Abstract
Chronic granulomatous disease is a rare inherited disorder caused by nonexistent or severely decreased phagocyte superoxide production that results in a severe defect in host defense and consequent predisposition to microbial infection. The enzyme responsible for generating the superoxide, NADPH oxidase, involves at least 5 protein components. The absence of, or a defect in, any 1 of 4 of these proteins (p22phox, p47phox, p67phox, or gp91phox) gives rise to the known types of chronic granulomatous disease. One of the rarest forms of the disease is due to defects in the CYBA gene encoding p22phox, which together with gp91phox forms flavocytochromeb558, the catalytic core of NADPH oxidase. To date, only 9 kindreds with p22phoxdeficiency have been described in the literature comprising 10 mutant alleles. Four polymorphisms in the CYBA gene have also been reported. Here we describe 9 new, unrelated kindreds containing 12 mutations, 9 of which are novel. In addition, we report 3 new polymorphisms. The novel mutations are (a) deletion of exons 2 and 3, (b) a missense mutation in exon 3 (T155→C), (c) a splice site mutation at the 5′ end of intron 3, (d) a missense mutation in exon 2 (G74→T), (e) a nonsense mutation in exon 1 (G26→A), (f) a missense mutation in exon 4 (C268→T), (g) a frameshift in exon 3 due to the insertion of C at C162, (h) a nonsense mutation in exon 2 (G107→A), and (i) a missense mutation in exon 2 (G70→A).
Chronic granulomatous disease (CGD) is a rare inherited disorder of the innate immune system caused by genetic defects in the superoxide-generating NADPH oxidase of phagocytes.1 In the absence of superoxide (O2−) production by these cells, microbial pathogens are not killed efficiently, and the host is left vulnerable to recurrent, life-threatening infections. NADPH oxidase activity requires the participation of at least 5 proteins. Two of them, gp91phox and p22phox, together form a heterodimeric flavin and heme-containing protein, flavocytochrome b558, the catalytic core of the enzyme. Flavocytochrome b558is present in the specific granule and plasma membranes of resting neutrophils. In contrast, p47phox, p67phox, and p40phox form a complex located in the cytosolic compartment of resting neutrophils. This complex translocates to the membrane and associates with flavocytochrome b558 during oxidase activation. The small GTP-binding protein, Rac2 is also required for oxidase activity, and associates with the membrane during the activation process (review by Clark2). Defects in the genes encoding 4 of thephox proteins (gp91phox, p22phox, p47phox, and p67phox) are known to cause CGD. The protein p40phox has been strongly implicated in NADPH oxidase regulation, but its role is unclear at present. No disorders have been recognized that are due to mutations in the p40phox gene (NCF-4), although p40phox levels are reduced in p67phox-deficient CGD.3,4 Until very recently, no genetic defects in Rac2 have been reported, which may be because such a defect produces a lethal phenotype, or because there are sufficient levels of the closely related Rac1 present in the phagocytes to compensate for any loss of Rac2. (Rac1 can substitute for Rac2 in cell-free systems of oxidase activation.)5-7 However, a single missense mutation in Rac2 has recently been reported to cause a CGD-like condition in 1 individual. The patient was heterozygous for the mutation but severely affected, suggesting that the amino acid substitution acts in a dominant-negative fashion. 8 9
In myeloid cells, the absence of p22phox protein because of genetic defects also results in the loss of gp91phox expression and vice versa, indicating that each of these proteins requires the other for mutual stability. However, this is apparently not true of all cell types, as gp91phox and p22phox are stably expressed in the absence of their partners in COS7 cells.10The primary structure of p22phox suggests it contains 4 membrane-spanning domains in the N-terminal two-thirds of the molecule, and a proline-rich domain in the C-terminal cytoplasmic tail. Such proline-rich regions can mediate protein-protein association by binding to SH3 domains that are found in a variety of proteins involved in signal transduction, including the cytosolic phoxproteins. The proline-rich domain of p22phox binds the N-terminal SH3 domain of p47phox, and this interaction is believed to play a dominant role in promoting the association of the cytosolic complex, containing p40phox, p47phox, and p67phox, with flavocytochromeb558 (reviewed in Heyworth et al11 and DeLeo and Quinn12).
The incidence of CGD is estimated to be approximately 1 in 200 000 to 250 000 individuals. The most common form (approximately 65%) is X-linked and is due to defects in the CYBB gene that codes for gp91phox. The remaining approximately 35% of cases are inherited in an autosomal recessive manner. A22 CGD (ie, CGD resulting from a defect in p22phox) is one of the rarest forms of the disease, accounting for only 6% of cases. The protein p22phox is encoded by the CYBAgene, located on chromosome 16q24.13 The approximately 600-base pair (bp) open reading frame is divided into 6 exons spanning about 8.5-kilobase (kb). Carriers of the autosomal recessive forms of CGD can be difficult to detect, as they typically appear normal by the nitroblue tetrazolium slide test and have rates of O2−production within the normal range. Identification of mutations in individuals with autosomally inherited forms of CGD provides the only effective basis for detecting carriers among family members or performing prenatal diagnoses. In the case of A22 CGD, patients from only 9 families (18 alleles, 10 different mutations) have been reported so far in the literature.13-17 Here we report an additional 9 families and describe 12 mutations, 9 of which are novel, and 3 new polymorphisms. These polymorphisms are of potential interest because of a recent report that the C214 wild-type genotype (His72) is associated with a higher risk of coronary heart disease than the rarer T214 (Tyr72) genotype18 although this association could not be confirmed by others.19-21 Another recent report describes a positive association of a polymorphism in the 3′ untranslated region with heart disease.22 These associations may be due to a putative role for p22phox in the activity of a recently described isoform of flavocytochrome b558 that is involved in mitogenic signaling 23.24
Patients, materials, and methods
Patients with chronic granulomatous disease and family members
Patient 1 is a 2-year-old Hispanic boy whose mother and father are second cousins. He has a history of recurrent cervical adenitis, inguinal lymphadenitis, and perirectal abscess from the age of 3 months. A male sibling died at the age of 4 months from a sudden onset of a gastrointestinal infection. A male cousin died at birth secondary to respiratory problems, and another male cousin is thought to have died of multiple infections associated with swollen joints.
Patient 2 is the son of unrelated parents with no family history of CGD. He had recurrent infections and liver abscesses by the age of 2, a kidney infection at age 4, and hepatic abscesses at age 8, at which time he was diagnosed with CGD. After treatment with prophylactic antibiotics, and with the exception of a liver abscess at age 12, he remained well until age 19. After discontinuing prophylactic antibiotic treatment, he developed 2 hepatic abscesses and a soft tissue abscess over his lower rib cage at age 25. He died at age 29 from gram-negative septicemia after a brief hospitalization.
Patient 3 is a 3-month-old girl with no family history of CGD. She was diagnosed after pneumonia caused by Aspergillus.
Patient 4 is a female with no clinical or family history available. Only DNA from the patient and her mother could be obtained.
Patient 5, a 2-year-old boy of unrelated parents, was referred after a history of chronic otitis media, recurrent skin infections, chronic dacryocystitis, intermittent diarrhea, and a leg abscess. There is no known family history of CGD.
Patient 6 is the 25-year-old daughter of first cousins from Southern India. Two siblings (1 male, 1 female) both died at age 6, probably as a result of CGD as determined from a review of pathology sections showing the presence of multiple granuloma in lung and lymph nodes. The patient had recurrent pulmonary infections and fever from the age of 10 months, and was diagnosed with CGD at age 8. She has continued to have recurrent lung infections, an episode of malaria, and a bone abscess from which Pseudomonas aeruginosa was isolated.
Patients 7 and 7a, are sisters, 15 and 19 years of age. The elder sister has had multifocal osteomyelitis from which Burkholderia gladioli was cultured, and also chronic immune thrombocytopenia.
Patient 8 is a 5-year-old girl with no known family history of CGD or intermarriage.
Patient 9 is a 6-year-old Hispanic girl diagnosed with CGD after an episode of Serratia osteomyelitis. A chest x-ray examination showed the presence of multiple tiny granuloma. A male sibling died in childhood. The parents were not known to be related.
Blood samples
Blood samples were obtained from patients and their family members (where available) by their physicians, following the procedures and appropriate consent protocols approved by the Human Subjects Committee of The Scripps Research Institute.
Neutrophil functional assays
Clinical diagnoses of CGD were confirmed by examination of the capacity of neutrophils to produce O2−using the nitroblue tetrazolium (NBT) slide test, spectrophotometric assay of cytochrome c reduction, and/or flow cytometry (using dihydrorhodamine [DHR] or dichlorofluorescein [DCF]), as previously described.25 The presence of flavocytochromeb558 was ascertained using reduced-minus-oxidized difference spectroscopy26; protein immunoblotting27 and/or by flow cytometry using monoclonal antibody 7D5.28,29
Preparation of DNA, amplification by polymerase chain reaction, and sequencing
Genomic DNA was isolated from whole blood using Puregene DNA Isolation Kits (Gentra Systems, Inc, Minneapolis, MN) or from EBV-transformed cell lines. In the latter case, 1 × 107 cells in phosphate-buffered saline (PBS) were pelleted, all but 200 μL of the supernatant removed and the remainder processed using the QIAamp Blood kit (Qiagen, Valencia, CA). In one patient, total RNA was isolated from whole blood using the RNeasy Blood Mini kit (Qiagen, Valencia, CA) and reverse transcriptase-polymerase chain reaction (RT-PCR) was performed using the Omniscript Reverse Transcriptase and Taq DNA Polymerase (Qiagen, Valencia, CA).
The following buffer was used for the amplification of each exon: 33.5 mmol/L Tris-HCl, (pH 8.8), 8.3 mmol/L (NH4)2SO4, 3.35 mmol/L MgCl2,85 μg/mL bovine serum albumin (BSA), 5% DMSO, 0.125 mmol/L each dNTP, 90 ng each specific primer, 2.5 units AmpliTaq polymerase, 500 ng genomic DNA. The primers used are shown in Table1.
Oligonucleotide primers used in this study for the amplification of the CYBA gene
| Intronic primers | |
| 1LA | ccagccgggttcgtgtc |
| 1RA2 | tggcgccccacttccccaccctgt |
| 2LA8 | ggtggcccacagtaggtagagaa |
| 2RA8 | gctcactgtgaagtggctcccca |
| 2RA6 | cgcccaccccagcctcag |
| 3LA | ctgagctgggctgttcctt |
| 3RA | ccacccaaccctgtgagc |
| 4LA | caaaggagtcccgagtgg |
| 4RA | gctccaagccctcctgag |
| 5LA | ccctgggtctgcagtctgcct |
| 5RA | cccaggctcacacttgctccca |
| 5LB | cctgagactttgttggcct |
| 5RB | ggcttcaagggccatgcgtgt |
| 6LA5 | cctctctgagtggcagtcaca |
| 6RA3 | cggccttcgctgcgttta |
| 6LB | cctgtcccagggccccta |
| 6RB | atgcaggtgggtgcacct |
| Exonic primers | |
| cDNA 1F | ATGGGGCAGATCGAGTGGGCCAT |
| cDNA 2F | CTCATCACCGGGGGCATCGT |
| cDNA 2R | GAAGCGCCCAGCTGTGGCCACGAT |
| cDNA 3R | GGGTACTCCAGCAGGCACACAA |
| cDNA 4R | GATGCAGGACGGCCCGAACAT |
| cDNA 5F | CTGCTGGCCACCATCCTTGGGA |
| cDNA 5F2 | CTGGCCATTGCGAGCGGCA |
| cDNA 6F | CAGATCGGAGGCACCATCA |
| cDNA 6R | TCACACGACCTCGTCGGTCAC |
| Intronic primers | |
| 1LA | ccagccgggttcgtgtc |
| 1RA2 | tggcgccccacttccccaccctgt |
| 2LA8 | ggtggcccacagtaggtagagaa |
| 2RA8 | gctcactgtgaagtggctcccca |
| 2RA6 | cgcccaccccagcctcag |
| 3LA | ctgagctgggctgttcctt |
| 3RA | ccacccaaccctgtgagc |
| 4LA | caaaggagtcccgagtgg |
| 4RA | gctccaagccctcctgag |
| 5LA | ccctgggtctgcagtctgcct |
| 5RA | cccaggctcacacttgctccca |
| 5LB | cctgagactttgttggcct |
| 5RB | ggcttcaagggccatgcgtgt |
| 6LA5 | cctctctgagtggcagtcaca |
| 6RA3 | cggccttcgctgcgttta |
| 6LB | cctgtcccagggccccta |
| 6RB | atgcaggtgggtgcacct |
| Exonic primers | |
| cDNA 1F | ATGGGGCAGATCGAGTGGGCCAT |
| cDNA 2F | CTCATCACCGGGGGCATCGT |
| cDNA 2R | GAAGCGCCCAGCTGTGGCCACGAT |
| cDNA 3R | GGGTACTCCAGCAGGCACACAA |
| cDNA 4R | GATGCAGGACGGCCCGAACAT |
| cDNA 5F | CTGCTGGCCACCATCCTTGGGA |
| cDNA 5F2 | CTGGCCATTGCGAGCGGCA |
| cDNA 6F | CAGATCGGAGGCACCATCA |
| cDNA 6R | TCACACGACCTCGTCGGTCAC |
For thermocycling, the following conditions were used: For exons 1 through 5, an initial denaturation at 94°C for 3 minutes then 30 cycles of 94°C for 5 seconds, 70°C for 1 minute, followed by a 7-minute extension at 72°C. For exon 6, an initial denaturation at 94°C for 3 minutes then 30 cycles of 94°C for 30 seconds, 63°C for 15 seconds, 72°C for 30 seconds, followed by a 7-minute extension at 72°C.
Amplified segments were purified using a QIAquick PCR purification kit (Qiagen, Valencia, CA) and sequenced in both directions using the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit (PE Applied Biosystems, Foster City, CA). Exons 1 through 5 were sequenced using reactions mixtures as follows: 2 μL Ready Reaction Premix, 3 μL 5X Sequencing Buffer, 10 ng primer, and 2 μL PCR product in a 20 μL total reaction volume. Exon 6 was sequenced using undiluted BigDye Terminators: 8 μL Ready Reaction Premix, 20 ng primer, and 4 μL PCR product in a 20 μL total reaction volume. Sequencing reactions were purified in 96-well MicroAmp Trays (PE Applied Biosystems, Foster City, CA) by precipitating with 80 μL 75% isopropanol.
The complementary DNA (cDNA) numbering system we have used here follows the standard convention that +1 is the A of the initiator ATG codon. This differs from the numbering of some sequences deposited in GenBank (accession numbers M21186 and J03774). Twenty-eight should be subtracted from the GenBank sequence number to make the initiator +1. We have followed the standard system of designating the CGD phenotype thus: A22+ represents normal levels of expression of nonfunctional p22phox protein; A22− represents diminished expression of p22phox, and A22° represents an absence of p22phox expression.
Results
Absence of NADPH oxidase activity and flavocytochromeb558in patients with CGD
The biochemical findings on the patients are reported in Table2. In all cases, no significant O2− production was detected by NBT staining, cytochrome c reduction, or by flow cytometry using DHR or DCF, or a combination of these methods. Patient 6 showed a small amount of O2− generation by DCF assay, but gp91phox or p22phox were undetectable by immunoblot, suggesting a complete, or near complete absence of flavocytochrome b558. Similarly, none of the other patients showed any evidence for gp91phox or p22phoxpolypeptides by immunoblot. Where possible, the parents of the patients were analyzed biochemically; values for O2−, and flavocytochromeb558 content were in the normal to low-normal range.
Biochemical characteristics of the patients with CGD
| Patient . | NBT % positive cells* . | DCF/DHR† . | nmol/min/107 cells‡ . | Flavocytochrome b558 . | |
|---|---|---|---|---|---|
| Heme content pmol/107 cells2-153 . | Protein content/ flow cytometry . | ||||
| Normal range (n > 100) | >90 | 100% | 122 ± 26 | 74 ± 13 | + |
| 1 (male) | nd | 0% | nd | negative | |
| 2 (male) | 0 | 0 | 0 | negative | |
| 3 (female) | nd | 0 | nd | negative | |
| 4 (female) | 0 | 0% | nd | negative | |
| 5 (male) | 0 | 0.2 | 0 | negative | |
| 6 (female) | 0 | 5.0% | <5 | negative | |
| 7 and 7a (female) | 0 | 0 | 0 | negative | |
| 8 (female) | 0 | nd | nd | negative | |
| 9 (female) | 0 | 0 | 0 | negative | |
| Patient . | NBT % positive cells* . | DCF/DHR† . | nmol/min/107 cells‡ . | Flavocytochrome b558 . | |
|---|---|---|---|---|---|
| Heme content pmol/107 cells2-153 . | Protein content/ flow cytometry . | ||||
| Normal range (n > 100) | >90 | 100% | 122 ± 26 | 74 ± 13 | + |
| 1 (male) | nd | 0% | nd | negative | |
| 2 (male) | 0 | 0 | 0 | negative | |
| 3 (female) | nd | 0 | nd | negative | |
| 4 (female) | 0 | 0% | nd | negative | |
| 5 (male) | 0 | 0.2 | 0 | negative | |
| 6 (female) | 0 | 5.0% | <5 | negative | |
| 7 and 7a (female) | 0 | 0 | 0 | negative | |
| 8 (female) | 0 | nd | nd | negative | |
| 9 (female) | 0 | 0 | 0 | negative | |
CGD = chronic granulomatous disease; NBT = nitroblue tetrazolium; DCF = dichlorofluorescein; DHR = dihydrorhodamine; nd = not done.
Using the Nitroblue Tetrazolium slide test.25
By flow cytometry using DHR or DCF. Values represent % fluorescence compared with PMA-stimulated normal cells.
Using the superoxide dismutase-inhibitable rate of cytochrome c reduction.25
Determined spectrophotometrically.26
Having demonstrated the absence of flavocytochromeb558 in these patients, genetic analysis was undertaken. Because the most common cause of CGD is X-linked and involves defects in the CYBB gene (coding for gp91phox), the male patients in this study (patients 1, 2, and 5) were initially analyzed for defects in the gp91phox gene by single strand conformational polymorphism.30 In each case, the results were normal. Conversely, it was considered most likely that female patients deficient in flavocytochrome b558 had a primary defect in CYBA. Except in individuals with a highly skewed X-chromosome inactivation, or with a XO karyotype, a defect inCYBB is unlikely to be the cause of CGD in females. Consequently, all 6 exons of the CYBA gene (coding for p22phox) were sequenced in each patient. Primers were chosen such that were greater than or equal to 16 intronic nucleotides at the 5′ ends, and at least 12 intronic nucleotides at the 3′ ends of each exon were sequenced, to increase the chances of detecting mutations that result in splicing errors (Table1).
Mutations in the CYBA gene identified in this study
| Patient . | Maternal allele . | Amino acid change . | Paternal allele . | Amino acid change . | CGD type . |
|---|---|---|---|---|---|
| 1 | Exons 2-3 deleted (homozygous) | Exons 2-3 deleted (homozygous) | A22° | ||
| 2 | T155 → C (homozygous) | Missense: Leu52 → Pro | T155 → C (homozygous) | Missense: Leu52 → Pro | A22° |
| 3 | C244 deleted3-150 (homozygous) | Frameshift | C244 deleted3-150 (homozygous) | Frameshift | A22° |
| 4 | intron 3 5′ gt → tt (homozygous) | Ssplice mutation | Intron 3 5′ gt → tt (homozygous) | Splice mutation | A22° |
| 5 | G74 → T | Missense: Gly25 → Val | G26 → A | Nonsense: Trp9 → Stop | A22° |
| 6 | C268 → T (homozygous) | Missense: Arg90 → Trp | C268 → T (homozygous) | Missense: Arg90 → Trp | A22° |
| 7 and 7a | Insertion C at C162 | Frameshift | C268 → T | Missense: Arg90 → Trp | A22° |
| 8 | G107 → A | Nonsense: Trp36 → Stop | G70 → A | Missense: Gly24 → Arg | A22° |
| 9 | C354 → A | Ser118 → Arg | C354 → A | Ser118 → Arg | A22° |
| Patient . | Maternal allele . | Amino acid change . | Paternal allele . | Amino acid change . | CGD type . |
|---|---|---|---|---|---|
| 1 | Exons 2-3 deleted (homozygous) | Exons 2-3 deleted (homozygous) | A22° | ||
| 2 | T155 → C (homozygous) | Missense: Leu52 → Pro | T155 → C (homozygous) | Missense: Leu52 → Pro | A22° |
| 3 | C244 deleted3-150 (homozygous) | Frameshift | C244 deleted3-150 (homozygous) | Frameshift | A22° |
| 4 | intron 3 5′ gt → tt (homozygous) | Ssplice mutation | Intron 3 5′ gt → tt (homozygous) | Splice mutation | A22° |
| 5 | G74 → T | Missense: Gly25 → Val | G26 → A | Nonsense: Trp9 → Stop | A22° |
| 6 | C268 → T (homozygous) | Missense: Arg90 → Trp | C268 → T (homozygous) | Missense: Arg90 → Trp | A22° |
| 7 and 7a | Insertion C at C162 | Frameshift | C268 → T | Missense: Arg90 → Trp | A22° |
| 8 | G107 → A | Nonsense: Trp36 → Stop | G70 → A | Missense: Gly24 → Arg | A22° |
| 9 | C354 → A | Ser118 → Arg | C354 → A | Ser118 → Arg | A22° |
CGD = chronic granulomatous disease.
Previously seen in a heterozygous patient.
Patient 1.
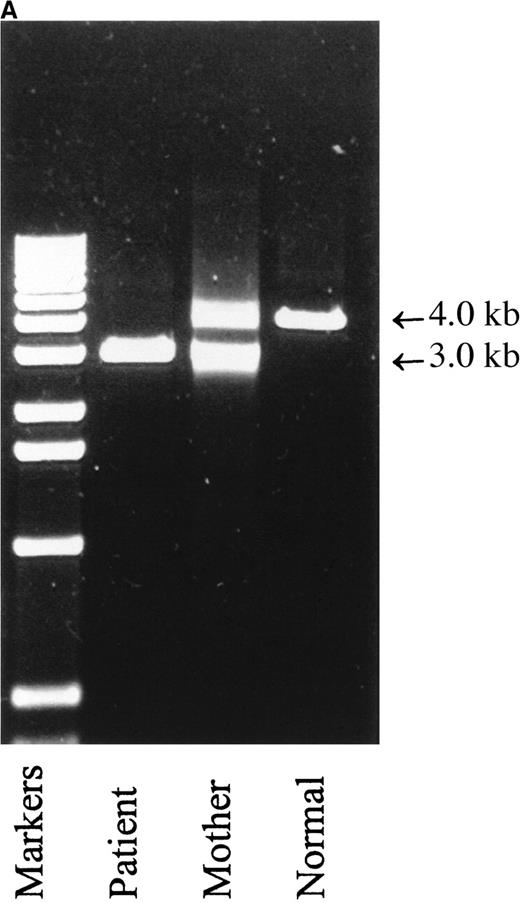
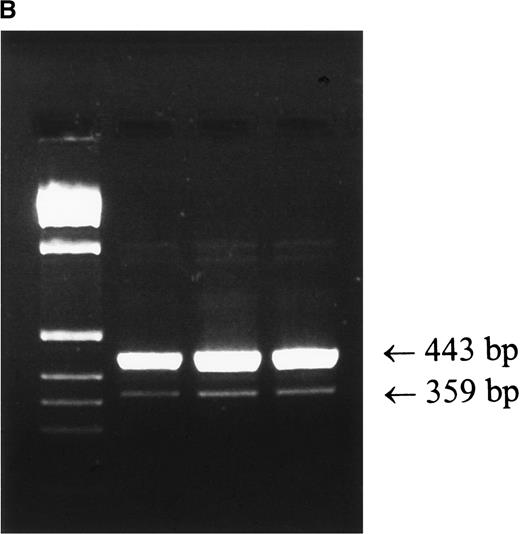
Attempts to amplify exons 2 and 3 individually from genomic DNA of patient 1 were unsuccessful. Exons 1, 4, 5, and 6 amplified normally, testifying to the integrity of the DNA and raising the possibility of a deletion within the gene. PCR amplification of the patient's genomic DNA from exon 1 to exon 4 (the shortest PCR practicable) produced a single fragment of approximately 3.0-kb, in contrast to the normal 4.0-kb fragment (Figure 1A). These results indicate that the patient is homozygous for a genomic deletion of exons 2 and 3. Products of both sizes were amplified from his mother (Figure 1A) and father (not shown), demonstrating that they are both carriers of this deletion. Analysis of the cDNA of the patient revealed 2 products, a major messenger RNA (mRNA) species of 443 bp, corresponding to the skipping of exons 2 and 3, and a minor species of 359 bp, corresponding to the skipping of exons 2, 3, and 4 (Figure 1B). The patient and his mother are also homozygous for a previously unreported polymorphism, the father is heterozygous. This novel polymorphism was the substitution of A for G at nucleotide 403 in exon 6. This causes the nonconservative substitution of lysine for glutamic acid at amino acid 135. Seventy-five normal individuals were sequenced without finding another allele of this type (Table 4), but in view of the mother's good health and positive DHR test, we conclude this represents a rare polymorphism.
Molecular mass analysis of PCR products derived from amplification of genomic DNA and of mRNA species derived by reverse transcriptase PCR.
(A) PCR amplification of exons 1 to 4 of genomic DNA from patient 1, his mother, and a normal control was performed as described in “Patients, materials, and methods.” The normal product is approximately 4.0-kb, and the product derived from amplification of exons 1 and 4 with exons 2 and 3 deleted is approximately 3.0-kb. (B) RT-PCR of mRNA prepared from whole blood was performed as described in “Patients, materials, and methods.” Left lane, markers; right lanes, 5, 10 and 13 μL of PCR product. The upper band is derived from a mRNA with exons 2 and 3 skipped, the lower band is derived from a mRNA species with exons 2, 3, and 4 skipped.
Molecular mass analysis of PCR products derived from amplification of genomic DNA and of mRNA species derived by reverse transcriptase PCR.
(A) PCR amplification of exons 1 to 4 of genomic DNA from patient 1, his mother, and a normal control was performed as described in “Patients, materials, and methods.” The normal product is approximately 4.0-kb, and the product derived from amplification of exons 1 and 4 with exons 2 and 3 deleted is approximately 3.0-kb. (B) RT-PCR of mRNA prepared from whole blood was performed as described in “Patients, materials, and methods.” Left lane, markers; right lanes, 5, 10 and 13 μL of PCR product. The upper band is derived from a mRNA with exons 2 and 3 skipped, the lower band is derived from a mRNA species with exons 2, 3, and 4 skipped.
Polymorphisms identified in the CYBA gene
| Nucleotide change . | Amino acid change . | . |
|---|---|---|
| −37 intron 1 3′ a → g | N/A | 1 normal homozygous g |
| 3 normal heterozygotes | ||
| 6 normal homozygous a | ||
| A179 → C | Lys60 → Thr | 1 normal heterozygote |
| 59 normal homozygous A | ||
| C214 → T | His72 → Tyr | Previously reported13 |
| G403 → A | Glu135 → Lys | Healthy mother of patient 1 homozygous |
| G480 → A | Silent | 2 normal homozygous A |
| 3 heterozygous | ||
| 42 normal homozygous G | ||
| Previously reported16 | ||
| C521 → T | Ala174 → Val | 3 normal homozygous T |
| 8 normal homozygous C | ||
| Previously reported13 | ||
| +24 of 3′ untranslated | N/A | 3 normal homozygous g |
| 1 heterozygote | ||
| 7 homozygous a | ||
| Previously reported16 |
| Nucleotide change . | Amino acid change . | . |
|---|---|---|
| −37 intron 1 3′ a → g | N/A | 1 normal homozygous g |
| 3 normal heterozygotes | ||
| 6 normal homozygous a | ||
| A179 → C | Lys60 → Thr | 1 normal heterozygote |
| 59 normal homozygous A | ||
| C214 → T | His72 → Tyr | Previously reported13 |
| G403 → A | Glu135 → Lys | Healthy mother of patient 1 homozygous |
| G480 → A | Silent | 2 normal homozygous A |
| 3 heterozygous | ||
| 42 normal homozygous G | ||
| Previously reported16 | ||
| C521 → T | Ala174 → Val | 3 normal homozygous T |
| 8 normal homozygous C | ||
| Previously reported13 | ||
| +24 of 3′ untranslated | N/A | 3 normal homozygous g |
| 1 heterozygote | ||
| 7 homozygous a | ||
| Previously reported16 |
Patient 2.
This patient was homozygous for a T-to-C transition at nucleotide 155 in exon 3. This missense mutation results in the substitution of proline for leucine at position 52. No other patient or normal subject has been found who carry this mutation indicating that it is unlikely to be a benign polymorphism. No other defects were found in the patient's CYBA gene. As expected, his mother was heterozygous for this substitution. DNA from the father was not available.
Patient 3.
This patient was homozygous for the deletion of C244 in exon 4. This frameshift results in a stop codon at amino acid position 190. The parents, who are not known to be related, were both found to be carriers of this single base pair deletion. This defect has been described previously in an unrelated patient who was heterozygous for this mutation.13
Patient 4.
Genomic sequencing of DNA from patient 4 revealed a homozygous splice-site mutation in intron 3, converting the 5′ gtgag→ttgag. This splicing error results in the loss of exon 3 as detected by RT-PCR. The patient's mother carries this mutation, the father was not available for study.
Patient 5.
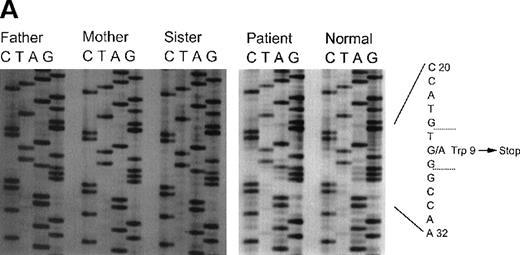
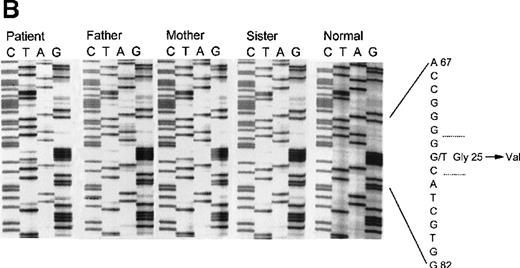
This patient was identified as a compound heterozygote for 2 mutations in his CYBA gene. The first allele contained a nonsense mutation, a transition of G26→A in exon 1, leading to a stop codon and truncation of the p22phox polypeptide at amino acid 9 (Figure 2A). This mutation was inherited from his father and was also found in his sister. The second allele had a G→T transition at nucleotide 74 in exon 2 (Figure 2B). This results in the substitution of glycine 25 with the larger and more hydrophobic valine. Although this is generally regarded as a relatively conservative substitution, it apparently leads to an unstable product, as no protein could be detected in the patient's neutrophils. As expected, the mother is a heterozygous carrier of this mutation and a normal allele. The patient's sister does not carry the mutated maternal allele and does not have CGD.
Sequence analysis of exon 1 and 2 in patient 5.
Panel A: Sequence analysis of exon 1 of the CYBA gene of patient 5 and his family members. The patient, his father, and sister are all heterozygous for A at nucleotide 26, producing a nonsense codon at amino acid 9. The normal control shows only G at position 26. Panel B: Sequence analysis of exon 2 of the CYBA gene of patient 5 and his family members. The patient is homozygous for T at nucleotide 74, causing the replacement of glycine 25 with valine. The patient's mother is heterozygous as seen by the presence of both G and T at position 74. The patient's father and sister do not carry this allele as seen by the appearance of only the G at this position.
Sequence analysis of exon 1 and 2 in patient 5.
Panel A: Sequence analysis of exon 1 of the CYBA gene of patient 5 and his family members. The patient, his father, and sister are all heterozygous for A at nucleotide 26, producing a nonsense codon at amino acid 9. The normal control shows only G at position 26. Panel B: Sequence analysis of exon 2 of the CYBA gene of patient 5 and his family members. The patient is homozygous for T at nucleotide 74, causing the replacement of glycine 25 with valine. The patient's mother is heterozygous as seen by the presence of both G and T at position 74. The patient's father and sister do not carry this allele as seen by the appearance of only the G at this position.
Patient 6.
Through genomic sequencing of DNA from patient 6, we discovered a homozygous C→T transition at nucleotide 268. This causes the nonconservative replacement of arginine 90 with tryptophan and the loss of p22phox. The patient's parents are first cousins, and both are carriers of this mutation.
Patients 7 and 7a.
Two sisters, patients 7 and 7a, were identified as compound heterozygotes. The maternal allele contained the insertion of C at nucleotide 162 in exon 3 causing a frameshift and a stop codon at amino acid 73. The paternal allele contains a missense mutation C268→T; predicting the nonconservative amino acid change arginine 90→ tryptophan. This is the same mutation found in patient 6, but the families are not known to be related.
Patient 8.
This patient is a compound heterozygote, with the allele inherited from her mother containing a nonsense mutation at nucleotide 107, G→A in exon 2, predicting tryptophan 36 being changed to a stop codon. The patient's second mutation, a transition of G→A at nucleotide 70, also in exon 2, causes a missense defect predicting the nonconservative substitution of an arginine residue for glycine at position 24. The father was not available for study.
Patient 9.
This patient is homozygous for the substitution of nucleotide 354 C→A, causing the replacement of serine 118 with arginine. As expected, both parents were carriers of this mutation. This defect has been described before in another homozygous patient also of Hispanic heritage.13
Polymorphisms in the CYBA gene
During the course of these studies, we identified a number of polymorphisms (listed in Table 4) in addition to those described above. These polymorphisms include 2 nonconservative amino acid substitutions: lysine 60→ threonine caused by an A→C transversion at nucleotide 179 (found 1 allele of 59 studied); and glutamic acid 135→ lysine caused by a G→A transition at nucleotide 403.
Discussion
Before this study, only 10 different mutations in the CYBAgene had been identified as causing A22 CGD. These mutations were found in patients from 9 kindreds encompassing 18 alleles (Cross et al31 and references therein). In addition, 4 polymorphisms in the gene had been documented. Here we report on 10 new patients from 9 families in whom we identified 12 mutations, 9 of which are novel. We also identified 3 previously unreported polymorphisms. Consistent with previous studies of mutations causing CGD, our results show a wide variety of defects, insertions, deletions, and substitutions leading to missense, nonsense, frameshift, and splice-site mutations, with no preponderance of common affected alleles or “hot-spots.” This heterogeneity has been seen in studies of the X-linked gp91phox gene (CYBB),14,30,32 the autosomal p67phox gene (NCF-2),33,34and the earlier studies of CYBA.31 The exception to this pattern is the overwhelming preponderance of a GT deletion (ΔGT) at the beginning of exon 2 of the NCF-1 gene, causing p47phox-deficiency, the second most common form of CGD. This unusual finding is explained by the presence of at least 1, and probably more, highly homologous pseudogenes that contain the ΔGT sequence.35,36 Relatively frequent recombination events between the wild-type gene and the pseudogene(s) account for the prevalent nature of the ΔGT genotype, and give rise to the single most common disease-causing allele in CGD.
Two of the mutations identified in this study have been described previously. The deletion of C244, which we found to be homozygous in the genomic DNA of patient 3, was first described in a compound heterozygote reported by Dinauer and coworkers.13 In that case, the other mutant allele carried a missense mutation G269→A, (arginine 90→glutamine). These patients are not known to be related. Patient 9 was found to be homozygous for C354→A, this mutation was found previously in another patient also heterozygous for this change.13 Although these patients are not thought to be related, both are of Hispanic origin and this may reflect the presence of a rare mutant allele in the Hispanic population.
Surprisingly, in the 17 unrelated kindreds with A22 CGD, 4 have changes causing missense mutations at arginine 90. The first of these patients was heterozygous for a G269→A transition, resulting in the replacement of arginine 90 with glutamine.13 This same mutation was found in a homozygous state in a patient whose parents were first cousins.16 We have found a second mutation that causes a change at this amino acid residue in 2 unrelated families. Patients 7 and 7a were heterozygous, and patient 6 was homozygous for a C→T transition at the adjacent nucleotide 268. This mutation results in the replacement of the arginine with tryptophan rather than glutamine. It is possible that this is a mutational hot spot, because these 4 events appear to have occurred independently. Both missense mutations result in the A22° phenotype.
Of the 9 different missense mutations known, including the 4 that are described here, only 1 results in the expression of stable protein (the A22+ phenotype). In that instance, the patient was homozygous for the substitution of proline 156 with glutamine.15 This particular substitution was very informative functionally, as biochemical analysis showed that it resulted in the failure of p47phox to translocate to the membrane. Proline 156 is within a short proline-rich region in the cytoplasmic tail of p22phox (amino acids 151-160) and the profound effect of its alteration to glutamine highlights the importance of this region of the p22phox molecule in interactions with an SH3 domain in p47phox.37
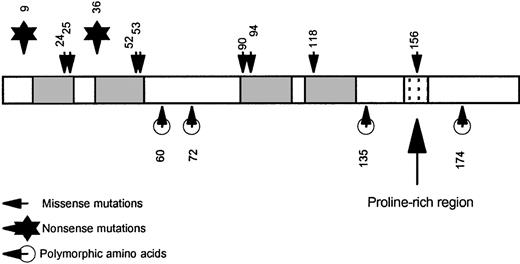
It is noteworthy that all the missense mutations reported so far that result in the A22° phenotype cause amino acid substitutions within the putative membrane-spanning domains of p22phox, whereas the mutation causing the A22+ phenotype and the 4 known polymorphic amino acid residues fall outside these regions (Figure3). We speculate that amino-acid substitutions in membrane-spanning domains are poorly tolerated and lead to unstable protein, particularly if they involve changes in charge or hydrophobicity. Outside the membrane-spanning domain, even nonconservative changes seem to be better tolerated. For example, the histidine 72→ tyrosine and glutamic acid 135→ lysine polymorphisms both lead to normal levels of functional p22phox.
Pictorial representation of p22phox.
The shaded areas represent the putative transmembrane regions. The N-terminus and C-terminus are both cytoplasmic. The stippled area is the proline-rich sequence (amino acids 151-160) that mediates a protein/protein interaction with p47phox. Note that all missense mutations that result in the complete loss of protein (the A22° phenotype) are located within the transmembrane regions, whereas the mutation causing the A22+ phenotype, as well as the polymorphic amino acids, are located outside the membrane.
Pictorial representation of p22phox.
The shaded areas represent the putative transmembrane regions. The N-terminus and C-terminus are both cytoplasmic. The stippled area is the proline-rich sequence (amino acids 151-160) that mediates a protein/protein interaction with p47phox. Note that all missense mutations that result in the complete loss of protein (the A22° phenotype) are located within the transmembrane regions, whereas the mutation causing the A22+ phenotype, as well as the polymorphic amino acids, are located outside the membrane.
During the course of this study, we found examples of all 4 of the polymorphisms previously reported in the literature (Table4);13,16 in addition, we also found 3 new polymorphisms, 2 of which produced amino acid substitutions. The first, lysine 60→ arginine, is a conservative substitution predicted to be located in a cytoplasmic loop between the central 2 membrane-spanning domains of p22phox. The second, glutamine 135→ lysine, is a less conservative change, located close to the cytoplasmic side of the membrane. Polymorphisms within p22phox may have significance for certain disease susceptibilities. Inoue and colleagues18 have recently reported a significant association between the C214 genotype (histidine 72) and coronary artery disease, compared with the T214 genotype (tyrosine 72), although these findings have not been confirmed by others.19-21 It has been postulated that p22phox forms part of an isoform of NADPH oxidase that may play a signaling role in nonphagocytic cells38 and very recently such an enzyme has been described.24 Polymorphic forms of p22phox might have differential effects in such systems.
The difficulty of detecting heterozygotes carrying exonic deletions is illustrated in the family of patient 1. Detection cannot be achieved by direct sequencing alone, because the primers will always amplify the normal sequence (for example, the parents of patient 1 appear normal by sequencing). Instead, analysis of the size of fragments generated by PCR amplification of genomic DNA or cDNA must be used to identify carriers as shown in Figure 1.
Carrier detection is of great importance for genetic counseling and prenatal diagnosis. In the case of X-linked CGD, this is usually relatively easy, because female carriers (who are mostly healthy) generally exhibit 2 populations of cells (due to X-chromosome inactivation), 1 positive for NADPH oxidase activity and 1 negative. These distinct populations can readily be distinguished biochemically using the NBT slide test or DHR flow cytometry. The situation with the autosomal recessive forms of CGD is quite different, however. Generally, carriers of autosomal recessive CGD have uniform populations of neutrophils that are capable of generating amounts of O2− within the normal range and the individuals concerned have no obvious clinical manifestations. In the relatively few heterozygotes for p22phox-deficiency in whom cellular flavocytochrome b558 concentrations have been measured, these too appear within normal limits. Consequently, the only reliable way of testing for such carriers is by molecular genetic analysis.
Acknowledgment
We are grateful to Valerie Moreau for her skilled secretarial assistance.
Supported by National Institutes of Health Grant Nos. RO1 AI 24838 (A.R.C), CA68276 (P.G.H.), and RR00833 (to the GCRC at TSRI).
Reprints:Andrew R. Cross, Department of Molecular & Experimental Medicine, MEM-241, The Scripps Research Institute, 10550 N Torrey Pines Rd, La Jolla, CA 92037; e-mail: scross@scripps.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal