht1 | +/− | p185 | B220/CD29 |
| ht2 | +/− | p185 | Mac1/Gr1 |
| ht3 | +/− | p160 | B220/CD19 |
| ht4 | +/− | p160 | B220/CD19 |
| ko1 | −/− | p160 | B220/CD19 |
| ko2 | −/− | p185 | B220/CD19 |
| ko3 | −/− | p210 | B220/CD19 |
| ko4 | −/− | p210 | B220/CD19 |
| ko5 | −/− | p210 | B220/CD19 |
| ko6 | −/− | p185 | B220/CD19 |
Leukemic blood samples were placed into culture. Ten days later cells were obtained under growth factor–independent conditions and could be amplified and characterized.
Analysis of cell lines derived from in vivo tumors.
(A) Expression of various Abl proteins in tumor-derived cell lines by direct Western blotting with an antiserum that detects the various Abl transforming proteins. (B) Northern blot analysis of the expression of the Stat5a/b target genes, CIS and OSM. Cell lines derived from wild-type (wt) mice or mice heterozygous for Stat5a/b and cell lines from Stat5a/b-deficient tumors were examined. GAPDH is included as a loading control. (C) The extent of activation of various Stats was examined by immunoprecipitation with Stat-specific antibodies and Western blotting (WB) with a monoclonal antibody against phosphotyrosine (4G10). Antisera specific for Stat1, Stat3, and Stat5 were used. The smaller form of Stat5 seen in one of the Stat5a/b-deficient tumor cells represents the N-terminal truncated form that is produced from the disrupted locus as previously described.6
Analysis of cell lines derived from in vivo tumors.
(A) Expression of various Abl proteins in tumor-derived cell lines by direct Western blotting with an antiserum that detects the various Abl transforming proteins. (B) Northern blot analysis of the expression of the Stat5a/b target genes, CIS and OSM. Cell lines derived from wild-type (wt) mice or mice heterozygous for Stat5a/b and cell lines from Stat5a/b-deficient tumors were examined. GAPDH is included as a loading control. (C) The extent of activation of various Stats was examined by immunoprecipitation with Stat-specific antibodies and Western blotting (WB) with a monoclonal antibody against phosphotyrosine (4G10). Antisera specific for Stat1, Stat3, and Stat5 were used. The smaller form of Stat5 seen in one of the Stat5a/b-deficient tumor cells represents the N-terminal truncated form that is produced from the disrupted locus as previously described.6
Discussion
IL-7 has been recognized as a major cytokine that stimulates long-term proliferation and differentiation of precursor B cells.1 Mice lacking IL-7 or the IL-7Rα chain show drastically reduced numbers of B-cell precursors and mature B cells. Interestingly, despite the lack of a functional IL-7R or IL-7, some B cells do undergo maturation, indicating that the block in B-cell maturation is incomplete.7-9 In this study, we demonstrate a partially overlapping phenotype in mice disrupted for the transcription factors Stat5a/b. Mice deficient for Stat5a/b have reduced numbers of peripheral B cells and of B-cell precursors in the bone marrow. Using distinct surface antigens (CD43, B220), FACS analysis allows the differentiation of several B-cell precursor subsets.10 11 These cellular subsets represent clearly defined states in B-cell development, all of which are under the control of IL-7. This wave of expansion is disrupted at the level of late pro-B and pre-B cells in Stat5a/b-deficient mice. Maturation and differentiation, however, seem largely undisturbed. Taken together, we conclude, that Stat5a/b-deficient B-cell precursors have an expansion, but not a differentiation defect at the late pro-B-cell and pre-B-cell stage of development. In contrast to mice lacking IL-7 or the IL-7Rα chain, numbers of B cells in the spleen and lymph nodes are normal in Stat5-deficient mice, but mature B cells are selectively reduced in the peripheral blood. The less severe phenotype suggests additional Stat5a/b–independent signaling pathways downstream of the IL-7R that compensate for the loss of Stat5a/b.
Stat5a/b are activated by a broad variety of cytokines, among them IL-2, IL-4, and IL-7.2 We have recently shown that T cells lacking Stat5a/b have a proliferation defect in response to stimulation with either IL-2 or IL-4.15,16 In contrast, mature Stat5a/b-deficient B cells proliferate normally in response to IL-4 or when challenged with other B-cell mitogens (Figure 2D). So, despite its clear role in IL-4–induced T-cell proliferation, Stat5a/b are not required for the proliferation of mature B cells. Stat5a/b were also implicated in immunoglobulin class switching and IgM production,18,19 33 however, we failed to obtain evidence for this hypothesis. Stat5a/b-deficient mice responded with normal IgM and IgG1 levels when challenged with T-cell–dependent or T-cell–independent antigens (Figure 2E,F).
Despite the large amount of literature,3-5,20-23,27-30 our results demonstrate that Stat5a/b are not essential for in vitro or in vivo transformation of B-cell precursors or myeloid cells by transforming derivatives of the abl proto-oncogene. It is well established that, depending on host and conditioning treatments,bcr-abl can induce T lineage disease,36,37however, our conditions favored the B lymphoid and myeloid diseases. The myeloid disease can range from a myeloproliferative disease to the clonal expansion of myeloid lineage transformed cells. Further studies are necessary to explore the role of Stat5a/b in the full spectrum of diseases associated with bcr-abl and to explain the reduced frequency of pure myeloid leukemias arising in mice transplanted with Stat5a/b-deficient bone marrow. Our data prove, that Stat5a/b are not essential or necessary for Ab-MuLV– or bcr-abl–induced transformation. The possible basis for the shift of phenotype observed in mice transplanted with Stat5a/b-deficientbcr-abl–transformed bone marrow is not known. Because the colony assays did not reveal any alteration in myeloid colony formation, one may speculate that the reduction of myeloid disease in the animals is related to altered or disturbed cytokine production. It has been shown that bcr-abl transduction results in the production of several cytokines, eg, granulocyte-macrophage colony-stimulating factor (GM-CSF) and IL-3, and that cytokine production contributes to the myeloproliferative disease.38 The reduced myeloid disease might therefore also be related to decreased levels of cytokines, in particular the Stat5a/b target gene oncostatin M (OSM).39OSM is shown to be expressed at high levels in the bone marrow.39
The lack of an effect on B lineage and myeloid transformation could be due to the utilization of pathways that can compensate for the absence of Stat5a/b. However, activation of Stat1 or Stat3 was not seen in the cell lines from Stat5a/b-deficient tumor cell lines. Moreover, although cell lines derived from wild-type mice expressed both the cytokine-inducible SH2-containing gene (CIS) andOSM, Stat5a/b target genes were not expressed in cell lines derived from tumors from Stat5a/b-deficient bone marrow cells or mice. Therefore, we have no evidence at this point that a redundant pathway is providing Stat5a/b functions. In summary, the results demonstrate that Stat5a/b are not critical substrates in bcr-abl– orabl-induced transformation of B cells or myeloid cells.
Acknowledgments
We thank M. Paktinat, S. Wingo, R. Cross, and R. Ashmun for technical assistance and J. Rehg, J. Downing, and M. Strain for help with the pathologic analysis of the diseased animals. We also thank Naomi Rosenberg and Owen Witte for the generous gift of A010 cells and the MSCV-210 and MSCV-p185 vectors, respectively.
Supported, in part, by Cancer Center CORE Grant (CA 21765) and ALSAC (American Lebanese Syrian Associated Charities).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
James N. Ihle, Howard Hughes Medical Institute, St. Jude Children's Research Hospital, 332 N Lauderdale, Memphis, TN 38105; e-mail: james.ihle@stjude.org.

![Fig. 2. Decreased numbers of B-cell precursors in the bone marrow and mature B cells in the blood of Stat5-deficient mice. / (A, B) FACS analysis of fresh bone marrow isolated from Stat5 (+/−) and Stat5 (−/−) mice. Data represent percent of IgM-negative lymphoid populations. B220 and CD43 staining allows the distinction in B-cell precursor subsets. (C) Peripheral blood was withdrawn from 3-month-old Stat5-deficient mice and their heterozygous littermates. Absolute numbers of B220-positive cells per microliter are shown. (D) B220-positive, Thy1.2-negative cells were sorted by FACS, plated in 96-well round bottom dishes and stimulated as indicated. 48 hours after stimulation, [3H]-thymidine was added. Thymidine incorporation in heterozygous cells (■) and in Stat5-deficient cells (▨) is shown. (E) Determination of the antibody response to the T-cell–dependent antigen ovalbumin. Ovalbumin-specific serum IgM and IgG1 titration curves are shown. Each dilution curve represents the average of 5 mice. ■, heterozygous mice; ▪, Stat5a/b-deficient mice. (F) measurement of the serum IgM response against the T-cell–independent antigen NP-Ficoll. Results are the average of 5 mice. Preimmunization titers, ▪; postimmunization titers, ▨.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/96/6/10.1182_blood.v96.6.2277/5/m_h81800174002.jpeg?Expires=1769081430&Signature=JiZ3nn~-beDntgXDCcnmtCZmfQRNB8N~juL6Xg5FREoms55I~xyTaIMRyLT477io7UJdC0YMsIsQM~VCz1F-c58wQ3ybsNus4Uxy5cQ~DV-XB7A~ooWxQk4WarqP06eXuRozMsSGs9iHehoC~Ssk98L~QY9agk4eVJFIZe2IQvxftYUjQ9tXLlGYxGmdM21aw06I5tWukuee1nlA2zLmxm7n6bcqfk~KRhxNNRZi1CNNcVa5vQl5JSiOpZeaVBmTV3W3KFlBQFb5xVh~0Wltev3nqsEoAK0rk47RRLRJU5q~TxEYyzdhJfh-Yc1EizRBtoHkustvkoBrQz10Nu5ofA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal