Abstract
To study the regulation of the early stages of hematopoiesis, cDNA representational difference analysis was used to isolate genes that were differentially expressed in primitive hematopoietic progenitors. The reasoning was that such genes were more likely to provide functions important to hematopoietic stem cells and progenitors. One of the genes identified through this approach encodes mouse Jagged2(mJagged2). Using quantitative reverse transcription–polymerase chain reaction, it was shown that mJagged2 was differentially expressed in c-kit+ hematopoietic progenitors, including those with the phenotypes of Lin− c-kit+Rhlo Holo and Lin−c-kit+ Rhhi Holo, and that they have been shown to be highly enriched for long-term and short-term repopulating hematopoietic stem cells, respectively. Western blot analyses showed that endothelial cells also expressed high levels of Jagged2, but stromal fibroblasts did not. Using a coculture system we found that exogenous, full-length mJagged2 promoted the survival and proliferation of hematopoietic progenitors, including the high-proliferative potential colony-forming cells. Direct cell-to-cell contact was required for this effect. Taken together, these findings indicate that both c-kit+ hematopoietic progenitors and endothelial cells express Jagged2 and that exogenous, full-length Jagged2 promotes the survival and proliferation of hematopoietic progenitors.
Although much progress has been made in understanding the regulation of the intermediate to late stages of hematopoiesis, relatively little is known about the control of hematopoietic stem cells (HSC). Part of the difficulty is the scarcity of HSC. To identify genes that might be important in the regulation of HSC, we capitalized on the availability of 2 complementary cell lines, EML C1 and MPRO. EML C1 is a stem cell factor (SCF)-dependent, multipotent hematopoietic cell line with erythroid, myeloid, and B-lymphoid potential.1 MPRO is a granulocyte macrophage–colony-stimulating factor (GM-CSF)-dependent neutrophilic promyelocyte cell line with a reversible block in terminal differentiation.2 Developmentally, EML C1 is relatively close to HSC, whereas MPRO is already committed to the neutrophil lineage. A subtraction of EML C1 cDNA by MPRO cDNA should yield genes that are differentially expressed in HSC or in progenitors committed to various hematopoietic lineages except the neutrophil lineage. The approach we used was cDNA representational difference analysis (RDA),3 which produces fewer false-positive results than most methods of subtraction.
Here we report the isolation and functional characterization of one such gene, E11-5, with the main focus on its roles in hematopoiesis. The cDNA represented by E11-5 encodes the mouse homologue of rat and human Jagged2 (rJagged2 and hJagged2)4,5 and is designated as mJagged2. mJagged2 belongs to the so-called DSL ligand family.6-8 All members of this ligand family are transmembrane proteins that serve as the ligands for Notch and related receptors.9 They are characterized by a conserved DSL motif in their extracellular domains. This motif was first noted inDrosophila Delta,10,11 Serrate,12,13and Caenorhabditis Lag27 and is essential for interaction with Notch and related receptors. In the embryos ofDrosophila and C. elegans, Notch and related receptors play important roles in cell fate decisions through the process of lateral inhibition or inductive signaling.8,9 Available evidence indicates that on binding of DSL ligands, the Notch receptor is cleaved at a site external to the transmembrane domain. The intermediate is further cleaved by presenilin or a presenilin-dependent γ-secretases within the transmembrane domain to release the intracellular domain.14,15 The freed intracellular domain forms a complex with the Drosophilatranscription factor Suppressor of Hairless (Su[H]) and the complex translocates to the nucleus where Su(H) binds to regulatory sequences of the Enhancer of Split [E(spl)] genes and up-regulates their expression. Besides Su(H), other Drosophilaproteins shown to bind the activated Notch cytoplasmic domain include EMB5 (a regulator of chromatin structure), Deltex (a ring zinc-finger–containing cytoplasmic protein), Numb (a signaling adaptor protein involved in the development of peripheral nervous system), Disheveled (an element of the Wingless signaling pathway), and Disabled (accessory protein of Abl kinase). In mammals, CBF1 (equivalent of Su[H]), Nur77 (a transcription factor involved in the apoptosis of thymocytes), Bcl3 (a member of the IkB family), and Deltex homologues have been shown to bind to activated cytoplasmic domains of Notch-1 (reviewed9). The diversity of signal transduction molecules capable of interacting with the activated Notch receptor underscores the diverse effects of Notch and DSL ligands.
Compared with Delta and Serrate of Drosophila, less is known about the functions of mammalian DSL ligands. To date, at least 4 mammalian DSL ligands have been described: Delta-like 1 (Dll-1),16 Delta-like 3 (Dll-3),17Jagged1,18,19 and Jagged2 (4,5 and this report). Four mammalian Notch receptors have been identified: Notch-1,20,21 Notch-2,22 Notch-3,23and Notch-4.24,25 The functions and the receptor usage of mammalian DSL ligands are largely unknown. Among the mammalian DSL ligands, rJagged14 was cloned first and has been studied more extensively. Earlier studies of mammalian DSL ligands focused on their involvement in differentiation.18,26,27 More recent studies indicate that the DSL–Notch signal transduction pathway may also regulate apoptosis28,29 and cellular proliferation.30 In mouse embryos, Jagged1 andJagged2 show both overlapping and unique patterns of expression. For example, Jagged2 is highly expressed in thymus, whereas Jagged1 is not.4,5Jagged2 is expressed in the basal layer of epidermis, whereas Jagged1 is expressed in the suprabasal layer (4,5 and our unpublished data). Thus, Jagged1 and Jagged2 may have nonredundant roles. Human CD34+ cells cocultured with hJagged2-expressing NIH3T3 cells in the presence of Flt3 ligand and PIXY 321, a hybrid of human IL-3 and GM-CSF, showed a slower decline of the CD34+cells.31 In the current study, we found that membrane-bound, full-length mJagged2 promoted the survival and proliferation of mouse hematopoietic progenitors in the absence of added hematopoietic growth factors. This effect required direct cell-to-cell contact. Furthermore, we found that endothelial cells expressed high levels of Jagged2 but that stromal fibroblasts did not. These new observations further implicate mJagged2 as a regulator of hematopoiesis.
Materials and methods
cDNA RDA
The details of cDNA RDA have been described.3 Briefly, EML C1 and MPRO cDNA were synthesized from poly(A+) RNAs using oligo d(T) as primers. Double-stranded cDNA was digested withDpnII (Biolab, Beverly, MA) and ligated with R-Bgl-12 and R-Bgl-24 linker/primers3 that allowed amplification by polymerase chain reaction (PCR) to generate a representation of cDNA. The MPRO cDNA representation (the driver) was subsequently digested with DpnII to remove the linker/primers and was heated to melt the double strands. The EML C1 cDNA representation (the tester) was digested with DpnII to remove the linker/primers and then was gel purified and re-ligated with a second set of linker/primers (J-Bgl-12 and J-Bgl-24).3 The J-Bgl-12/24-ligated EML C1 cDNA representation was mixed with a 100-fold excess of melt MPRO cDNA representation and was allowed to hybridize at 67°C for 24 hours. Common sequences formed tester:driver duplexes that contained the linker/primer at only one end and, therefore, could be amplified only in a linear fashion by PCR. Unique sequences formed tester:tester duplexes that contained the linker/primer at each end and could be amplified exponentially. This resulted in a kinetic enrichment of the unique sequences in EML C1 representation further enriched by mung bean nuclease (Stratagene, La Jolla, CA) digestion of the single-stranded, linearly amplified DNA of the tester:driver duplexes. The resultant DNA was then reintroduced to the beginning of next round of RDA. The ratios of tester-to-driver DNA were 1:8000 and 1:40 000 for the second and third rounds, respectively. The RDA products were purified and cloned into pBluescript SK(+) (Stratagene) for sequencing.
cDNA library screening and 5′ RACE
A Swiss Webster/NIH mouse embryo cDNA library in λgt11 was screened with radiolabeled E11-5 DNA. Eleven positive clones representing 2 overlapping fragments were picked. After secondary screening, phage DNA was purified for cycle sequencing. For 5′ RACE (rapid amplification of cDNA ends), 1 μg EML C1 mRNA was primed with a gene-specific primer (5′-GCTGAAGTGCAGGCTCTT) using sequence data from the λgt11 clones. First-strand synthesis and d(C)-tailing were carried out per manufacturer's instruction (Gibco BRL, Grand Island, NY) and amplified by PCR using a gene-specific primer (5′-GGTTGATCATGCCAGCGTGCGACACC) and the abridged anchor primer. The product was reamplified with a second nested, gene-specific primer (5′-CGCTCAATCAGCAGCTCCTCATCTGG) and the abridged universal amplification primer. The complete cDNA sequence of E11-5 was compiled from the λgt11 cDNA clones and 5′ RACE product and was verified in full-length cDNA from the EML C1 cell line.
RT-PCR and Northern blot analyses
All primers chosen for reverse transcription (RT)-PCR span the intron–exon junctions to prevent amplification of genomic DNA. The correct PCR product was 477 bp in length. To prepare total RNA for RT-PCR, cells were pelleted and lysed on ice in diethylpyrocarbonate-treated water containing RNasin (0.5 U/μL; Stratagene) at a ratio of 10 μL/1000 cells. Serial 5-fold dilutions (up to 625-fold) were made from this master lysate and used as a set in RT-PCR. The first-strand synthesis and the primary PCR were performed in a volume of 50 μL in the same tube containing 10 μL cell lysate, 25 pmol each of antisense primer (5′-CCCATGCTGACACTGCCCAT) and sense primer (5′-GTGTGGAGCCCTGGCACTGT), 1.5 mmol/L MgCl2, 50 mmol/L KCl, 10 mmol/L Tris-HCl (pH 9 at 25°C), 0.1% Triton X-100, 200 μmol/L dNTP, 1.25 U Taq polymerase (Qiagen, Valencia, CA), 10 U RNasin, and 5 U SuperScript II (Boehringer Mannheim, Indianapolis, IN). The mixture was incubated for 30 minutes at 42°C (RT reaction), heated to 95°C for 5 minutes, and amplified using 25 cycles at 94°C for 30 seconds, 65°C for 30 seconds, and 72°C for 1 minute, followed by a final elongation of 4 minutes at 72°C. For secondary PCR, 5 μL of the primary PCR was added to a new tube containing 25 pmol each of nested antisense primer (5′-GGCAATCACAGTAATAGCCGCCAAT) and the nested sense primer (5′-GGCACTGTGACTGTGAGACCAACT), 1.5 mmol/L Mg Cl2, 50 mmol/L KCl, 10 mmol/L Tris-HCl (pH 9 at 25°C), 0.1% Triton X-100, 1 mmol/L dNTP, and 1.25 U Taq polymerase in a volume of 50 μL. Samples were denatured at 94°C for 30 seconds and amplified using 25 cycles at 94°C for 30 seconds, 65°C for 30 seconds, and 72°C for 1 minute, followed by a final extension at 72°C for 4 minutes. Five microliters of the secondary PCR products was resolved on a 2% agarose gel and stained with ethidium bromide. In all mJagged2 RT-PCR, only one band was observed. The sequence of the single PCR product was verified by direct cycle sequencing. Primers used in mouse β-actin RT-PCR were antisense primer for cDNA synthesis and primary PCR (5′-GCCACGCTCGGTCAGGATCTT), sense primer (5′-GCCCAGAGCAAGAGAGGTATCCT), nested antisense primer (5′-TTTGATGTCACGCACGATTTCC), and nested sense primer (5′-TGTGATGGTGGGAATGGGTCAG). For Northern blot analyses, RNA was resolved on 1% agarose gels and blotted onto HyBond-N (Amersham, Buckinghamshire, UK) and hybridized with random-primed, P32-labeled probes at 65°C in Rapid Hyb buffer (Amersham).
Retroviral vectors and transduction of Rab-9 cells
The entire coding region of mJagged2 was cloned into the LXSN vector.32 The sequence preceding the first ATG was modified to conform to the Kozak consensus sequence. This vector was named LMJSN. Similarly, a 3.3-kb DNA encoding the entire extracellular domain (ECD) of mJagged2 (aa 1-1083) plus the Kozak consensus sequence and a stop codon was cloned into the LXSN vector. The resultant vector was named LECDSN. Amphotropic producer cell lines (PA317/LXSN, PA317/LMJSN, and PA317/LECDSN) were established as described.33 Rab-9 cell strain was obtained from American Type Culture Collection and maintained in Dulbecco's modified Eagle's medium (DMEM)/10% fetal calf serum (FCS). This diploid cell strain was established from the skin of a rabbit and was not transformed or immortalized. A clonal strain, Rab-9 C.14, was established and infected with 1 mL supernatant from each viral producer line for 24 hours and was selected with G418 (0.75 mg/mL) for 10 days. The resultant cell strains, designated as Rab-9/LXSN, Rab-9/LMJSN, and Rab-9/LECDSN, were subcultured at a 1:4 ratio every 3 to 5 days.
Immunofluorescence staining and Western blot analyses
The entire intracellular domain (ICD) (aa 1109-1247) of mJagged2 was expressed as GST fusion proteins per standard protocol (Gibco BRL) and was used to immunize a New Zealand White rabbit. Indirect immunofluorescence staining of Rab-9 cells grown on glass coverslips was performed using rabbit preimmune and immune sera (1:200 dilution) as the first antibody and fluorescein isothiocyanate-conjugated donkey antirabbit IgG (H+L) (1:400 dilution; PharMingen, San Diego, CA) as the second antibody. For Western blot analysis, cells were lysed in lysis buffer (50 mmol/L Tris, pH 7.5, 100 mmol/L NaCl, 5 mmol/L EDTA, 1 mmol/L phenylmethylsulfonyl fluoride, 1% Triton X-100, 0.5% sodium dodecyl sulfate [SDS]). From each cell line, 3 to 10 μg total protein was run on 8% SDS–polyacrylamide gel electrophoresis (PAGE) and blotted onto nitrocellulose (HyBond-ECL; Amersham). Western blot was probed with preimmune or immune rabbit sera at 1:2000 dilution, followed by horseradish peroxidase-conjugated donkey antirabbit IgG (Amersham), and visualized by the enhanced chemiluminescence method (Amersham).
Fractionation and enrichment of bone marrow
Fractionation and enrichment of bone marrow were performed according to published methods with modification.34 35 Briefly, low-density mononuclear bone marrow cells from B6SJL mice were separated by Nycodenz (1.077 mg/mL; Nycomed) density centrifugation. For further purification, mononuclear cells (MNC) were stained with a cocktail of monoclonal antibodies (anti-7/4, anti-B220, anti-YW25.12.7, Mac-1, Gr-1, Ter-119) (PharMingen) spun over an FCS cushion and rosetted twice with magnetic beads coated with sheep antirat IgG (Dynabeads M-450; Dynal, Oslo, Norway) at a bead:cell ratio of 10:1. Unstained cells were separated by the Dynal magnet and collected as the Lin− (lineage marker-negative) fraction. The Lin− cells were stained with Hoechst 33342 (Ho) (Sigma, St Louis, MO) and rhodamine (Rh) (Sigma), then with phycoerythrin-labeled antic-kit antibody (PharMingen) and propidium iodide (Sigma) and sorted with a FACStar Plus flow cytometer (Becton Dickinson, Franklin Lakes, NJ) equipped with dual argon lasers. The long-term repopulating potential of the Lin− c-kit+ RhloHolo cells was verified by single-cell transplantation (together with supporting cells).
Coculture of bone marrow cells with Rab-9 cells
Monolayers of Rab-9/LXSN, LMJSN, and LECDSN were established in 12-well plates 1 week before coculture experiments. Bone marrow MNC or Lin− c-kit+ cells were cocultured with Rab-9 cells in DMEM/10%FCS (4 mL/well). Half the medium was replaced every 2 to 4 days. Subculturing was performed at each colony assay and at additional time points.
Hematopoietic colony assays
Lin− c-kit+ RhloHolo cells were cultured in Iscove's modified Dulbecco's medium (IMDM) supplemented with FCS (12.5% vol/vol), horse serum (12.5% vol/vol), SCF (100 ng/mL), mouse IL-3 (100 ng/mL), mouse IL-6 (20 ng/mL), and mouse GM-CSF (5 ng/mL). For colony assays, cells were plated in 1% methylcellulose medium supplemented with 20% horse serum and the 4 growth factors specified above at the same concentrations. Colony assays of nonadherent cells harvested from Rab-9 cocultures were carried out in 1% methylcellulose medium supplemented with 30% FCS, 10% bovine serum albumin (wt/vol), SCF (100 ng/mL), IL-3 (10 ng/mL), and GM-CSF (5 ng/mL).
Stromal cell lines and human umbilical vein endothelial cells
STW, STV, STS, and STD are spontaneously immortalized fibroblast cell lines established from long-term bone marrow cultures ofW(+/+), W/Wv, Sl (+/+), andSl/Sld mice, respectively.36The S17 cell line37 was a gift from Robert Rottapel (University of Toronto, Toronto, Canada). Human umbilical vein endothelial cells (HUVEC) were kindly provided by Nasserine Haque and Peter Harpel (Mount Sinai School of Medicine, New York, NY).
Results
Cloning of mJagged2 from a multipotent hematopoietic cell line
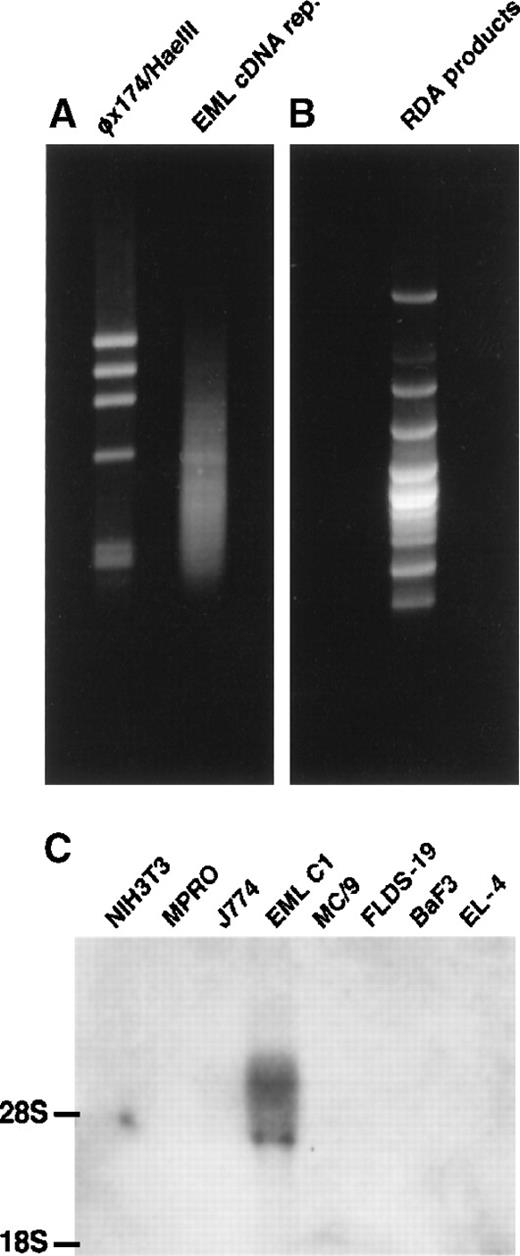
cDNA RDA was performed as described3 with some modifications. The EML C1 cDNA representation appeared as a smear of DNA fragments ranging from 0.2 to 1.2 kb on the gel (Figure1A) and was used as the “tester” in RDA. The MPRO cDNA representation also appeared as a smear (not shown) and served as the “driver” in RDA. After 3 rounds of RDA, the products were resolved on an agarose gel (Figure 1B). Individual DNA was cloned and sequenced. Among the genes identified by EML C1-MPRO cDNA RDA were CD34 and c-kit (both are expressed by HSC and progenitors), Flt-1, mFrizzled-2, andPax-6. Another gene, designated E11-5, exhibited EML C1-specific expression among a panel of hematopoietic cell lines and was further characterized (Figure 1C).
Isolation of E11-5 by cDNA RDA.
(A) The right lane contains EML C1 cDNA representation. The left lane contains the φχ174/HaeIII markers. (B) The result of EML C1-MPRO cDNA RDA was resolved on 2% agarose and stained with ethidium bromide. Some bands contained more than one DNA species. (C) Northern blot analysis of the expression of E11-5 in cell lines representing different hematopoietic lineages. Each lane contains 5 μg total RNA from NIH3T3 (fibroblast), MPRO (neutrophil), J774 (macrophage), EML C1 (stem cell-like), MC/9 (mast cell), FLDS-19 (erythroblast), BaF3 (B-cell), and EL-4 (T-cell). The blot was hybridized with a radiolabeled, nick-translated E11-5 (mJagged2) probe.
Isolation of E11-5 by cDNA RDA.
(A) The right lane contains EML C1 cDNA representation. The left lane contains the φχ174/HaeIII markers. (B) The result of EML C1-MPRO cDNA RDA was resolved on 2% agarose and stained with ethidium bromide. Some bands contained more than one DNA species. (C) Northern blot analysis of the expression of E11-5 in cell lines representing different hematopoietic lineages. Each lane contains 5 μg total RNA from NIH3T3 (fibroblast), MPRO (neutrophil), J774 (macrophage), EML C1 (stem cell-like), MC/9 (mast cell), FLDS-19 (erythroblast), BaF3 (B-cell), and EL-4 (T-cell). The blot was hybridized with a radiolabeled, nick-translated E11-5 (mJagged2) probe.
The E11-5 DNA fragment contained 412 bp. The complete coding sequence, including parts of the 5′ and 3′ untranslated regions, was compiled from 2 overlapping cDNA clones and the 5′ RACE product and was submitted to GenBank (accession no. AF038572). Homology search revealed high degrees of similarity to rJagged2 (95% identity at DNA level) and hJagged2 (87% identity at DNA level) and lower degrees of homology to rJagged1 (50% at DNA level) and hJagged1 (46% at DNA level). The open reading frame of mJagged2 encoded a protein of 1247 amino acids. Hydropathy analysis revealed 2 hydrophobic domains: a predicted signal peptide (a.a. 1-22) and a transmembrane domain (a.a. 1084-1108). Amino acids 195-243 constitute the conserved DSL domain. Like rJagged2, mJagged2 contains 16 tandem repeats of epidermal growth factor-like domains downstream of the DSL domain. After these epidermal growth factor-like repeats are a cysteine-rich region, a transmembrane domain, and a 139-amino acid intracellular segment.
Normal hematopoietic progenitors express mJagged2
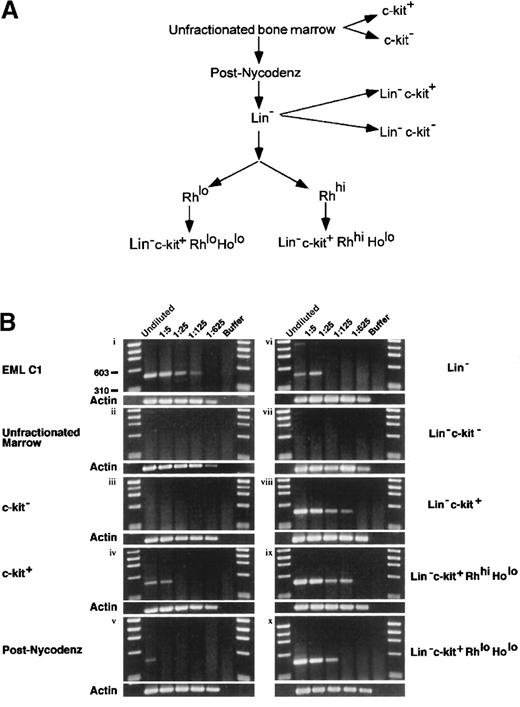
Although mJagged2 was initially cloned from the multipotent hematopoietic cell line EML C1, it was important to demonstrate that normal HSC and progenitors also expressed mJagged2. To determine whether normal mouse HSC expressed mJagged2, we performed RT-PCR using mJagged2-specific primers on Lin− c-kit+ RhloHolo (lineage markers-negative, c-kit [SCF receptor]-positive, Rh 123-low uptake, and Ho-low uptake) mouse bone marrow cells. The Lin− c-kit+Rhlo Holo fraction is highly enriched for LTR-HSC.34,35 For comparison, RT-PCR was also performed on other fractions of mouse marrow enriched or depleted of HSC and progenitors. The fractionation of bone marrow is outlined in Figure2A. To allow comparison of expression levels, RT-PCR was performed on undiluted and serial 5-fold diluted lysates. Results are shown in Figure 2B. EML C1 lysate served as a positive control for RT-PCR (Figure 2Bi). The unfractionated bone marrow showed no detectable mJagged2 expression (Figure 2Bii). When bone marrow was directly separated into c-kit−and c-kit+ fractions, only the c-kit+ fraction (enriched for progenitors) showed mJagged2 expression (Figure2Biii-iv). When marrow was first fractionated by density-gradient centrifugation over Nycodenz to remove mature cells, low-level expression of mJagged2 was detected in MNC (Figure 2Bv). Purification of the MNC by antibody depletion of lineage markers–positive cells yielded the Lin− fraction, which showed slightly increased levels of mJagged2 expression (Figure 2Bvi). Further enrichment for HSC and progenitors by positive selection for c-kit expression yielded the Lin−c-kit+ fraction that showed markedly increased mJagged2 expression (Figure 2Bviii). The Lin−c-kit− fraction, devoid of HSC and progenitors, did not express mJagged2 (Figure 2Bvii). The Lin−c-kit+ Rhhi Holo fraction contained spleen colony-forming units (CFU-S8-12) capable of short-term repopulation of lethally irradiated mice (STR-HSC).34,38 39 This fraction also expressed high levels of mJagged2 (Figure 2Bix). The Lin−c-kit+ Rhlo Holo fraction (containing LTR-HSC) showed relatively high levels of mJagged2(Figure 2Bx). Overall, the expression of mJagged2 appears to be restricted to the c-kit+ hematopoietic progenitors. In all mJagged2 RT-PCR experiments (except Figure 2Bvi), there was only one visible PCR product (477 bp) whose identity was verified by direct sequencing of the gel-purified DNA. (The faint, 1.4-kb PCR product in panel vi of Figure 2B resulted from amplification of the genomic mJagged2 DNA.)
Expression of mJagged2 in various bone marrow fractions.
(A) Fractionation of bone marrow for RT-PCR. To purify LTR-HSC, total bone marrow was first fractionated by Nycodenz density centrifugation, followed by depletion of lineage-marker–bearing cells using monoclonal antibodies, then by FACS sorting of Rh 123-stained cells, and finally by sorting of c-kit–bearing, Ho-low cells. Additional fractions (c-kit−, c-kit+, Lin−c-kit−, Lin− c-kit+) were sorted directly from total bone marrow or Lin−cells based on c-kit expression. See details in “Materials and methods.” (B) Quantitative RT-PCR for mJagged2. Serial 5-fold dilutions (up to 1:625 dilution) of cell lysates prepared from 1000 cells were used as a set in RT-PCR with nested primers. See “Materials and methods” for details of RT-PCR. Five-microliter aliquots of RT-PCR products were resolved on 2% agarose and stained with ethidium bromide. Results of β-actin RT-PCR are shown below each panel. First and last lanes of each gel contain the φχ174/HaeIII markers. (i) EML C1; (ii) unfractionated bone marrow; (iii) c-kit− fraction (devoid of stem cells); (iv) c-kit+ fraction (containing progenitors); (v) post-Nycodenz density centrifugation (ie, MNC); (vi) Lin− fraction; (vii) Lin-− c-kit+ fraction (enriched for progenitors); (viii) Lin− c-kit−fraction (devoid of stem cells); (ix) Lin−c-kit+ Rhhi Holo fraction (STR-HSC); (x) Lin− c-kit+Rhlo Holo fraction (LTR-HSC).
Expression of mJagged2 in various bone marrow fractions.
(A) Fractionation of bone marrow for RT-PCR. To purify LTR-HSC, total bone marrow was first fractionated by Nycodenz density centrifugation, followed by depletion of lineage-marker–bearing cells using monoclonal antibodies, then by FACS sorting of Rh 123-stained cells, and finally by sorting of c-kit–bearing, Ho-low cells. Additional fractions (c-kit−, c-kit+, Lin−c-kit−, Lin− c-kit+) were sorted directly from total bone marrow or Lin−cells based on c-kit expression. See details in “Materials and methods.” (B) Quantitative RT-PCR for mJagged2. Serial 5-fold dilutions (up to 1:625 dilution) of cell lysates prepared from 1000 cells were used as a set in RT-PCR with nested primers. See “Materials and methods” for details of RT-PCR. Five-microliter aliquots of RT-PCR products were resolved on 2% agarose and stained with ethidium bromide. Results of β-actin RT-PCR are shown below each panel. First and last lanes of each gel contain the φχ174/HaeIII markers. (i) EML C1; (ii) unfractionated bone marrow; (iii) c-kit− fraction (devoid of stem cells); (iv) c-kit+ fraction (containing progenitors); (v) post-Nycodenz density centrifugation (ie, MNC); (vi) Lin− fraction; (vii) Lin-− c-kit+ fraction (enriched for progenitors); (viii) Lin− c-kit−fraction (devoid of stem cells); (ix) Lin−c-kit+ Rhhi Holo fraction (STR-HSC); (x) Lin− c-kit+Rhlo Holo fraction (LTR-HSC).
Expression of mJagged2 is down-regulated on differentiation
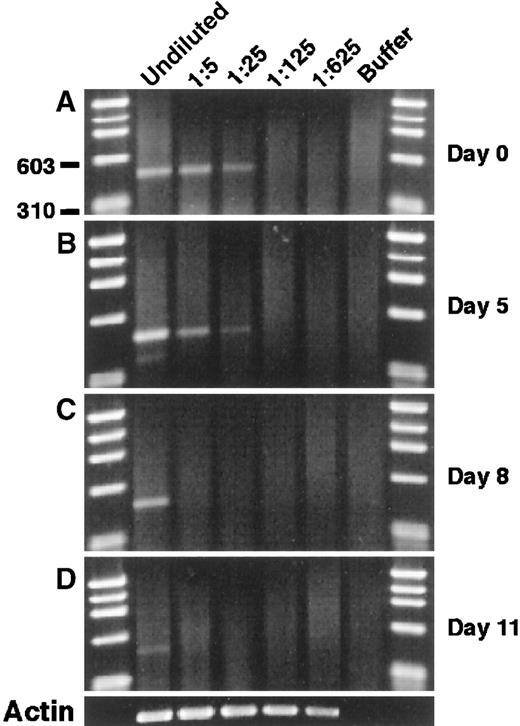
To examine the modulation of mJagged2 during hematopoiesis, we cultured Lin− c-kit+ RhloHolo cells in a medium containing SCF, IL-3, IL-6, and GM-CSF and performed mJagged2 RT-PCR on cells sequentially harvested from the liquid culture. The results showed that the level of mJagged2 expression was high in the starting population and was down-regulated during subsequent proliferation and differentiation (Figure 3A-C) and was almost undetectable after 11 days (Figure 3D). Wright-stained cytospin preparations of cells harvested on days 8 and 11 showed increasing and predominantly mature neutrophils, macrophages, and mast cells (not shown).
Modulation of mJagged2 expression during the proliferation and differentiation of HSC in vitro. Five hundred Lin− c-kit+ RhloHolo cells were cultured in liquid media containing SCF, IL-3, IL-6, and GM-CSF.
Cells were harvested sequentially, and RT-PCR of mJagged2 was performed on serial 5-fold dilutions of cell lysates prepared from 1000 cells. (A) Day 0; (B) day 5; (C) day 8; (D) day 11. The result of β-actin RT-PCR on day 11 is shown at the bottom. First and last lanes contain the φχ174/HaeIII markers.
Modulation of mJagged2 expression during the proliferation and differentiation of HSC in vitro. Five hundred Lin− c-kit+ RhloHolo cells were cultured in liquid media containing SCF, IL-3, IL-6, and GM-CSF.
Cells were harvested sequentially, and RT-PCR of mJagged2 was performed on serial 5-fold dilutions of cell lysates prepared from 1000 cells. (A) Day 0; (B) day 5; (C) day 8; (D) day 11. The result of β-actin RT-PCR on day 11 is shown at the bottom. First and last lanes contain the φχ174/HaeIII markers.
Endothelial cells express high levels of Jagged2 whereas fibroblasts do not
Preliminary in situ hybridization studies revealed that the endothelial cells lining the lumen of the dorsal aortae of day 8.5 and day 12.5 mouse embryos expressed mJagged2, whereas most of the mesenchymal cells did not (Queva C, Tsai S, unpublished data). Furthermore, the hJagged2 cDNA was initially cloned from a HUVEC cDNA library.5 These 2 observations suggested that endothelial cells could be an important source of mJagged2 in the hematopoietic microenvironment. Therefore, we examined the expression of mJagged2 protein in primary HUVEC and in 5 independently derived stromal fibroblast cell lines (S17, STW, STV, STS, STD)36 37 by Western blot analysis using the anti-ICD antibodies (recognizing the intracellular domain of both human and mouse Jagged2). The results are shown in Figure4. Although HUVEC expressed high levels of Jagged2 (Figure 4A), none of the stromal fibroblast cell lines examined showed detectable levels of Jagged2 expression (Figure 4B). These findings were corroborated by the results of Northern blot analysis (Figure 4C). Stimulation of stromal fibroblast cell lines with tumor necrosis factor did not result in the induction of Jagged2 (not shown). Western and Northern blot analyses of additional fibroblast cell lines or strains such as mouse NIH3T3 (Figure 1C, lane 1), primary human foreskin fibroblasts (not shown), and primary rabbit skin fibroblasts (Figure 4A, lane 1) also failed to detect Jagged2 expression.
Expression of Jagged2 in endothelial cells but not in fibroblasts.
Total cell lysates were resolved by SDS-PAGE, blotted onto membranes, and probed with anti-ICD antibodies and horseradish peroxidase-conjugated second antibodies. (A) Expression of Jagged2 protein in HUVEC. Lane 1, Rab-9/LXSN (negative control; see below); lane 2, Rab-9/LMJSN (positive control; see below); lane 3, HUVEC; lane 4, S17. The position of the approximately 150-kd Jagged2 protein is indicated. (B) Lack of Jagged2 protein expression in marrow stromal fibroblast cell lines. Lane 1, Rab-9/LXSN (negative control; see below); lane 2, Rab-9/LMJSN (positive control; see below); lane 3, stromal cell line STS; lane 4, stromal cell line STD; lane 5, stromal cell line STW; lane 6, stromal cell line STV; lane 7, stromal cell line S17. The position of the approximately 150-kd Jagged2 protein is shown. (C) Northern blot analysis of Jagged2 expression in endothelial cells and stromal fibroblasts. Each lane contained 5 μg total RNA. The blot was hybridized to a 1:1 mixture of full-length h-Jagged2 and mJagged2 probes. Lane 1, EML C1 (positive control); lane 2, HUVEC; lane 3, HUVEC treated with apolipoprotein-a [apo(a)] for 12 hours; lane 4, STS; lane 5, STD; lane 6, STW; lane 7, STV. The position of 28S rRNA is marked.
Expression of Jagged2 in endothelial cells but not in fibroblasts.
Total cell lysates were resolved by SDS-PAGE, blotted onto membranes, and probed with anti-ICD antibodies and horseradish peroxidase-conjugated second antibodies. (A) Expression of Jagged2 protein in HUVEC. Lane 1, Rab-9/LXSN (negative control; see below); lane 2, Rab-9/LMJSN (positive control; see below); lane 3, HUVEC; lane 4, S17. The position of the approximately 150-kd Jagged2 protein is indicated. (B) Lack of Jagged2 protein expression in marrow stromal fibroblast cell lines. Lane 1, Rab-9/LXSN (negative control; see below); lane 2, Rab-9/LMJSN (positive control; see below); lane 3, stromal cell line STS; lane 4, stromal cell line STD; lane 5, stromal cell line STW; lane 6, stromal cell line STV; lane 7, stromal cell line S17. The position of the approximately 150-kd Jagged2 protein is shown. (C) Northern blot analysis of Jagged2 expression in endothelial cells and stromal fibroblasts. Each lane contained 5 μg total RNA. The blot was hybridized to a 1:1 mixture of full-length h-Jagged2 and mJagged2 probes. Lane 1, EML C1 (positive control); lane 2, HUVEC; lane 3, HUVEC treated with apolipoprotein-a [apo(a)] for 12 hours; lane 4, STS; lane 5, STD; lane 6, STW; lane 7, STV. The position of 28S rRNA is marked.
Full-length mJagged2 promotes the survival and proliferation of mouse bone marrow progenitors
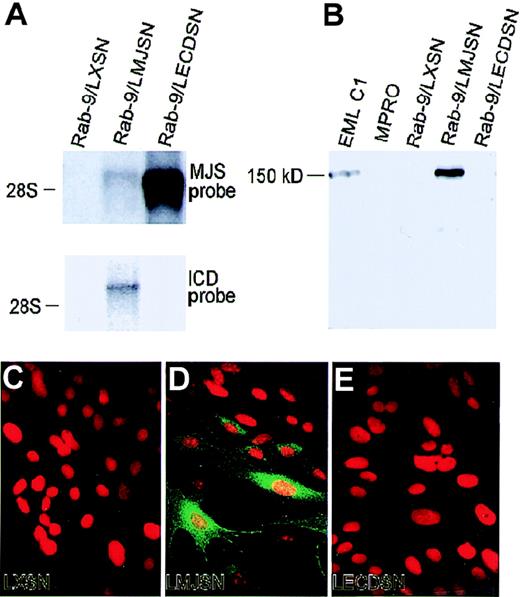
To determine the role of mJagged2 in the survival, proliferation, or differentiation of hematopoietic progenitors, we first expressed mJagged2 in a rabbit-skin fibroblast cell strain (Rab-9) through retroviral vector-mediated gene transfer. Mouse hematopoietic progenitors were then cocultured with Rab-9 cells expressing ectopic mJagged2. Mouse fibroblasts or mouse stromal cell lines were not used for expressing mJagged2 because they produce multiple mouse hematopoietic growth factors that could overshadow or antagonize the action of mJagged2 on mouse hematopoietic progenitors. For the same reason, no exogenous hematopoietic growth factors were added to the cocultures. The Rab-9 fibroblast cell strain differs from fibroblast cell lines in that it is not transformed or immortalized and behaves like normal fibroblasts. To ensure homogeneity, we established a clonal strain, Rab-9 C.14, which was then transduced with retroviral vectors LXSN (empty vector control), LMJSN (expressing full-lengthmouse Jagged2; a.a 1-1247), and LECDSN (expressing the entire extracellulardomain of mJagged2; a.a. 1-1083) and selected with G418. Northern blot analyses showed the expression of mJagged2 RNA of predicted sizes in Rab-9/LMJSN and Rab-9/LECDSN but not in Rab-9/LXSN. The level of the full-length mJagged2 message was less than one tenth that of truncated mJagged2 (Figure5A), indicating that the sequence downstream from the extracellular domain might be involved in transcriptional or post-transcriptional regulation of the gene. Western blot analyses using anti-ICD antibodies verified the production of the approximately 150-kd mJagged2 protein in the multipotent hematopoietic cell line EML C1 (positive control) and Rab-9/LMJSN but not in Rab-9/LXSN or Rab-9/LECDSN (Figure 5B). Immunofluorescence staining with anti-ICD antibodies also verified the expression of mJagged2 in Rab-9/LMJSN but not in Rab-9/LXSN or Rab-9/LECDSN (Figure 5C-E).
Retroviral vector-mediated expression of mJagged2 in the Rab-9 cells.
Rab-9 cells were infected with retroviral vectors LXSN (empty vector control), LMJSN (expressing full-length mJagged2), and LECDSN (expressing the extracellular domain of mJagged2) and selected with G418. (A) Northern blot analysis using 5 μg total RNA in each lane and probed with either the full-length mJagged2 probe (MJS; upper panel) or the intracellular domain probe (ICD; lower panel). The position of the 28S rRNA is shown. (B) Western blot analysis using antisera against the intracellular domain of mJagged2. Ten micrograms total cell lysate from each cell line was used in each lane. EML C1 served as the positive and size (150-kd) controls, whereas MPRO served as the negative control. Rab-9/LMJSN expressed mJagged2 of correct size. No specific bands were detected when the blot was probed with preimmune serum (not shown). (C-E) Indirect immunofluorescence staining using rabbit anti-ICD antibodies against the intracellular domain of mJagged2, followed by fluorescein isothiocyanate-conjugated donkey antirabbit IgG (green fluorescence). Cells were double stained with propidium iodide (red fluorescence) to reveal the nuclei. (C) Rab-9/LXSN. (D) Rab-9/LMJSN. (E) Rab-9/LECDSN. Only Rab-9/LMJSN showed positive staining with clonal variation in the level of expression. Staining with preimmune or anti-GST sera was negative (not shown).
Retroviral vector-mediated expression of mJagged2 in the Rab-9 cells.
Rab-9 cells were infected with retroviral vectors LXSN (empty vector control), LMJSN (expressing full-length mJagged2), and LECDSN (expressing the extracellular domain of mJagged2) and selected with G418. (A) Northern blot analysis using 5 μg total RNA in each lane and probed with either the full-length mJagged2 probe (MJS; upper panel) or the intracellular domain probe (ICD; lower panel). The position of the 28S rRNA is shown. (B) Western blot analysis using antisera against the intracellular domain of mJagged2. Ten micrograms total cell lysate from each cell line was used in each lane. EML C1 served as the positive and size (150-kd) controls, whereas MPRO served as the negative control. Rab-9/LMJSN expressed mJagged2 of correct size. No specific bands were detected when the blot was probed with preimmune serum (not shown). (C-E) Indirect immunofluorescence staining using rabbit anti-ICD antibodies against the intracellular domain of mJagged2, followed by fluorescein isothiocyanate-conjugated donkey antirabbit IgG (green fluorescence). Cells were double stained with propidium iodide (red fluorescence) to reveal the nuclei. (C) Rab-9/LXSN. (D) Rab-9/LMJSN. (E) Rab-9/LECDSN. Only Rab-9/LMJSN showed positive staining with clonal variation in the level of expression. Staining with preimmune or anti-GST sera was negative (not shown).
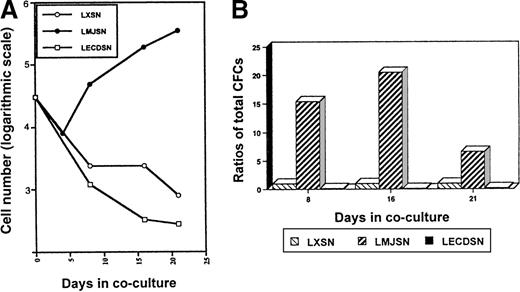
In the initial experiments to determine whether exogenous mJagged2 could support the survival or proliferation of hematopoietic progenitors, or both, mouse bone marrow MNC were cocultured with monolayers of Rab-9/LXSN or Rab-9/LMJSN or Rab-9/LECDSN in DMEM supplemented with 10% FCS. No exogenous hematopoietic growth factors were added. Phase-contrast microscopy revealed that most MNC died in the Rab-9/LXSN and Rab-9/LECDSN cocultures after 1 week. In contrast, many MNC survived in the Rab-9/LMJSN cocultures. The surviving MNC in Rab-9/LMJSN cocultures proliferated slowly (Figure6A). Colony assays performed on nonadherent cells harvested on days 8 and 16 of cocultures revealed the presence of large numbers of hematopoietic progenitors in Rab-9/LMJSN cocultures. On day 8 of coculture, the combined number of high-proliferative potential colony-forming cells (HPP-CFC), CFU-GM, CFU-G, and CFU-M in Rab-9/LMJSN cocultures was 15.4-fold higher than that of Rab-9/LXSN coculture. On day 16, the corresponding figure was 20.5 (Figure6B). The numbers of progenitors in Rab-9/LECDSN cocultures were consistently lower than those of the control cocultures (P = .0006 for day 8; P = .0002 for day 16) (Figure 6B), suggesting a possible negative effect.
Survival and proliferation of hematopoietic progenitors (MNCs) in Rab-9 cocultures.
(A) Growth curves of nonadherent hematopoietic cells in Rab-9/LXSN, Rab-9/LMJSN, and Rab-9/LECDSN cocultures. In each well, 3 × 105 MNC was cocultured with Rab-9 cells in DMEM/10% FCS in 12-well plates and subcultured at 1:2 to 1:4 ratios every 3 to 4 days. No exogenous hematopoietic growth factors were added. Cell numbers have been corrected for subculture ratios and shown on a logarithmic scale (eg, 3 = 103 and 4 = 104). Open circle, LXSN; filled circle, LMJSN; open square, LECDSN; 1 of 4 similar experiments. Each point represents the mean of duplicate measurements. (B) Results of colony assays performed on nonadherent cells recovered from cocultures on days 8, 16, and 21. Cells were plated in methylcellulose culture medium supplemented with SCF, IL-3, and GM-CSF. Data are expressed as the ratios of total CFC (HPP-CFC, CFU-GM, CFU-G, and CFU-M combined) recovered from Rab-9/LMJSN or Rab-9/LCEDSN cocultures to those recovered from Rab-9/LXSN cocultures. At each assay point, the total number of CFC recovered from Rab-9/LXSN coculture is treated as 1. Results are means of triplicates.
Survival and proliferation of hematopoietic progenitors (MNCs) in Rab-9 cocultures.
(A) Growth curves of nonadherent hematopoietic cells in Rab-9/LXSN, Rab-9/LMJSN, and Rab-9/LECDSN cocultures. In each well, 3 × 105 MNC was cocultured with Rab-9 cells in DMEM/10% FCS in 12-well plates and subcultured at 1:2 to 1:4 ratios every 3 to 4 days. No exogenous hematopoietic growth factors were added. Cell numbers have been corrected for subculture ratios and shown on a logarithmic scale (eg, 3 = 103 and 4 = 104). Open circle, LXSN; filled circle, LMJSN; open square, LECDSN; 1 of 4 similar experiments. Each point represents the mean of duplicate measurements. (B) Results of colony assays performed on nonadherent cells recovered from cocultures on days 8, 16, and 21. Cells were plated in methylcellulose culture medium supplemented with SCF, IL-3, and GM-CSF. Data are expressed as the ratios of total CFC (HPP-CFC, CFU-GM, CFU-G, and CFU-M combined) recovered from Rab-9/LMJSN or Rab-9/LCEDSN cocultures to those recovered from Rab-9/LXSN cocultures. At each assay point, the total number of CFC recovered from Rab-9/LXSN coculture is treated as 1. Results are means of triplicates.
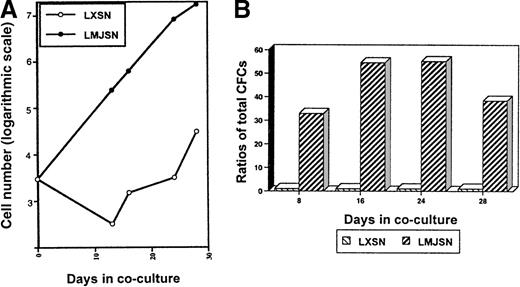
When the above experiment was repeated with Lin−c-kit+ cells (3000 cells/well), more proliferation of the hematopoietic cells was observed (Figure7A). The numbers (corrected for subculture ratios) of nonadherent cells on day 28 were 38.4 × 103 (Rab-9/LXSN cocultures; negative controls) and 1.01 × 107 (Rab-9/LMJSN cocultures) (Figure 7A). Examination of Wright-stained cytospin preparations of cells harvested from the Lin−c-kit+–Rab-9/LMJSN cocultures on day 16 revealed the presence of undifferentiated blasts (21.7%), promyelocytes (40.6%), metamyelocytes (7.7%), segmented neutrophils (19.3%), monocytes (8.3%), mast cells (1.4%), and basophilic megakaryoblasts (less than 1%). The ratios of total CFC in Rab-9/LMJSN cocultures to those in Rab-9/LXSN cocultures on days 8, 16, 24, and 28 were 32, 54, 55, and 37, respectively (Figure 7B). Classification of CFC according to the size and morphology of the colonies they formed in semisolid media revealed that approximately 40% to 60% of all colonies were macroscopic, HPP-CFC type colonies containing 3 or more subcolonies (Table 1). The calculated numbers of HPP-CFC (corrected for subculture ratios) on day 8 were 20 ± 8 (Rab-9/LXSN cocultures) and 651 ± 23 (Rab-9/LMJSN cocultures). On day 28, the corresponding numbers were 1280 ± 576 (Rab-9/LXSN cocultures) and 48 128 ± 14 272 (Rab-9/LMJSN co-cultures). Although the calculated numbers of CFC on day 28 are undoubtedly overestimated because of the preferential retention of primitive progenitors in stroma early and release into the nonadherent fractions later, these figures nevertheless reflect greater increases in total CFC and HPP-CFC in the Rab-9/LMJSN cocultures.
Survival and proliferation of Lin−c-kit+ hematopoietic progenitors in Rab-9 cocultures.
(A) Growth curves of nonadherent hematopoietic cells in Rab-9 cocultures. This experiment is similar to that described in Figure 6except that Lin− c-kit+ cells (3000 per well) were used. Cell numbers have been corrected for subculture ratios and represent means of duplicate measurements. Open circle, LXSN; filled circle, LMJSN. (B) Results of colony assays performed on nonadherent cells harvested from the cocultures on day 8, 16, 24, and 28. Cells were plated in methylcellulose culture medium supplemented with SCF, IL-3, and GM-CSF. Data are expressed as the ratios of total CFCs (HPP-CFC, CFU-GM, CFU-G, and CFU-M combined) recovered from Rab-9/LMJSN cocultures to those recovered from Rab-9/LXSN cocultures. At each assay point, the total number of CFC recovered from Rab-9/LXSN coculture is treated as 1. Results are means of triplicates.
Survival and proliferation of Lin−c-kit+ hematopoietic progenitors in Rab-9 cocultures.
(A) Growth curves of nonadherent hematopoietic cells in Rab-9 cocultures. This experiment is similar to that described in Figure 6except that Lin− c-kit+ cells (3000 per well) were used. Cell numbers have been corrected for subculture ratios and represent means of duplicate measurements. Open circle, LXSN; filled circle, LMJSN. (B) Results of colony assays performed on nonadherent cells harvested from the cocultures on day 8, 16, 24, and 28. Cells were plated in methylcellulose culture medium supplemented with SCF, IL-3, and GM-CSF. Data are expressed as the ratios of total CFCs (HPP-CFC, CFU-GM, CFU-G, and CFU-M combined) recovered from Rab-9/LMJSN cocultures to those recovered from Rab-9/LXSN cocultures. At each assay point, the total number of CFC recovered from Rab-9/LXSN coculture is treated as 1. Results are means of triplicates.
Classification of hematopoietic progenitors recovered from Rab-9 cocultures
| . | CFC per culture* . | |||
|---|---|---|---|---|
| HPP-CFC . | CFU-GM . | CFU-G . | CFU-M . | |
| Day 8 of coculture | ||||
| Rab-9/LXSN | 20 ± 8 | 11 ± 5 | 0 ± 0 | 0 ± 0 |
| Rab-9/LMJSN | 615 ± 23 | 295 ± 18 | 64 ± 7 | 27 ± 14 |
| Day 28 of coculture | ||||
| Rab-9/LXSN | 40 ± 18 | 51 ± 17 | 4 ± 4 | 7 ± 6 |
| Rab-9/LMJSN | 1504 ± 446 | 1608 ± 473 | 400 ± 246 | 304 ± 73 |
| . | CFC per culture* . | |||
|---|---|---|---|---|
| HPP-CFC . | CFU-GM . | CFU-G . | CFU-M . | |
| Day 8 of coculture | ||||
| Rab-9/LXSN | 20 ± 8 | 11 ± 5 | 0 ± 0 | 0 ± 0 |
| Rab-9/LMJSN | 615 ± 23 | 295 ± 18 | 64 ± 7 | 27 ± 14 |
| Day 28 of coculture | ||||
| Rab-9/LXSN | 40 ± 18 | 51 ± 17 | 4 ± 4 | 7 ± 6 |
| Rab-9/LMJSN | 1504 ± 446 | 1608 ± 473 | 400 ± 246 | 304 ± 73 |
Experimental conditions were identical to those described in the legend to Figure 7. Lin− c-kit+ cells (3000 per well) were used to initiate the cocultures. Colony assays were performed in methylcellulose supplemented with SCF, IL-3, and GM-CSF. Colonies were counted on day 8 of colony assays. Numbers shown have not been corrected for subculture ratios, but all cultures were handled in the same way (eg, same feeding frequencies and subculture ratios) in parallel.
Mean ± SD; n = 3.
Survival effect of membrane-bound mJagged2 requires direct cell-to-cell contact
To ascertain whether direct cell-to-cell contact was necessary for the survival effect of mJagged2 in the Rab-9 cocultures, we analyzed additional cultures in which mouse bone marrow MNCs were placed in Transwells (Costar, Corning, NY) separated from the Rab-9/LXSN, Rab-9/LMJSN and Rab-9/LECDSN under layers by a 0.4-μm membrane. Direct phase-contrast microscopy revealed that most cells in the Transwells died in 1 week. Colony assays of surviving cells in the Transwells on day 8 revealed that few were CFCs (Table2). This result indicated that the survival effect of Rab-9/LMJSN was not attributable to diffusable factors and required direct cell-to-cell contact. This conclusion was also supported by the finding that no colony-stimulating activity was detected in the conditioned medium of Rab-9/LMJSN in colony assays (not shown).
Survival effects of full-length mJagged2 required cell-to-cell contact
| Types of monolayer . | Total CFCs recovered per culture . | |
|---|---|---|
| Without transwell . | With transwell . | |
| Rab-9/LXSN | 248 ± 35 | 0 ± 0 |
| Rab-9/LMJSN | 3812 ± 139 | 1 ± 2 |
| Rab-9/LECDSN | 34 ± 13 | 0 ± 0 |
| Types of monolayer . | Total CFCs recovered per culture . | |
|---|---|---|
| Without transwell . | With transwell . | |
| Rab-9/LXSN | 248 ± 35 | 0 ± 0 |
| Rab-9/LMJSN | 3812 ± 139 | 1 ± 2 |
| Rab-9/LECDSN | 34 ± 13 | 0 ± 0 |
Experimental conditions were identical to those described in the legend to Figure 6. In Transwell cultures, bone marrow MNC were placed within the Transwell inserts and separated from the Rab-9 monolayers by a 0.4-μm nylon membrane. Transwell inserts were not toxic to hematopoietic progenitors because the latter were able to proliferate vigorously in Transwells in the presence of IL-3 and GM-CSF (not shown). Colony assays were performed on day 8 of coculture in methylcellulose supplemented with SCF, IL-3, and GM-CSF. Total CFC (HPP-CFC, CFU-GM, CFU-G, CFU-M) were counted on day 8 of colony assays. For reference, the plating efficiency (or frequency of CFC) of cells harvested from the Rab-9/LMJSN coculture was 4%.
Mean ± SD; n = 3.
Discussion
In this report, we describe the cloning of mJagged2 by cDNA RDA from EML C1, an SCF-dependent, multipotent hematopoietic cell line.1 mJagged2 encodes a single-pass membrane protein characterized by the DSL motif in its extracellular domain. Using quantitative RT-PCR, we showed that mJagged2 was expressed in freshly isolated c-kit+ mouse hematopoietic progenitors, including those in the Lin−c-kit+ Rhlo Holo, Lin− c-kit+ RhhiHolo, and the Lin− c-kit+fractions (Figure 2Bviii-x). The expression was lower in fractions with lower contents of HSC, progenitors, or both (Figure 2Biv-vi), and it was undetectable in fractions depleted of c-kit+ cells (Figure 2Biii and vii). Overall, the expression of mJagged2correlates with c-kit expression and with the presence of HSC and progenitors in the hematopoietic compartment.
Like CD34, Flk-1/KDR,40 and AA4.1, it seems thatJagged2 is also expressed by HSC and endothelial cells. Our in situ hybridization studies showed hybridization of an mJagged2antisense probe in the endothelial cells lining the lumen of dorsal aorta and in vessels surrounding the neural tube (Queva C, Tsai S, unpublished data). Western and Northern analyses revealed that primary human endothelial cells expressed high levels of Jagged2 (Figure 4A, C). Because bone marrow stromal fibroblasts, like endothelial cells, are an integral part of the hematopoietic microenvironment and have been shown to express a related protein, Jagged1,19 we surveyed the expression of mJagged2 in 5 different mouse stromal fibroblast cell lines. Despite repeated effort, we failed to detect mJagged2 expression in any of these stromal fibroblast cell lines by Northern or Western analysis (Figure 4B-C). Additional fibroblast cell line or strains also showed no detectable levels of Jagged2. Thus,Jagged2 appears to be differentially expressed between fibroblasts and endothelial cells. At present, the biologic significance of this finding is unclear and is under investigation.
To investigate the influence of exogenous mJagged2 on hematopoiesis, we cocultured mouse bone marrow MNC or Lin−c-kit+ progenitors with Rab-9 cells engineered to express high levels of full-length, membrane-bound mJagged2 in the absence of exogenous hematopoietic growth factors. Direct observation indicated that most mouse marrow cells in Rab-9/LXSN (negative control) cocultures died quickly, probably because of the lack of hematopoietic growth factors. In contrast, many hematopoietic progenitors in Rab-9/LMJSN (expressing full-length mJagged2) cocultures survived and proliferated slowly (Figures 6A, 7A). Preliminary comparison indicates that the proliferative effect of mJagged2 is relatively weak compared with that of GM-CSF or IL-3 (approximately 2 orders of magnitude lower; not shown). Many HPP-CFC were still present in the Rab-9/LMJSN cocultures after 4 weeks of coculturing (Table 1). This finding raises the possibility that mJagged2 may promote the survival and proliferation of even more primitive hematopoietic progenitors. Alternatively, it may delay their differentiation. These are not mutually exclusive possibilities.
Consistent with its status as a membrane-bound protein, mJagged2 exerted its effects through direct cell-to-cell contact. Separation of the Rab-9/LMJSN monolayers from bone marrow progenitors by a 0.4-μm nylon membrane abolished the survival effect (Table 2). This observation made it unlikely that the survival effect was mediated by soluble factors. It is possible that the survival effect resulted from insoluble, membrane-bound molecules produced by Rab-9/LMJSN cells in response to activation of their Notch receptors by ectopically expressed mJagged2. To evaluate this possibility, we repeated the coculture experiments using Rab-9 cells transfected with a constitutively activated Notch-1 construct (a gift from R. Kopan).41 No survival effect on hematopoietic progenitors was detected (not shown).
The Rab-9 coculture experiments described above revealed the survival and proliferative effects of exogenous mJagged2 on mouse hematopoietic progenitors. In the hematopoietic microenvironment, endothelial cells may be an important source of Jagged2 stimuli. The significance of endogenous mJagged2 in hematopoietic progenitors, on the other hand, is unknown. It is conceivable that endogenous mJagged2 may have the same effects on hematopoietic progenitors as exogenous mJagged2 through an autocrine mechanism or, alternatively, on adjacent hematopoietic progenitors through a juxtacrine mechanism. We cannot rule out a priori the possibility that the endogenous Jagged2 of hematopoietic progenitors may have other functions, such as lineage development or interaction with other cell types.
Acknowledgments
We thank Peter Harpel, Nasserine Haque, Ewa Sitnicka, Raphael Kopan, and Robert Rottapel for sharing research materials, Gordon Keller for critical reading of the manuscript, Carl Storey and Lily Kiang for expert technical assistance, and Doris Damian for advice on statistical analysis.
Supported by Public Health Service grants DK48622 (S.T.) and DK 48708 (S.B.).
Reprints:Schickwann Tsai, The Mount Sinai School of Medicine, Box 1496, 1425 Madison Avenue, Room 13-23B, New York, NY 10029; e-mail:tsais01@doc.mssm.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Fig. 4. Expression of Jagged2 in endothelial cells but not in fibroblasts. / Total cell lysates were resolved by SDS-PAGE, blotted onto membranes, and probed with anti-ICD antibodies and horseradish peroxidase-conjugated second antibodies. (A) Expression of Jagged2 protein in HUVEC. Lane 1, Rab-9/LXSN (negative control; see below); lane 2, Rab-9/LMJSN (positive control; see below); lane 3, HUVEC; lane 4, S17. The position of the approximately 150-kd Jagged2 protein is indicated. (B) Lack of Jagged2 protein expression in marrow stromal fibroblast cell lines. Lane 1, Rab-9/LXSN (negative control; see below); lane 2, Rab-9/LMJSN (positive control; see below); lane 3, stromal cell line STS; lane 4, stromal cell line STD; lane 5, stromal cell line STW; lane 6, stromal cell line STV; lane 7, stromal cell line S17. The position of the approximately 150-kd Jagged2 protein is shown. (C) Northern blot analysis of Jagged2 expression in endothelial cells and stromal fibroblasts. Each lane contained 5 μg total RNA. The blot was hybridized to a 1:1 mixture of full-length h-Jagged2 and mJagged2 probes. Lane 1, EML C1 (positive control); lane 2, HUVEC; lane 3, HUVEC treated with apolipoprotein-a [apo(a)] for 12 hours; lane 4, STS; lane 5, STD; lane 6, STW; lane 7, STV. The position of 28S rRNA is marked.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/96/3/10.1182_blood.v96.3.950/5/m_bloo01543004aw.jpeg?Expires=1770582173&Signature=oTRjIOnEL3B584a6WzIMg8N5WykZJMzcJqLc7WBh7zwQPaLx1tBNKIQHV4l7lfLkFbmMaIW0BWU9v5odBOh~dUCv6-C3pC9083yHmfR~edYudOn79EbB7VGbXBa7yi2vLNafRz8-qKlfKwouPuHeokG3trntFKT6RjJeNPyB37au5nWcpqghN3YJVhBxWvhjOPY~DZhnLnyOPvPqudxFYsvj67IQ9jinBjMXI-q3qf~UNcVsFjiSTfT0Es2AwVl8DMbrNQMAHlaY9LYc573sUy4hymo3LAnuse5VCl~yPaFP5j7QuPR6Fucws0T3AFlrIzsI74ZAYbym5homszIoqw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 4. Expression of Jagged2 in endothelial cells but not in fibroblasts. / Total cell lysates were resolved by SDS-PAGE, blotted onto membranes, and probed with anti-ICD antibodies and horseradish peroxidase-conjugated second antibodies. (A) Expression of Jagged2 protein in HUVEC. Lane 1, Rab-9/LXSN (negative control; see below); lane 2, Rab-9/LMJSN (positive control; see below); lane 3, HUVEC; lane 4, S17. The position of the approximately 150-kd Jagged2 protein is indicated. (B) Lack of Jagged2 protein expression in marrow stromal fibroblast cell lines. Lane 1, Rab-9/LXSN (negative control; see below); lane 2, Rab-9/LMJSN (positive control; see below); lane 3, stromal cell line STS; lane 4, stromal cell line STD; lane 5, stromal cell line STW; lane 6, stromal cell line STV; lane 7, stromal cell line S17. The position of the approximately 150-kd Jagged2 protein is shown. (C) Northern blot analysis of Jagged2 expression in endothelial cells and stromal fibroblasts. Each lane contained 5 μg total RNA. The blot was hybridized to a 1:1 mixture of full-length h-Jagged2 and mJagged2 probes. Lane 1, EML C1 (positive control); lane 2, HUVEC; lane 3, HUVEC treated with apolipoprotein-a [apo(a)] for 12 hours; lane 4, STS; lane 5, STD; lane 6, STW; lane 7, STV. The position of 28S rRNA is marked.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/96/3/10.1182_blood.v96.3.950/5/m_bloo01543004bw.jpeg?Expires=1770582173&Signature=4J4u2BBR3tp1FSeunpCxuNUUdD6T8iYNleq9dJ~Tdqos99JCcDrWeV0FDhxEIGFrac8Zi1VfKL2BfdXpcPmygY-ruSbGNUYCXYWbrnAo2pO0vJcYIiEHJcUS50djHSsxpIpyZNYIJGYrq27BBZ9wsRY-G2t3gT10h9ktqi2vBJSEpmZM1XvVZKmEFyTfjZLr1G7jeicpKcjkOqlgKJKFtWHbiwVy8dR5Fz8Rz5CsmKbU7vOwdJMiCY58RIoQ6n6wsLptKcXRBHymO19lc-0omz1CcLMrCmvnA2YQe764~WyalycVEY~d42hB5Wa-J3YSVIaGP83xn3WbL8xgHtYs4w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 4. Expression of Jagged2 in endothelial cells but not in fibroblasts. / Total cell lysates were resolved by SDS-PAGE, blotted onto membranes, and probed with anti-ICD antibodies and horseradish peroxidase-conjugated second antibodies. (A) Expression of Jagged2 protein in HUVEC. Lane 1, Rab-9/LXSN (negative control; see below); lane 2, Rab-9/LMJSN (positive control; see below); lane 3, HUVEC; lane 4, S17. The position of the approximately 150-kd Jagged2 protein is indicated. (B) Lack of Jagged2 protein expression in marrow stromal fibroblast cell lines. Lane 1, Rab-9/LXSN (negative control; see below); lane 2, Rab-9/LMJSN (positive control; see below); lane 3, stromal cell line STS; lane 4, stromal cell line STD; lane 5, stromal cell line STW; lane 6, stromal cell line STV; lane 7, stromal cell line S17. The position of the approximately 150-kd Jagged2 protein is shown. (C) Northern blot analysis of Jagged2 expression in endothelial cells and stromal fibroblasts. Each lane contained 5 μg total RNA. The blot was hybridized to a 1:1 mixture of full-length h-Jagged2 and mJagged2 probes. Lane 1, EML C1 (positive control); lane 2, HUVEC; lane 3, HUVEC treated with apolipoprotein-a [apo(a)] for 12 hours; lane 4, STS; lane 5, STD; lane 6, STW; lane 7, STV. The position of 28S rRNA is marked.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/96/3/10.1182_blood.v96.3.950/5/m_bloo01543004cw.jpeg?Expires=1770582173&Signature=4AkFriP5GVZAh8XjnJwBJZkabyp2COazPYzi5erVZl~w0fs0NMnoIHi5XsupWfOc08UJp-Ub56KIWOjQ5mtZ8wEOyRKNhsmC7W1GztJWt7XIcPgz4G6CnLSRKnYSBHaNt7zlJpKqeyf4c4kpHmHpbtnVFregnkifdy21upmgVWKCWyDV01EPgDYH6daSwOMeMtuc2saRY7uwBIsAvCa6mhhPGnTvg~txnuXjUo7bTGU6RLpw4538JrbR9XRyAe2W0-MP1zj51ppEjijKFLgiN-Eko2O3oOKMTF4ILrYptJTXd0zSfU6LE04V8FSY69u2ZaLm3Qg5QZXhDHMRzrMGJg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal