Abstract
Hematopoietic stem cell transplantation is characterized by a prolonged period of humoral immunodeficiency. We have previously shown that the deficiencies are probably not due to the failure to utilize the appropriate V regions in the pre-immune repertoire. However, a striking observation, which correlated with the absence of immunoglobulin IgD− cells and was consistent with a defect in antigen-driven responses, was that rearrangements in bone marrow transplant (BMT) recipients exhibited much less somatic mutation than did rearrangements obtained from healthy subjects. In this paper, we present evidence suggesting that naive B cells obtained from BMT recipients lack the capacity to accumulate somatic mutations in a T-cell–dependent manner compared with healthy subjects. This appears to be a B-cell–autonomous deficit because T cells from some patients, which were not able to support the accumulation of mutations in autologous naive B cells, were able to support accumulation of mutations in heterologous healthy-subject naive B cells.
Hematopoietic stem cell transplantation is a recognized treatment for certain leukemias, other blood diseases, and some inborn errors of metabolism and has potential as a vehicle for gene therapy. However, hematopoietic stem cell transplant, using either bone marrow or mobilized peripheral stem cells, is characterized by a prolonged period of immunodeficiency affecting both B-cell and T-cell compartments (reviewed in Storek et al1). B-cell counts are low but usually approach normalcy by 1 year after transplantation.1 Coincident with recovering B cells, serum immunoglobulin IgM, IgG1 and IgG3 levels but not IgG2 and IgA levels return to normal by 1 year posttransplantation.2-5 Thus, marrow recipients who survive the initial postgrafting period do not always become fully immunocompetent. Many recipients are deficient in generating specific antibody responses to exogenous stimuli. The complete reconstitution of B-cell immunity in recipients can take years.
The nature of the B-cell defect(s) leading to this specific humoral immunodeficiency is uncertain. Normal levels of serum IgM, IgG1, and IgG35 indicate that immunodeficiency is not due to a general failure to produce immunoglobulin or an overt lack of T-cell help (although impaired T-cell function may be an important factor). Evidence indicates that the processes involved in generating and selecting the primary antibody repertoire are largely functional within the first year following bone marrow transplant (BMT) and that the immunodeficiencies common among BMT recipients are probably not due to the failure to utilize appropriate V region genes in generating the pre-immune antibody repertoire.6,7 The complexity of the CDR3 (third complementarity determining region) and DH and JH utilization is similar in BMT recipients and healthy subjects 1 year posttransplantation, further supporting the conclusion that the primary antibody repertoire is generated normally following BMT.8,31 However, a striking observation was that rearrangements in BMT recipients exhibited much less somatic mutation than did rearrangements obtained from healthy subjects.6-8The failure in the BMT recipients to accumulate somatic mutations in rearranged VH genes is consistent with a maturational arrest at a fairly late stage of differentiation. This deficit could be a consequence of either an intrinsic B-cell deficit or a lack of adequate T-cell help. In this paper, we present evidence suggesting that, in contrast to healthy-subject B cells, B cells obtained from transplantation patients 1 year posttransplantation lack the capacity to accumulate somatic mutations in a T-cell–dependent manner. This appears to be a B-cell–autonomous deficit, because T cells from some patients were able to support accumulation of mutations in heterologous healthy-subject B cells but not in autologous B cells.
Patients, materials, and methods
Patients and donors
Blood samples were obtained under Institutional Review Board–approved protocols, and written consent was always obtained. Blood mononuclear cells were separated by density-gradient centrifugation, with the use of Ficoll-Hypaque (1.077 kg/L) (Amersham Pharmacia Biotech, Piscataway, NJ). We studied 9 recipients of allogeneic hematopoietic cell transplants at approximately 1 year after grafting (median, 378 days; range, 354-432 days) in combination with 6 healthy subjects. The median age at transplantation was 42 years (range, 29-52 years). No patient had a history of splenectomy. All patients were transplanted for hematological malignancies. They were usually conditioned with cyclophosphamide (120 mg/kg) and fractionated total body irradiation (12.0 to 13.2 Gy). The hematopoietic cell donors for BMT1, 2, 3, 7, 8, and 9 were siblings matched for HLA-A, HLA-B, and HLA-DR, and the donors for patients BMT4, 5, and 6 were unrelated volunteers matched for HLA-A, HLA-B, and HLA-DR. Seven patients (BMT1, 4, 5, 6, 7, 8, 9) received unmodified marrow; 1 patient (BMT3) received unmodified filgrastim-mobilized blood progenitor cells; and 1 patient (BMT2) received filgrastim-mobilized blood progenitor cells positively enriched for CD34+ cells. Graft-versus-host disease (GVHD) prophylaxis typically consisted of methotrexate (day 1, 3, 6, and 11) and cyclosporine (day −1 through 180).9 Grade 2 to 3 acute GVHD occurred in 7 patients (BMT2, 3, 5, 6, 7, 8, and 9); it was usually treated with oral prednisone (1 to 2 mg/kg/d). Prior to the 1-year posttransplantation evaluation, clinical limited chronic GVHD developed in 4 patients (BMT2, 5, 6, and 8) and clinical extensive chronic GVHD in 3 patients (BMT1, 3, and 9).
At the time of the 1-year posttransplantation evaluation, the patients were thoroughly tested for potential relapse of the hematological malignancy and for chronic GVHD status. Eight patients were in complete remission; 1 patient (BMT2) was in early relapse (patient with IgG-lambda multiple myeloma who had 12% plasma cells in marrow and 16.0 g/L monoclonal IgG-lambda in serum at 1 year posttransplantation). All 9 patients were complete chimeras defined by more than 99% donor cells in marrow and/or blood, with the use of Y-chromosome in situ hybridization or variable nucleotide tandem repeats.10 11Two patients had clinical extensive chronic GVHD (BMT3 and 9), and the remaining 7 patients had no clinical GVHD at the 1-year posttransplantation blood draw. One patient (BMT1) was on oral prednisone (50 mg/d), and the remaining 8 patients were on no systemic immunosuppressive drugs. Patients had received no biological response modifiers such as interferon and no intravenous immunoglobulin (IVIG) within 2 months prior to the 1-year posttransplantation evaluation, except for 1 patient (BMT2) who received IVIG approximately 6 weeks prior to the 1-year posttransplantation evaluation.
Each patient sample was tested in parallel with a volunteer control (age 20 to 50) recruited from employees of the Virginia Mason Research Center or the Fred Hutchinson Cancer Research Center.
Flow cytometry and sorting
The enumeration of B cells and CD4 T cells was done with the use of 3-color flow cytometry as described.12 13 For sorting, blood mononuclear cells were stained with fluorescein isothiocyanate (FITC)–conjugated goat-antihuman IgD antibody (F(ab′)2) (Caltag, Burlingame, CA) and phycoerythrin (PE)-conjugated mouse-antihuman CD4 antibody (Becton Dickinson, Franklin Lakes, NJ, or Coulter-Immunotech, Fullerton, CA) and sorted on a FACS Vantage (Becton Dickinson). Forward- versus side-scatter gate was set to encompass primarily lymphoid cells and only a small fraction of monocytoid cells. CD4 T cells were defined as CD4highcells; IgD+ B cells were defined as IgDhighcells. Sorted fractions were more than 91% pure.
For the major histocompatibility complex (MHC) control experiments, B cells were isolated from peripheral blood lymphocytes (PBL) by anti-CD19–coated immunomagnetic beads (DynaBeads, Dynal, Lake Success, NY) according to the manufacturer's protocol. Naive human B-cells (CD19+/IgD+) were further purified from the CD19+ B cells by anti-IgD (FITC-conjugated) sorting on a FACS Vantage. The resulting population was more than 95% CD19+IgD+IgM+. Autologous CD4+ T cells were isolated with the use of anti-CD4–coated immunomagnetic beads (Dynal) according to the manufacturer's protocol. The purity of CD4 T cells as determined by FACS analysis was greater than 99% following isolation.
Cell culture
In vitro cultures for analysis of somatic mutation were performed as described.14 In brief, B cells (103 per well) were cultured in flat-bottomed microplates in RPMI 1640 medium supplemented with interleukin (IL)–4 (100 U/mL) and anti-IgM (1 μg/mL). Other additions, depending on the experiment, were CD40L (1 μg/mL), CDw32L cells (L cells) (104 per well), resting or activated CD4+ T cells (105per well). L cells and T cells were irradiated before initiation of the cultures (70 and 30 Gy, respectively). To activate T cells, wells were precoated with 64.1 antibody, a murine monoclonal antihuman CD3.
Complementary DNA library construction
Complementary DNA (cDNA) libraries were constructed as described.14 15 Polymerase chain reactions (PCRs) used for constructing the libraries were performed with the use of the family-specific 5′ primers, E310 (VH3-L), 5′-CTGAATTCCATGGAGTTTGGGCTGAGCTG-3′, corresponding to the 5′ ends of the leader sequence of VH3 family, and a 70:15:15 mixture of the 3′ primers E311, 5′-GACTCTAGACT(CT)ACCTGAGGAGACGGTGACC-3′, complementary to the 3′ ends of JH1, 4, 5, and 6 gene sequences; E312, 5′-GACTCTAGACT(CT)ACCTGAGGAGACAGTGACC-3′, complementary to the 3′ end of JH2 gene sequence; and E313, 5′-GAC-TCTAGACT(CT)ACCTGAAGAGACGGTGACC-3′, complementary to the 3′ end of JH3 gene sequence. Restriction sites (EcoRI for 5′ primers; XbaI for 3′ primers) included in the primers are underlined. For amplification of VH3 transcripts Pfu or Taq DNA polymerase (Promega Corp, Madison, WI) was used in a 30-cycle program.
Detection of somatic mutation
Semiquantitative PCR
cDNA was amplified with the use of Taq DNA polymerase (Promega) and primers for VH3 expressing IgM (E310 and E213, 5′-AATTCTAGATCACAGGAGACGAGGGGGAAAAG- 3′) and IgG transcripts (E310 and E212, 5′-AATTCTAGAGGGGAAGTAGTCCTTGACCAGGCA-3′). As an internal control, β-actin primers were included (E376, 5′-GGTGGGCATGGGTCAGAAGGATT-3′ and E377, 5′-CCAGAGGCGTACAGGGATAGCAC-3′). The PCR products were size-fractionated on a 1.5% agarose gel and stained with ethidium bromide.
Results
It was not certain if the previously reported lack of somatically mutated B cells in peripheral blood of transplant recipients was a result of an actual deficit in the somatic mutational process or if it was the product of population dynamics. Our approach to investigating this issue was suggested by the observation that a high incidence of mutations was observed in VH transcripts obtained from healthy-subject B cells following a 14-day coculture with activated CD4+ T cells.14 We hypothesized that if there was an actual deficit in the mutational process, then BMT B cells would not accumulate mutations in cocultures with activated CD4+ T cells. Furthermore, we reasoned that if an intrinsic B-cell deficit existed, then neither healthy-subject T cells nor BMT-recipient T cells would support the accumulation of somatic mutations in BMT-recipient B cells in culture. Alternatively, if the deficit was only in the T-cell compartment, then healthy-subject T cells should support the accumulation of mutations among BMT-recipient B cells. To test these hypotheses, B cells and CD4+ T cells obtained from transplant recipients and from healthy subjects were cocultured, and accumulation of somatic mutations was assessed.
In this study, 9 recipients of allogeneic bone marrow transplants were studied. At the time of the 1-year posttransplantation evaluation, the number and percentage of B cells and T cells in peripheral blood were determined (Table 1). The median CD4+ T-cell count was 265 × 106/L, which was below the normal range. The B-cell counts were normal or supranormal, except for 1 patient in whom the B-cell count was subnormal; the median count was 253 × 106/L, which fell in the normal range. The percentage of cells expressing membrane IgD was determined as described in Figure1; it tended to be above normal, although the median fell within the normal range (Table 1).
Lymphocyte subsets by flow cytometry 1 year following transplantation
| Sample identification . | Abs MNC (×106/L) . | % B cells . | Abs B (×106/L) . | % IgD+ . | % T cells . | % CD4 . | % CD8 . | Abs CD4 (×106/L) . | Abs CD8 (×106/L) . |
|---|---|---|---|---|---|---|---|---|---|
| BMT1 | 3070 | 7 | 215 | 91 | 57 | 13 | 53 | 227 | 927 |
| BMT2 | 550 | 3 | 17 | 87 | 30 | 48 | 23 | 79 | 38 |
| BMT3 | 2750 | 22 | 605 | 93 | 46 | 53 | 44 | 670 | 557 |
| BMT4 | 1120 | 26 | 291 | 95 | 50 | 47 | 42 | 263 | 235 |
| BMT5 | 1940 | 33 | 640 | 95 | 30 | 57 | 34 | 332 | 198 |
| BMT6 | 810 | 18 | 146 | 98 | 36 | 53 | 30 | 155 | 87 |
| BMT7 | 1510 | 14 | 211 | 93 | 57 | 31 | 51 | 267 | 439 |
| BMT8 | 2010 | 28 | 563 | 96 | 41 | 54 | 25 | 445 | 206 |
| BMT9* | |||||||||
| Median | 1725 | 20 | 253 | 94 | 44 | 51 | 38 | 265 | 221 |
| Normal range† | 94-561 | 62-95 | 400-1313 |
| Sample identification . | Abs MNC (×106/L) . | % B cells . | Abs B (×106/L) . | % IgD+ . | % T cells . | % CD4 . | % CD8 . | Abs CD4 (×106/L) . | Abs CD8 (×106/L) . |
|---|---|---|---|---|---|---|---|---|---|
| BMT1 | 3070 | 7 | 215 | 91 | 57 | 13 | 53 | 227 | 927 |
| BMT2 | 550 | 3 | 17 | 87 | 30 | 48 | 23 | 79 | 38 |
| BMT3 | 2750 | 22 | 605 | 93 | 46 | 53 | 44 | 670 | 557 |
| BMT4 | 1120 | 26 | 291 | 95 | 50 | 47 | 42 | 263 | 235 |
| BMT5 | 1940 | 33 | 640 | 95 | 30 | 57 | 34 | 332 | 198 |
| BMT6 | 810 | 18 | 146 | 98 | 36 | 53 | 30 | 155 | 87 |
| BMT7 | 1510 | 14 | 211 | 93 | 57 | 31 | 51 | 267 | 439 |
| BMT8 | 2010 | 28 | 563 | 96 | 41 | 54 | 25 | 445 | 206 |
| BMT9* | |||||||||
| Median | 1725 | 20 | 253 | 94 | 44 | 51 | 38 | 265 | 221 |
| Normal range† | 94-561 | 62-95 | 400-1313 |
Abs, absolute; MNC, mononuclear cells.
No flow data available for BMT9.
Normal ranges were defined as 5th to 95th percentiles of at least 91 adult healthy volunteers analyzed at the Fred Hutchinson Cancer Research Center.
Flow cytometric analysis and gating of blood B cells showing the relative lack of IgD− B cells in a BMT recipient 1 year after grafting.
Both specimens, normal and patient, were processed concurrently. Ficoll-isolated mononuclear cells (MNCs) were stained with anti-IgD–FITC (goat F(ab′)2 antihuman delta chain), anti-(CD4, CD8, CD14, CD16)–PE, and anti-(CD19, CD20)–peridinin chlorophyll protein. Data were acquired on FACSCAN cytometer. For analysis, first the MNC gate (R1, R4) was drawn on the forward- versus side-scatter dot plots (left). Then, B cells were gated on the CD19/CD20 versus CD4/CD8/CD14/CD16 dot plots (R2, R5), excluding non-B cells binding anti-CD19/CD20 nonspecifically, “B cell + non-B cell” doublets, and CD20low T cells (middle). Finally, CDl9/CD20 versus IgD dot plots were created exclusively of the cells falling within the MNC gate and the B-cell gate, ie, R1 and R2 in the normal and R4 and R5 in the patient (right). To calculate the percentage of IgD− B cells, regions R3 and R6 were set so as to encompass the B cells showing only background FITC fluorescence. In this example, 18% of B cells in the normal were IgD− versus 4% of B cells in the patient.
Flow cytometric analysis and gating of blood B cells showing the relative lack of IgD− B cells in a BMT recipient 1 year after grafting.
Both specimens, normal and patient, were processed concurrently. Ficoll-isolated mononuclear cells (MNCs) were stained with anti-IgD–FITC (goat F(ab′)2 antihuman delta chain), anti-(CD4, CD8, CD14, CD16)–PE, and anti-(CD19, CD20)–peridinin chlorophyll protein. Data were acquired on FACSCAN cytometer. For analysis, first the MNC gate (R1, R4) was drawn on the forward- versus side-scatter dot plots (left). Then, B cells were gated on the CD19/CD20 versus CD4/CD8/CD14/CD16 dot plots (R2, R5), excluding non-B cells binding anti-CD19/CD20 nonspecifically, “B cell + non-B cell” doublets, and CD20low T cells (middle). Finally, CDl9/CD20 versus IgD dot plots were created exclusively of the cells falling within the MNC gate and the B-cell gate, ie, R1 and R2 in the normal and R4 and R5 in the patient (right). To calculate the percentage of IgD− B cells, regions R3 and R6 were set so as to encompass the B cells showing only background FITC fluorescence. In this example, 18% of B cells in the normal were IgD− versus 4% of B cells in the patient.
Autologous combinations of BMT-recipient lymphocytes had poor cell growth, and as shown in Figure 2 (Group 1), yielded a low incidence of somatic mutation. As expected, autologous combinations of healthy-subject lymphocytes yielded a high incidence of mutation (Figure 2, Group 2). These results indicate a deficit among BMT-recipient lymphocytes that might be a consequence of poor growth or a failure to activate BMT-recipient T cells.
Incidence of somatically mutated VHtranscripts in lymphocyte cocultures is dependent primarily on the source of B cells.
Accumulation of somatic mutation was assessed by sequential hybridization as described.7,14-16 The results are presented as the percentage of VH transcripts that have acquired 1 or more mutations in either of two 21–base-pair (bp) target sequences.7,14 15 In the Figure, h indicates healthy subject; p, BMT patient, (−), T cells replaced by CD40 ligand and L cells. Closed circles indicate sources of transcripts selected for sequence analysis (see also Figure 4).
Incidence of somatically mutated VHtranscripts in lymphocyte cocultures is dependent primarily on the source of B cells.
Accumulation of somatic mutation was assessed by sequential hybridization as described.7,14-16 The results are presented as the percentage of VH transcripts that have acquired 1 or more mutations in either of two 21–base-pair (bp) target sequences.7,14 15 In the Figure, h indicates healthy subject; p, BMT patient, (−), T cells replaced by CD40 ligand and L cells. Closed circles indicate sources of transcripts selected for sequence analysis (see also Figure 4).
Heterologous combinations of BMT-recipient B cells and healthy-subject T lymphocytes yielded background levels ofmutation in all but 1 experiment (Group 3). Thus, neither healthy-subject T cells nor BMT-recipient T cells were consistently able to induce the accumulation of somatic mutation in BMT-recipient B cells, suggesting that BMT B cells had an intrinsic inability to be driven to accumulate somatic mutations. The inability of healthy-subject T cells to induce mutation in heterologous culture combinations is probably not due to MHC mismatch between BMT-recipient B cells and healthy-subject T cells; B cells in heterologous combinations of lymphocytes from HLA-disparate healthy donors, shown in Table 2, accumulate mutations as well as B cells in autologous combinations (Figure 3). That resting T cells, even if supplemented with CD40L, did not induce somatic mutations in these cultures suggests that the accumulation of somatic mutation in this system is not driven by an allogeneic reaction.
HLA-class II typing of healthy donors
| Sample ID . | DRB1* . | DQA1* . | DQB1* . | DPB1* . |
|---|---|---|---|---|
| HS1 | 0101, 0404 | 0101, 0301 | 0501, 0302 | 0402, 0601 |
| HS2 | 0701, 0701 | 0201, 0201 | 0201, 0201 | 0402, 1101 |
| HS3 | 0701, 0101 | 0201, 0101 | 0201, 0501 | 0201, 0402 |
| HS4† | ||||
| HS5† | ||||
| HS6† |
| Sample ID . | DRB1* . | DQA1* . | DQB1* . | DPB1* . |
|---|---|---|---|---|
| HS1 | 0101, 0404 | 0101, 0301 | 0501, 0302 | 0402, 0601 |
| HS2 | 0701, 0701 | 0201, 0201 | 0201, 0201 | 0402, 1101 |
| HS3 | 0701, 0101 | 0201, 0101 | 0201, 0501 | 0201, 0402 |
| HS4† | ||||
| HS5† | ||||
| HS6† |
HLA-class II loci.
MHC typing unavailable.
Incidence of somatically mutated VHtranscripts in lymphocyte cocultures of healthy subjects with disparate MHC loci.
Each data point represents the average of duplicate cultures. For each point, an average of 724 VH transcripts were analyzed (range 476-885, median 731). Accumulation of somatic mutation was assessed as in Figure 2.
Incidence of somatically mutated VHtranscripts in lymphocyte cocultures of healthy subjects with disparate MHC loci.
Each data point represents the average of duplicate cultures. For each point, an average of 724 VH transcripts were analyzed (range 476-885, median 731). Accumulation of somatic mutation was assessed as in Figure 2.
In 3 of 6 experiments, T cells from BMT recipients were able to support mutation in healthy-subject B cells, indicating that T lymphocytes from some BMT recipients can give adequate T-cell help (Figure 2). This finding is consistent with previous reports that posttransplantation CD4 T cells appear qualitatively normal.17-20
To estimate the accumulation of somatic mutation more accurately, we analyzed nucleotide sequences among transcripts selected from 2 B-cell cultures (Figure 4). In 1 culture, BMT-recipient B cells (BMT5) had been cocultured with autologous T cells (Table 3, exp 4). In the other culture, healthy-subject B cells (HS4) had been cultured with BMT-recipient T cells (BMT5) (Table 3, exp 4). These cultures are also indicated by filled circles in Figure 2. The sequences from the first set have an average of 0.2% mutation (median 0.17%). In contrast, the sequences from the second set have on average 3.1% mutation (median 1.7%), more than 10-fold higher. The different percentages of mutated VH transcripts seen in Figure 2 reflect both quantitative differences in the number of mutations among the VHtranscripts and differences in the incidence of transcripts with any mutation. The correlation between mutation detected by hybridization and that detected by nucleotide sequence analysis has been reported.14 15
DNA sequence analysis of V3-23 transcripts from T-cell–activated B-cell cultures.
B-cell cultures from BMT-recipient B cells supported by autologous activated T cells (sequences 1-1-1-5) and healthy-subject B cells supported by BMT-recipient T cells (sequences 4-1-4-7). Repeated isolation of the same cDNA clone is indicated by parentheses. Sequences 1-1-1-5 are taken from BMT5; sequences 4-1-4-7 are taken from HS4 (indicated by the filled circles in Figure 2).
DNA sequence analysis of V3-23 transcripts from T-cell–activated B-cell cultures.
B-cell cultures from BMT-recipient B cells supported by autologous activated T cells (sequences 1-1-1-5) and healthy-subject B cells supported by BMT-recipient T cells (sequences 4-1-4-7). Repeated isolation of the same cDNA clone is indicated by parentheses. Sequences 1-1-1-5 are taken from BMT5; sequences 4-1-4-7 are taken from HS4 (indicated by the filled circles in Figure 2).
Incidence of somatically mutated VHtranscripts in lymphocyte cocultures is dependent primarily on the B-cell donor
| Experiment . | Source of B cells . | Source of T cells . | Number of transcripts . | Mutated transcripts3-150 (%) . | Nucleotide substitutes3-151 (%) . | |
|---|---|---|---|---|---|---|
| VH3 . | V3-23 . | |||||
| Exp 1 | BMT1 | BMT1 | nd3 | 161 | 12 | |
| BMT1 | HS1 | nd | 100 | 8 | ||
| HS1 | BMT1 | nd | 138 | 13 | ||
| HS1 | HS1 | nd | 155 | 57 | ||
| Exp 2 | BMT2 | HS1 | 344 | 151 | 8 | |
| Exp 3 | BMT3 | BMT3 | nd | 53 | 15 | |
| BMT3 | HS3 | 374 | 99 | 1 | ||
| BMT4 | HS3 | >354 | 54 | 86 | ||
| HS3 | BMT3 | 712 | 37 | 86 | ||
| HS3 | HS3 | >375 | 11 | 65 | ||
| BMT3 | — | 726 | 206 | 1 | ||
| BMT4 | — | 482 | 59 | 1 | ||
| HS3 | — | 381 | 20 | 10 | ||
| Exp 4 | BMT6 | BMT6 | 461 | 74 | 1 | |
| BMT5 | BMT5 | 922 | 254 | 13 | 0.2 | |
| BMT6 | HS4 | 412 | 109 | 10 | ||
| BMT5 | HS4 | >742 | 98 | 1 | ||
| HS4 | BMT5 | 620 | 273 | 64 | 3.1 | |
| HS4 | HS4 | 491 | 74 | 60 | ||
| BMT6 | — | 570 | 155 | 11 | ||
| BMT5 | — | 789 | 163 | 12 | ||
| HS4 | — | 86 | 12 | 0 | ||
| Exp 5 | BMT7 | BMT7 | 647 | 152 | 7 | |
| BMT7 | HS5 | 751 | 212 | 6 | ||
| HS5 | BMT7 | 601 | 171 | 14 | ||
| HS5 | HS5 | 754 | 137 | 19 | ||
| BMT7 | — | 745 | 139 | 11 | ||
| HS5 | — | 820 | 168 | 7 | ||
| Exp 6 | BMT8 | BMT8 | 353 | 90 | 28 | |
| BMT8 | HS6 | 664 | 87 | 21 | ||
| HS6 | BMT8 | 681 | 90 | 88 | ||
| HS6 | HS6 | 736 | 164 | 75 | ||
| BMT8 | — | 784 | 130 | 18 | ||
| HS6 | — | 683 | 153 | 19 | ||
| Exp 7 | BMT9 | BMT9 | 345 | 58 | 33 | |
| BMT9 | HS4 | 331 | 70 | 23 | ||
| HS4 | BMT9 | 288 | 142 | 20 | ||
| HS4 | HS4 | >1320 | 194 | 82 | ||
| BMT9 | — | 684 | 98 | 7 | ||
| HS4 | — | 697 | 176 | 33 | ||
| Experiment . | Source of B cells . | Source of T cells . | Number of transcripts . | Mutated transcripts3-150 (%) . | Nucleotide substitutes3-151 (%) . | |
|---|---|---|---|---|---|---|
| VH3 . | V3-23 . | |||||
| Exp 1 | BMT1 | BMT1 | nd3 | 161 | 12 | |
| BMT1 | HS1 | nd | 100 | 8 | ||
| HS1 | BMT1 | nd | 138 | 13 | ||
| HS1 | HS1 | nd | 155 | 57 | ||
| Exp 2 | BMT2 | HS1 | 344 | 151 | 8 | |
| Exp 3 | BMT3 | BMT3 | nd | 53 | 15 | |
| BMT3 | HS3 | 374 | 99 | 1 | ||
| BMT4 | HS3 | >354 | 54 | 86 | ||
| HS3 | BMT3 | 712 | 37 | 86 | ||
| HS3 | HS3 | >375 | 11 | 65 | ||
| BMT3 | — | 726 | 206 | 1 | ||
| BMT4 | — | 482 | 59 | 1 | ||
| HS3 | — | 381 | 20 | 10 | ||
| Exp 4 | BMT6 | BMT6 | 461 | 74 | 1 | |
| BMT5 | BMT5 | 922 | 254 | 13 | 0.2 | |
| BMT6 | HS4 | 412 | 109 | 10 | ||
| BMT5 | HS4 | >742 | 98 | 1 | ||
| HS4 | BMT5 | 620 | 273 | 64 | 3.1 | |
| HS4 | HS4 | 491 | 74 | 60 | ||
| BMT6 | — | 570 | 155 | 11 | ||
| BMT5 | — | 789 | 163 | 12 | ||
| HS4 | — | 86 | 12 | 0 | ||
| Exp 5 | BMT7 | BMT7 | 647 | 152 | 7 | |
| BMT7 | HS5 | 751 | 212 | 6 | ||
| HS5 | BMT7 | 601 | 171 | 14 | ||
| HS5 | HS5 | 754 | 137 | 19 | ||
| BMT7 | — | 745 | 139 | 11 | ||
| HS5 | — | 820 | 168 | 7 | ||
| Exp 6 | BMT8 | BMT8 | 353 | 90 | 28 | |
| BMT8 | HS6 | 664 | 87 | 21 | ||
| HS6 | BMT8 | 681 | 90 | 88 | ||
| HS6 | HS6 | 736 | 164 | 75 | ||
| BMT8 | — | 784 | 130 | 18 | ||
| HS6 | — | 683 | 153 | 19 | ||
| Exp 7 | BMT9 | BMT9 | 345 | 58 | 33 | |
| BMT9 | HS4 | 331 | 70 | 23 | ||
| HS4 | BMT9 | 288 | 142 | 20 | ||
| HS4 | HS4 | >1320 | 194 | 82 | ||
| BMT9 | — | 684 | 98 | 7 | ||
| HS4 | — | 697 | 176 | 33 | ||
Nd indicates not determined; BMT, bone marrow transplant; HS, healthy subject. Dash indicates that no T cells were added but that B-cell cultures were supported by L cells supplemented with CD40L (1 μg/mL).
Percentage of V3-23 transcripts with mutation detectable by hybridization.
Incidence (%) of nucleotide substitution per base pair (bp) based on 290 bp/transcript of sequence shown in Figure 4.
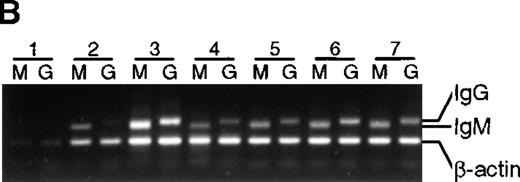
To determine if the lack of somatic mutation in BMT recipients correlated with the inability to differentiate in culture, IgG messenger RNA (mRNA) production (an indicator of differentiation21) was estimated with the use of a semiquantitative PCR (Figure 5). Except for lane 1 in Figure 5A and 5B, in which poor cell growth was observed, an IgG PCR product was obtained from all healthy subjects as well as all BMT recipients, suggesting that in these cultures isotype switching has occurred. As expected, no IgG PCR product was obtained from preculture IgD+ cells (data not shown). In addition, secreted IgG was detected by enzyme-linked immunosorbent assay in culture supernatants from T-cell–supported cultures irrespective of somatic mutation (data not shown).
Presence of IgM and IgG mRNA in 14-day B-cell cultures of BMT recipients and healthy subjects.
cDNA from B-cell cultures from 9 recipients and 7 healthy subjects was amplified with the use of 5′ primers specific for VH3 and 3′ primers specific for either IgM or IgG. Results are representative of analysis of triplicate cultures. As an internal control, β-actin mRNA was used. Lanes M were amplified with the use of Cμ primers, lanes G with the use of Cγ primers. (A) BMT recipients. (B) Healthy subjects.
Presence of IgM and IgG mRNA in 14-day B-cell cultures of BMT recipients and healthy subjects.
cDNA from B-cell cultures from 9 recipients and 7 healthy subjects was amplified with the use of 5′ primers specific for VH3 and 3′ primers specific for either IgM or IgG. Results are representative of analysis of triplicate cultures. As an internal control, β-actin mRNA was used. Lanes M were amplified with the use of Cμ primers, lanes G with the use of Cγ primers. (A) BMT recipients. (B) Healthy subjects.
Discussion
We have previously shown that BMT recipients fail to acquire somatic mutations in rearranged VH genes in PBL6-8 and have low memory B-cell counts.22 In the current study, we wished to determine the intrinsic capacity of transplant recipient naive B cells to acquire mutations. Several models have been described that mimic germinal center reactions in which somatic mutation takes place.14,23-26 We used an in vitro system in which activated CD4+ T cells drive B-cell differentiation over a 14-day culture period. In this system, healthy-subject B cells accumulate large numbers of V-segment mutations, presumably as a consequence of activation of the somatic mutator mechanism.14 In contrast, we found that B cells obtained from BMT recipients 1 year posttransplantation failed to accumulate mutations. This deficit could not be overcome by coculture with healthy-subject T cells, although T cells from certain patients were able to drive the accumulation of mutations in healthy-subject B cells.
Among the patients studied here, the only striking difference was in the capacity of patient T cells to support mutation in healthy-subject B cells. All 3 of these patients (BMT3, 5, 8) had normal levels of CD4+ T cells and supranormal levels of B cells, suggesting that recovery of the immune system was more robust in these patients.
Unmutated, naive B cells have the phenotype CD19+IgM+ IgD+. This phenotype is exhibited among healthy subjects by approximately 80% of B cells, and among BMT recipients by more than 95% of B cells.12 Thus, by both cell-surface phenotype and the extent of somatic mutation, the B-cell repertoire post-BMT resembles the pre-immune component of the B-cell repertoire of a healthy adult. Simply because of population dynamics, it would seem logical that the B-cell repertoire post-BMT would be primarily naive. However, our finding that these B cells cannot be driven to acquire somatic mutations suggests that additional processes are at work. The failure of the cells to accumulate somatic mutations did not seem to parallel a failure to switch class. Taken together, the data are most easily explained by postulating that there is a deficit in the capacity of BMT-recipient B cells to respond to signals to activate the somatic mutator mechanism.
Because the marrow donors are themselves healthy subjects whose B cells are capable of acquiring mutations, this posttransplantation deficit must be developmentally determined. All patients received cyclosporine for the first 180 days after transplantation, and this immunosuppressive treatment might be expected to delay recovery of immunocompetency. However, ongoing immunosuppressive therapy cannot explain the results because cyclosporine was terminated at least 180 days prior to our studies for all but 1 patient (BMT1), who was receiving immunosuppressive therapy at the time of this study.
Another possibility is that during the pretransplantation conditioning regimen, a critical cellular function is disrupted and is not restored by marrow transplant. One such function, for example, could be the delivery of survival signals. In normal B-cell differentiation, newly formed transitional B cells are recruited into a long-lived pre-immune B-cell pool, in a process thought to be dependent on survival signals delivered in secondary lymphoid organs.27,28 Because the pretransplantation conditioning regimen may disrupt lymphoid tissue architecture (particularly follicular dendritic cells),29the survival signals may be missing in the transplant recipients. As a result, BMT recipients may fail to select a long-lived naive B-cell compartment. The implication of this is that the B-cell compartment in BMT recipients is composed primarily of transitional B cells, which are short lived and have newly emerged from the bone marrow.30We postulate that this transitional B-cell population can participate in primary immune responses and can be driven to differentiate into plasma cells, but does not participate in a germinal center reaction and does not acquire mutations.
Acknowledgments
Drs R. J. Armitage, M. K. Spriggs, and W. C. Fanslow for kindly providing recombinant CD40 ligand and Dr E. Vitetta for antibody 64.1.
Supported in part by National Institutes of Health grants AI41714, AR39918, and CA68496.
Reprints:Eric C. B. Milner, 1201 Ninth Ave, Seattle, WA 98101; e-mail: emilner@vmresearch.org.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal