Abstract
The role of fusion proteins in acute myeloid leukemia (AML) is well recognized, but the leukemic target cell and the cellular mechanisms generating the AML phenotype are essentially unknown. To address this issue, an in vitro model to study the biologic activity of leukemogenic proteins was established. Highly purified human hematopoietic progenitor cells/stem cells (HPC/HSC) in bulk cells or single cells are transduced with retroviral vectors carrying cDNA of the fusion protein and the green fluorescent protein (GFP), purified to homogeneity and induced into multilineage or unilineage differentiation by specific hematopoietic growth factor (HGF) combinations. Expression of PML/RARα fusion protein in human HPC/HSC dictates the acute promyelocytic leukemia (APL) phenotype, largely through these previously unreported effects: rapid induction of HPC/HSC differentiation to the promyelocytic stage, followed by maturation arrest, which is abolished by retinoic acid; reprogramming of HPC commitment to preferential granulopoietic differentiation, irrespective of the HGF stimulus (transduction of single sibling HPC formally demonstrated this effect); HPC protection from apoptosis induced by HGF deprivation. A PML/RARα mutated in the co-repressor N-CoR/histone deacetylase binding region lost these biologic effects, showing that PML/RARα alters the early hematopoietic program through N-CoR–dependent target gene repression mechanisms. These observations identify the cellular mechanism underlying development of the APL phenotype, showing that the fusion protein directly dictates the specific lineage and differentiation stage of leukemic cells.
Introduction
Acute myeloid leukemias (AML) are characterized by the accumulation of hematopoietic precursors blocked at different stages of differentiation. The cellular target for the oncogenic event and the mechanisms responsible for the heterogeneous phenotype of the leukemic blasts are, however, unclear. Transformation may occur at a specific differentiation stage, eg, early differentiated precursors, causing developmental arrest. Alternatively, it may take place in multipotent hematopoietic progenitor cells (HPC) or in hematopoietic stem cells (HSC), allowing differentiation followed by a maturation block. The second model implies the existence of a leukemic HSC population,1 as suggested by the finding of CD34+ CD38− cells from AML transplant leukemia in NOD/SCID mice.2 Conversely, acute promyelocytic leukemia (APL) blasts are unable to survive in the NOD/SCID system; it is, hence, unclear whether promyelocytic blasts originate from leukemic HSC or HPC.2
Current transformation model systems—that is, the expression of oncogenes into cell lines and transgenic animals—do not allow analysis of the early cellular events induced by oncogenic proteins. Specifically, these models do not provide insight into the relationship between a single oncogenic event and blast accumulation, the early phases of leukemogenesis, and the effects of oncogenes on HSC/HPC commitment and differentiation through hematopoietic lineages apparently unaffected by leukemic transformation.
A detailed study of leukemogenesis can be accomplished by oncogene transfer into normal human HPC/HSC, in which commitment and differentiation along each hematopoietic lineage can be studied in depth.3-7 However, data on the biologic effect of fusion protein expression in human HPC/HSC are limited to the demonstration of the transforming activity of the TLS-ERG protein.8 This approach has been hindered by the inefficient transfer of leukemic fusion genes in primary human HPC/HSC and the unsatisfactory purification and in vitro manipulation of HPC/HSC.
We have established a methodology to purify human HPC/HSC by the negative selection of cells that express lineage and proliferation markers (Lin). The final highly clonogenic immature CD34+Lin− population can be induced into unilineage erythroid (E), megakaryocytic (MK), granulocytic (G), or monocytic (M) differentiation/maturation by specific hematopoietic growth factor (HGF) combinations.3-7 Purified HPC/HSC are efficiently infected by retroviral vectors expressing the green fluorescent protein (GFP); this allows, as early as 24 hours after the infection, FACS purification to homogeneity of the transduced HPC that maintain a normal proliferation/differentiation potential.9
APL is an ideal model in which to investigate the cellular effects of fusion protein on hematopoietic proliferation and differentiation. The APL phenotype strictly correlates with the expression of RARα fusion proteins.10,11 APL blasts are blocked at the level of promyelocytes, implying that HPC/HSC commitment and initial differentiation have occurred.12 In addition they terminally mature in response to retinoic acid (RA).13,14The transforming activity of PML/RARα fusion protein has been shown in cell lines and transgenic mice.15-20 This chimeric transcription factor recruits to target gene promoters histone deacetylase (HD) through the transcriptional co-repressors N-CoR and SMRT.21-24 In hematopoietic cell lines this leads to blocking of terminal differentiation and apoptosis and to an increased differentiation response to RA.15 PML/RARα transgenic mice show myeloid differentiation alterations and, in a variable percentage, a promyelocytic leukemia that responds to the differentiation action of RA.18-20 However, animal models do not allow the exploration of early transformation events and of how leukemic commitment affects lineage distribution.25 26
We show here that expression of the PML/RARα fusion protein in human HPC/HSC dictates the APL phenotype by 2 previously unreported effects, the rapid induction of HPC/HSC differentiation to the promyelocytic stage, followed by maturation arrest, and the reprogramming of HPC commitment characterized by preferential granulopoietic differentiation to promyelocytes, irrespective of the HGF stimulus. Formal demonstration of HPC reprogramming was obtained by analysis of transduced single sibling HPC.27 These data indicate that fusion protein expression alters the entire early hematopoietic program and directly dictates the lineage- and differentiation-specific leukemic phenotype.
Materials and methods
Purification, culture, and infection of HPC/HSC
Purification of peripheral blood HPC/HSC was performed from normal donor buffy coats by the negative selection of cells expressing Lin as described.3,6 Typically, the final population is resting, consists of more than 95% CD34+Lin−, does not express CD 71, and has undifferentiated blastic morphology on May–Grünwald–Giemsa-stained cytospin slides.3 All progenitor cell cultures were grown in serum-free, retinoid-free media, except for unilineage monocyte-differentiating cultures to which 20% fetal calf serum was added. For the infection, purified HPC/HSC were stimulated with IL-3 (100 U/mL), FLT3 ligand (50 ng/mL), and stem cell factor (50 ng/mL) for 48 hours (days −4 and −3). Viral supernatant was produced by transient transfection of packaging cells with retroviral constructs, as described. Infection was obtained by 4 cycles of centrifugation of the cells in viral supernatant at 1800 rpm for 40 minutes (days −2 and −1). PINCO retroviral vectors (GFP) or PINCO vectors containing PML/RARα and PML/RARα-AHT cDNA were used for this study.21
Expression of exogenous genes and FACS purification of transduced cells
GFP expression was verified 24 to 36 hours after the last infection cycle by microscopic examination and FACS analysis. Fluorescent cells were sorted by a FACS Vantage (Becton Dickinson) with a 488-nm laser emission using the maximal purity “normal C” program gating the mononuclear nongranular cells.9 PML/RARα protein expression was studied by immunofluorescence with the PGM3 α-PML monoclonal antibody as described.28 Expression of the fusion proteins delocalized PML to the typical micro-speckled nuclear distribution pattern.
Phenotypic analysis of transduced cells
Multilineage and unilineage cultures of HPC were grown as described in liquid and methylcellulose cultures.3-7 The numbers of seeded cells/plate were 100 for BFU-E and CFU-G, 500 for CFU-MK, and 1000 for CFU-Mo. The multilineage cocktail contained saturating concentrations of erythropoietin (3 U/mL), TPO (50 ng/mL), granulocyte macrophage–colony]stimulating factor (GM-CSF; 10 ng/mL), M-CSF (250 U/mL), G-CSF (500 U/mL), IL-6 (10 ng/mL), stem cell factor (100 ng/mL), FLT3 ligand (100 ng/mL), and IL-3 (100 U/mL).3-7 Cells in liquid cultures were counted every 48 hours, and their morphology was studied on May-Grünwald-Giemsa-stained cytospin slides. Blast cells were defined as morphologically undifferentiated cells. Their number corresponded to that of CD34+ cells. Promyelocytes were defined on morphologic basis; their number corresponded to the cells that expressed CD15, CD9, and CD33 (not shown). In selected experiments, expression of surface markers was studied by immunofluorescence with TRITC-conjugated monoclonal antibody against the antigens CD34, CD33, CD15, CD9, CD11b, and glycophorin A, confirming the morphologic data. Methylcellulose colonies were counted, identified microscopically, picked, and studied for morphology on cytospin slides.
Single HPC culture, infection, and analysis
Single HPCs were isolated with a glass micropipette and cultured in 96-well plates. After the first cell division, the siblings were separated and infected with control or PML/RARα vector. Infection was obtained as for bulk cultures, but with 3 infection cycles in 50 μL viral supernatant. Fluorescent cells were identified after 48 hours by microscopic examination. The couples of sibling cells in which both control and PML/RARα-infected cells were fluorescent were isolated in single wells in a medium containing erythropoietin, IL-3, and GM-CSF as described.7 27 Differentiated colonies were analyzed on Wright–Giemsa-stained cytospin slides
Results
Transduction of HPC/HSC and expression of PML/RARα
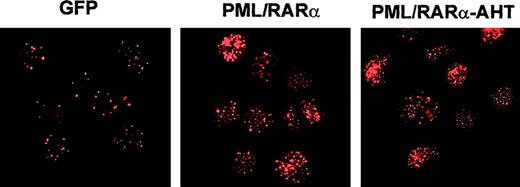
HPCs were induced to proliferate and were transduced (see “Materials and Methods”) with control retrovirus or vectors carrying a PML/RARα cDNA or the PML/RARα-AHT cDNA, mutated in the N-CoR binding region. Infection efficiency was 45% to 50% for the control vector and 20% to 25% for the fusion protein vectors (not shown). Twenty-four to 36 hours after infection, GFP+ cells were FACS sorted to obtain 95% or more pure population of GFP+ cells.9 Sorted PML/RARα and PML/RARα-AHT HPC revealed a nuclear micro-speckled pattern by α-PML immunofluorescence28 (Figure1), indicating a strong expression of the fusion proteins. Therefore, the PML/RARα protein is highly expressed during the HPC lineage commitment and initial differentiation that take place in the first days of culture.3-7
Fusion protein expression in HPC/HSC.
Transduced cells were purified by FACS and analyzed by indirect immunofluorescence with α-PML monoclonal antibody,26 as described.9 In control cells (GFP), PML is expressed in a nuclear speckled distribution. Both PML/RARα and AHT mutant are expressed in the typical nuclear microspeckled pattern.
Fusion protein expression in HPC/HSC.
Transduced cells were purified by FACS and analyzed by indirect immunofluorescence with α-PML monoclonal antibody,26 as described.9 In control cells (GFP), PML is expressed in a nuclear speckled distribution. Both PML/RARα and AHT mutant are expressed in the typical nuclear microspeckled pattern.
PML/RARα expression induces HPC differentiation followed by promyelocyte maturation block
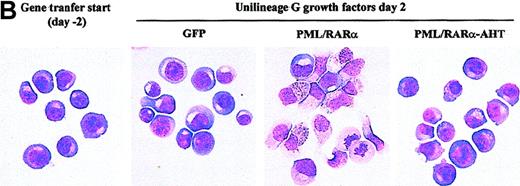
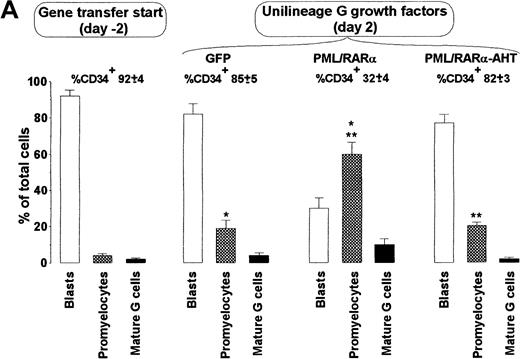
APL cells are relatively differentiated. It is unclear, however, whether the oncogenic event occurs in promyelocytes or HPC/HSC allowed to differentiate into promyelocytes.29 To address this issue, we analyzed PML/RARα+ HPC differentiation in unilineage G liquid suspension cultures (supplemented with saturating level of G-CSF and low dosages of IL-3 and GM-CSF).21PML/RARα had a dual effect. After 2 days from the end of gene transduction, as many as 60% of the cells had a hypergranular promyelocytic morphology, whereas control cells retained a blast morphology and expressed the CD34 marker (Figure2). Because total cell counts and percentages of dead or apoptotic cells did not show important changes in PML/RARα samples with respect to controls (Figure 2B, 3A, and unpublished results), promyelocytic differentiation occurred without a significant increase in cell proliferation. PML/RARα expression, therefore, had an unexpected differentiation effect on HPCs. During the following culture days, control cells fully matured to neutrophilic granulocytes5 (Figure3B); conversely, PML/RARα+HPCs displayed impaired maturation with an accumulation of promyelocytes, followed by incomplete maturation (Figure 3B). At lower HGF concentrations, the early differentiation effect was unchanged, whereas the differentiation block was more pronounced (Figure 3B). Confirming the specificity of this effect, PML/RARα-induced differentiation impairment was abolished by 10−6 mol/L RA. In these conditions, PML/RARα HPCs differentiated more rapidly than control cells (Figure 3B).
PML/RARα induces promyelocytic differentiation of HPC.
(A) The morphology of the cells was analyzed on cytospin slides stained with May-Grünwald-Giemsa. (first left panel) HPCs stimulated with IL-3, FLT3 ligand, and stem cell factor (see “Materials and methods”) before retroviral infection. (right panels) HPCs transduced with the control (GFP) or the indicated fusion protein vector. Mature G cells comprise metamyelocytes, myelocytes, and granulocytes. Mean ± SD values from 5 experiments. The percentage of CD34+cells in the different samples is analyzed by FACS and is indicated above the columns. Single and double asterisks above the columns indicate statistical significance (P < .0001) of the differences in GFP vs. PML/RARα cells and PML/RARα vs. PML/RARα-AHT cells, respectively, as calculated from Student 2-tailed t test. (B) Representative cytospins of the samples described in A.
PML/RARα induces promyelocytic differentiation of HPC.
(A) The morphology of the cells was analyzed on cytospin slides stained with May-Grünwald-Giemsa. (first left panel) HPCs stimulated with IL-3, FLT3 ligand, and stem cell factor (see “Materials and methods”) before retroviral infection. (right panels) HPCs transduced with the control (GFP) or the indicated fusion protein vector. Mature G cells comprise metamyelocytes, myelocytes, and granulocytes. Mean ± SD values from 5 experiments. The percentage of CD34+cells in the different samples is analyzed by FACS and is indicated above the columns. Single and double asterisks above the columns indicate statistical significance (P < .0001) of the differences in GFP vs. PML/RARα cells and PML/RARα vs. PML/RARα-AHT cells, respectively, as calculated from Student 2-tailed t test. (B) Representative cytospins of the samples described in A.
Unilineage granulopoietic (G) proliferation and differentiation of HPC transduced with the indicated vector.
These findings are from 1 of 5 representative experiments. (A) Cell growth induced by G HGF at standard concentration,21 1:100 or 1:1000 dilutions. (B) Cell differentiation as assessed by morphologic analysis of liquid cultures in the presence of standard concentrations of G HGF, standard concentrations of G HGFs with the addition of 10−6 mol/L RA, or 1:100 diluted G HGF. As described in “Materials and Methods,” the HPCs were isolated and stimulated with growth factors on days −4 and −3, infected on day −2 and −1, and FACS purified on day 0.
Unilineage granulopoietic (G) proliferation and differentiation of HPC transduced with the indicated vector.
These findings are from 1 of 5 representative experiments. (A) Cell growth induced by G HGF at standard concentration,21 1:100 or 1:1000 dilutions. (B) Cell differentiation as assessed by morphologic analysis of liquid cultures in the presence of standard concentrations of G HGF, standard concentrations of G HGFs with the addition of 10−6 mol/L RA, or 1:100 diluted G HGF. As described in “Materials and Methods,” the HPCs were isolated and stimulated with growth factors on days −4 and −3, infected on day −2 and −1, and FACS purified on day 0.
In HGF-deprived liquid suspension culture, control HPCs rapidly ceased proliferation (Figure 3A) and died by apoptosis (not shown), whereas PML/RARα+ cells displayed a marked resistance to starvation and continued their expansion, albeit at a reduced rate (Figure 3A).
These data were confirmed by clonogenic assays (Table1). In unilineage G culture conditions, most of the colonies generated by PML/RARα+ HPC had a promyelocytic morphology. In diluted G growth factor, only the PML/RARα+ HPC resisted growth factor deprivation and generated promyelocytic colonies.
Number and type of colonies generated by 100 transduced HPC in methylcellulose culture under standard or diluted G HGF conditions
| . | GFP . | PML/RARα . | PML/RARα-AHT . | |||
|---|---|---|---|---|---|---|
| Promyelocytic . | Granulocytic . | Promyelocytic . | Granulocytic . | Promyelocytic . | Granulocytic . | |
| Standard G HGF | 0 | 12 ± 3 | 11.3 ± 3.5 | 2.3 ± 1.5 | 0 | 10 ± 3 |
| Diluted G HGF (1:100) | 0 | 2.7 ± 2 | 6.7 ± 3 | 1.3 ± 0.6 | 0 | 3.7 ± 1.5 |
| Diluted G HGF (1:1000) | 0 | 0 | 4.7 ± 2.5 | 1.0 ± 1 | 0 | 1.7 ± 1.1 |
| . | GFP . | PML/RARα . | PML/RARα-AHT . | |||
|---|---|---|---|---|---|---|
| Promyelocytic . | Granulocytic . | Promyelocytic . | Granulocytic . | Promyelocytic . | Granulocytic . | |
| Standard G HGF | 0 | 12 ± 3 | 11.3 ± 3.5 | 2.3 ± 1.5 | 0 | 10 ± 3 |
| Diluted G HGF (1:100) | 0 | 2.7 ± 2 | 6.7 ± 3 | 1.3 ± 0.6 | 0 | 3.7 ± 1.5 |
| Diluted G HGF (1:1000) | 0 | 0 | 4.7 ± 2.5 | 1.0 ± 1 | 0 | 1.7 ± 1.1 |
Values are mean ± SD values from 3 experiments.
Taken together, these results showed that PML/RARα dictated the promyelocytic blast phenotype by inducing HPC differentiation followed by maturation blockade and by protecting HPC from growth factor deprivation.
PML/RARα forces HPC to differentiate through granulopoietic lineage
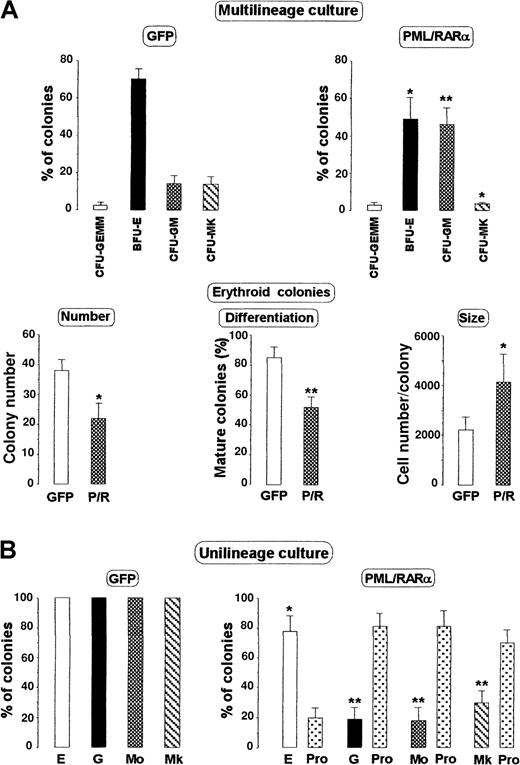
Because PML/RAR rapidly induces HPC to differentiate into promyelocytes, we asked whether the fusion protein could force HPC with multipotent (CFU-GEMM) or nongranulopoietic (eg, BFU-E) differentiation capacity to promyelocytic differentiation by reprogramming the HPC lineage commitment. We analyzed the effects of PML/RARα on HPC/HSC differentiation in methylcellulose clonogenic cultures (Figure4A) supplemented with saturating concentrations of an HGF cocktail promoting multilineage differentiation (see “Materials and methods”). Despite the multilineage differentiation stimulus, PML/RARα+ HPCs generated a much higher proportion of CFU-GM/CFU-G colonies than control cells. CFU-GEMM/BFU-E/CFU-MK colonies were conversely reduced; thus, the total number of colonies did not change, which suggested that PML/RARα did not affect the HPC clonogenic capacity. In addition, cytospin slides showed that the myeloid cells within CFU-GEMM and CFU-GM/CFU-G colonies were primarily promyelocytes (Figure 4 legend). Furthermore, BFU-E–derived colonies were larger and more immature than control colonies (Figure 4A).30 These results are in line with those in unilineage methylcellulose cultures (Figure 4B): control cells produced a single colony type (BFU-E, CFU-MK, CFU-G, or CFU-M colonies in E, Mk, G, or Mo culture, respectively), depending on the HGF cocktail,3-7 whereas PML/RARα+ HPCs generated a high number of promyelocytic colonies independently of the unilineage HGF stimulus (Figure 4B).
Clonogenesis by HPC transduced with the indicated vector.
(A, top) Morphology of the colonies generated by 100 HPCs stimulated with multilineage HGFs (see “Materials and Methods”), expressed as a percentage of the total number. Mean ± SD values from 5 experiments. The total number of colonies ± SD was 52 ± 7.1 for control (GFP) and 50 ± 5.5 for PML/RARα cells. (A, bottom) Analysis of the erythroid colonies in multilineage cultures: 40% of the cells within CFU-GEMM colonies and 62% of the cells within CFU-GM colonies were promyelocytes. (B) Morphology of the colonies generated by unilineage HGF stimuli (see “Materials and methods” and Grignani et al21). E, BFU-E; G, CFU-G; Mo, CFU–monocytic; Mk, CFU–megakaryocytic; Pro, promyelocytic colonies. Mean ± SD values from 5 experiments. The numbers of seeded cells/plate were 100 for BFU-E and CFU-G, 500 for CFU-MK, and 1000 for CFU-Mo. The mean number of colonies/plate ± SD generated by GFP cells were 25.4 ± 4.4 BFU-E, 12.5 ± 2.3 CFU-G, 15.6 ± 2.1 CFU-MK, and 17.8 ± 1.8 CFU-Mo. The total number of colonies generated by PML/RARα cells was 19.6 ± 1.8 BFU-E/promyelocytic, 13.6 ± 2.1 for CFU-G/promyelocytic, 18.4 ± 3.2 CFU-MK/promyelocytic, and 19.5 ± 3.1 CFU-Mo/promyelocytic. Asterisks above the columns indicate statistically significant differences between GFP and PML/RARα samples as calculated from a Student 2-tailed t test. *P < .01; **P< .001.
Clonogenesis by HPC transduced with the indicated vector.
(A, top) Morphology of the colonies generated by 100 HPCs stimulated with multilineage HGFs (see “Materials and Methods”), expressed as a percentage of the total number. Mean ± SD values from 5 experiments. The total number of colonies ± SD was 52 ± 7.1 for control (GFP) and 50 ± 5.5 for PML/RARα cells. (A, bottom) Analysis of the erythroid colonies in multilineage cultures: 40% of the cells within CFU-GEMM colonies and 62% of the cells within CFU-GM colonies were promyelocytes. (B) Morphology of the colonies generated by unilineage HGF stimuli (see “Materials and methods” and Grignani et al21). E, BFU-E; G, CFU-G; Mo, CFU–monocytic; Mk, CFU–megakaryocytic; Pro, promyelocytic colonies. Mean ± SD values from 5 experiments. The numbers of seeded cells/plate were 100 for BFU-E and CFU-G, 500 for CFU-MK, and 1000 for CFU-Mo. The mean number of colonies/plate ± SD generated by GFP cells were 25.4 ± 4.4 BFU-E, 12.5 ± 2.3 CFU-G, 15.6 ± 2.1 CFU-MK, and 17.8 ± 1.8 CFU-Mo. The total number of colonies generated by PML/RARα cells was 19.6 ± 1.8 BFU-E/promyelocytic, 13.6 ± 2.1 for CFU-G/promyelocytic, 18.4 ± 3.2 CFU-MK/promyelocytic, and 19.5 ± 3.1 CFU-Mo/promyelocytic. Asterisks above the columns indicate statistically significant differences between GFP and PML/RARα samples as calculated from a Student 2-tailed t test. *P < .01; **P< .001.
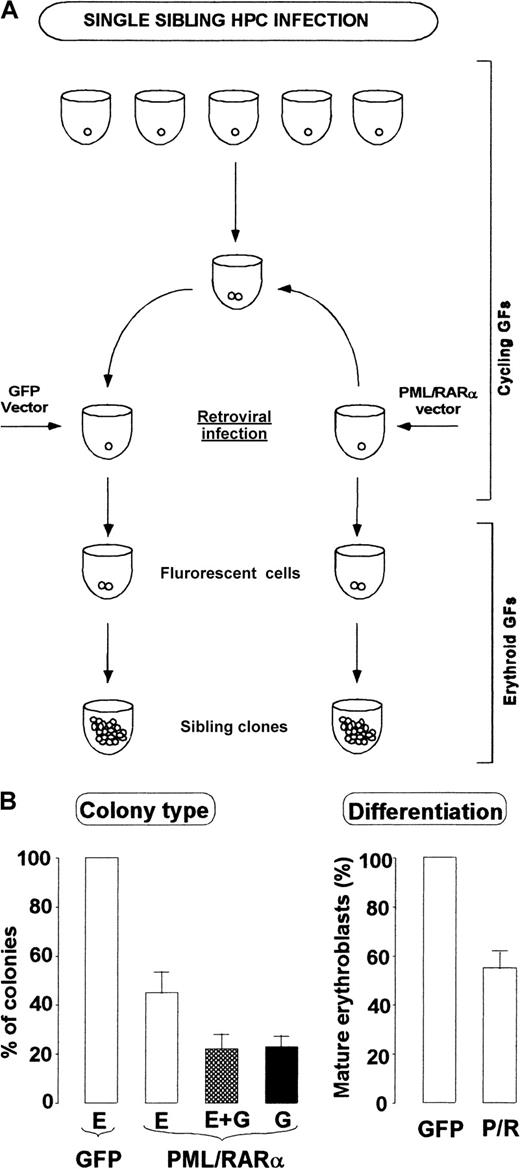
To prove formally that PML/RARα expression alters the commitment program, we isolated single HPCs and stimulated their proliferation to generate each into 2 cells.27 Sibling cells were isolated and infected with PML/RARα or control retrovirus. After 48 hours fluorescent cells were scored, and control or PML/RARα-transduced siblings were induced to differentiate along the E pathway in liquid suspension culture (Figure 5A). At day 10 the clones were individually collected, and their morphology was analyzed. Although all control cells formed mature erythroid clones, PML/RARα+ cells produced 26% pure granulocytic clones, 24% produced mixed erythroid granulocytic clones, and only 50% produced erythroid clones whose differentiation stage was more immature than that of control ones (Figure 5B). Overall these data show that PML/RARα expression can force the HPC program into preferential granulopoietic differentiation through the promyelocytic stage.
HPC commitment reprogramming by PML/RARα: single-cell analysis.
(A) Scheme of sibling HPCs generation, infection and analysis. (B) Morphologic analysis of the clones generated by single sibling HPC infected with control (GFP) or PML/RARα vector. (left panel) E, erythroid clones; E + G mixed clones with erythroid and myeloid cells, mostly represented by promyelocytes; G, granulocytic clones, primarily promyelocytic. (right panel) Differentiation levels of erythroid clones, as evaluated from percentages of orthochromatic erythroblasts. Mean ± SD values from 3 experiments in which at least 20 sibling colonies/vector were studied.
HPC commitment reprogramming by PML/RARα: single-cell analysis.
(A) Scheme of sibling HPCs generation, infection and analysis. (B) Morphologic analysis of the clones generated by single sibling HPC infected with control (GFP) or PML/RARα vector. (left panel) E, erythroid clones; E + G mixed clones with erythroid and myeloid cells, mostly represented by promyelocytes; G, granulocytic clones, primarily promyelocytic. (right panel) Differentiation levels of erythroid clones, as evaluated from percentages of orthochromatic erythroblasts. Mean ± SD values from 3 experiments in which at least 20 sibling colonies/vector were studied.
PML/RARα mutant devoid of N-CoR binding activity does not alter HPC commitment and myeloid maturation
The ability of PML/RARα to block differentiation requires the recruitment of a co-repressor/HD complex. To ascertain which of the biologic effects of the fusion protein depend on this complex, we studied HPCs infected with a retrovirus carrying a PML/RARα AHT mutant unable to bind the N-CoR/HD complex. Cells expressing the mutant protein at levels similar to those of PML/RARα (Figure 1) showed neither the initial differentiation wave nor the subsequent maturation block. HGF-induced G differentiation was slightly accelerated when compared to controls (Figure 2, 3B). In multilineage and unilineage clonogenic assays, HPC expressing PML/RARα+ AHT generated the same number, type, and size of colonies as control cells (not shown). Expression of the mutant protein partially protected HPCs from cell death induced by growth factor deprivation in a clonogenic assay, though the native PML/RARα protein showed stronger activity (Table 1).
Discussion
We show here that the expression of the oncogenic fusion protein PML/RARα in normal human HPC/HSC induces the development of a leukemic promyelocytic phenotype through 3 cellular mechanisms: reprogramming of HPC commitment toward preferential granulocytic differentiation, induction of HPC differentiation through the promyelocytic stage, and inhibition of promyelocyte differentiation. The first 2 effects are previously unreported. They indicate that a single oncogenic event may have a complex dual effect. Before maturation block, commitment and differentiation are actively induced and force the cells to a particular hematopoietic lineage and maturation stage by affecting the entire hematopoiesis. Our results in bulk cell and single cell cultures show that PML/RARα expression not only affects granulopoiesis-committed HPCs, it directs the entire spectrum of HPCs toward the granulocytic lineage. These findings identify the cellular mechanism underlying the development of a specific leukemic phenotype.
Current knowledge of APL target cells is mainly based on experiments showing that CD34+ CD38− progenitor cells from patients with APL do not express PML/RARα29 and are unable to transplant leukemia in NOD/SCID mice,2thereby suggesting that the APL target cell is a nontransplantable, mature promyelocyte. Our data, instead, indicate that PML/RARα exerts its effects on early hematopoietic progenitors. These cells are actually most of our retroviral infection targets, on which PML/RARα has a strong biologic effect. However, PML-RARα expression did not alter the proliferation/differentiation program of Dexter type 12-week extended long-term culture-initiating cells (LTC-IC), representative of stem cells (Hao et al31 and our unpublished results). Therefore, we propose that the APL target cell are early progenitors, probably more differentiated than CD34+ CD38− cells.
The mechanism through which PML/RARα reprograms HPC commitment awaits further studies. Retinoic acid, acting on its normal receptors, exerts a dual effect on normal hematopoietic progenitors: it delays the differentiation of primitive hematopoietic progenitors and induces maturation of committed myeloid progenitors.32 These 2 effects are opposite to those exerted by PML/RARα. Thus, both differentiation induction and differentiation block by PML/RARα may be due to its dominant negative effect on the RARα molecular pathway. Because PML cooperates with RARα,33,34 a negative activity on PML may contribute to this effect. The constitutive transcriptional repression exerted by PML/RARα20-23 may induce lineage commitment and the initial differentiation phase by decreasing the expression of critical genes that keep the cells in an undifferentiated state. The subsequent differentiation block is likely to result from the impaired induction of RARα target genes in more mature cells.20-23 32
Unlike murine progenitors,35 PML/RARα+ human HPC/HSC are seemingly incapable of indefinite proliferation in culture. In liquid suspension culture with saturating KL, FLT3 ligand, and IL-3, PML/RARα expression persisted, though at reduced levels, and the cells acquired a promyelocytic phenotype and rapidly stopped proliferating (not shown). This may reflect intrinsic differences between mouse and human cells.36 Development of a frank human leukemia in vivo may require a further genetic lesion. However, APL blasts have a very low proliferation rate, and, in most cases, the patients are leukopenic.12 Promyelocyte differentiation block and protection from apoptosis, coupled with altered hematopoiesis, may be responsible for blast accumulation in vivo. The balance between leukemic HPC self-renewal and differentiation may be the basis for the poor cellularity of APL. Promyelocytic differentiation induced by PML/RARα may limit expansion of the leukemic HPC pool, thus explaining both the leukopenia in patients with APL and the difficulty in transplanting APL into NOD/SCID mice.
The PML/RARα protein was detected by immunofluorescence in all the transduced cells purified by FACS, indicating a good correlation between GFP and fusion gene expression. Notably, biologic effects similar to those described above were obtained with a retrovirus encoding a GFP-PML/RARα fusion protein ensuring that fluorescent cells express the oncogenic protein (not shown). Therefore, most biologic changes in HPCs, affecting commitment and the first days of differentiation, are coupled with high levels of fusion protein expression. In differentiating HGF cultures, PLM/RARα protein expression decreased after a week. The late differentiation observed in standard G culture5 may be attributed to reduced expression of the fusion protein. Alternatively, this may be the consequence of the saturating G HGF stimulus. Indeed, this HGF cocktail also induces a partial differentiation of APL blasts in vitro (our unpublished results), and PML/RARα HPC differentiation was almost blocked at lower HGF concentrations (Fig. 3D).
These data reflect the potential of the model system established here. We used a gene transfer strategy that allowed isolation of purified transduced HPC/HSC,9 followed by their differentiation along each hematopoietic lineages.3-7 The apoptotic effect of PML-RARα in permanent packaging cell lines37 is bypassed by an efficient transient production of retroviral particles.9 Furthermore, the promyelocytic phenotype induced by PML/RARα was unambiguously distinguishable from that of the transduced blasts, allowing analysis of the initial differentiation phase. Finally, the reprogramming of HPC commitment induced by the fusion protein was explored at the single HPC level by sibling cell analysis in unilineage culture. Altogether, this strategy permits analysis of the transduced primary HPC/HSC from the earliest phases after oncogene transfer through differentiation/maturation along each hematopoietic pathway. Our approach allowed the PML/RARα effects so far detected in cell lines (ie, maturation block, protection from apoptosis, sensitization to RA differentiation stimulus) to reproduce in primary HPC culture, while it unveiled novel biologic actions of the fusion protein (ie, HPC reprogramming and differentiation induction to the promyelocytic stage).
PML/RARα exerts its pathogenetic activity by recruiting HD through the co-repressors N-CoR and SMRT.21-24 We show here that the PML/RARα-AHT protein, mutated in the co-repressor N-CoR binding site, lost the potentially oncogenic biologic effects (Fig. 2, 3, 4). The PML/RARα-AHT mutant does not block myeloid differentiation and does not modify lineage commitment, indicating that, most likely, PML/RARα alters lineage choice and cell maturation through an N-CoR/HD–dependent repression of target gene transcription. We cannot exclude, however, that other functions of the PML/RARα protein, such as binding to co-activators or other co-factor proteins, could be altered in the AHT mutant. Expression of the mutant protein moderately increased HPC differentiation rate, possibly by competing out the RA-independent transcriptional repressor activity of the normal RARα protein. The mutant had a limited protective effect on starvation, suggesting that this effect is partially independent of transcriptional repression. Actually, a contribution to the PML/RARα-induced phenotype could also derive from RXR sequestration and altered RAR-RXR heterodimerization, similar to what happens in hematopoietic cells with RARα overexpression.16 38 Overall, our results are the first demonstration in normal primary cells that recruitment of the N-CoR/HD complex by PML/RARα is required to modify the genetic program regulating both commitment and differentiation and to determine the cellular effects generating the APL phenotype.
F.G., P.G.P., and C.P. are supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC). L.L. is a recipient of an AIRC fellowship.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
F. Grignani and C. Peschle, Department of Hematology and Oncology, Istituto Superiore di Sanità, V. le Regina Elena, 299; 00161 Rome, Italy; e-mail:fragrig@unipg.it;c.peschle@ema.net.iss.it.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal