Appropriate monitoring of patients after allogeneic stem cell transplantation (SCT) for chronic myeloid leukemia (CML) is crucial, because the response to donor lymphocyte infusions is superior in the case of cytogenetic or molecular relapse compared with hematological relapse.1 Chomel et al recently reported that fluorescence in situ hybridization (FISH) performed on interphase nuclei (IP-FISH) detects between 2% and 11% BCR-ABL–positive cells in the blood or bone marrow of CML patients in molecular remission (RT-PCR–negative) after allogeneic bone marrow transplantation.2 The authors concluded that the BCR-ABL fusion gene was transcriptionally silent in the FISH-positive cells.
We analyzed our database for specimens that fulfilled the following criteria: (1) classical allogeneic SCT (ie, conventional conditioning) for CML; (2) RT-PCR for BCR-ABL negative; (3) IP-FISH performed with LSI bcr/abl ES kit (Vysis, Stuttgart, Germany). This probe has a low false-positive rate since it detects an additional red signal on the derivate chromosome 9. Fourteen patients after sex-mismatched transplantations and 17 patients after sex-matched transplantations for Ph-positive CML were identified (Table1). RT-PCR was performed as described.3 The false-positive rate of the PI-FISH system was determined in patients with Ph-negative disorders and found to be in the range of 0.1% (Table 2). In the patients after allografting, a mean of 360 (range, 135-500) nuclei per specimen were analyzed. Chimerism was determined by sex chromosome specific probes (SO CEP X /SG CEP Y, Vysis), with a mean of 496 (range, 100-1000) nuclei per sample analyzed. If mixed chimerism was detected, cohybridization with BCR-ABL probes (LSI bcr/abl dual color translocation probe, Vysis) and sex chromosome probes was carried out; the respective signals, although of identical colors, could be distinguished based on their different sizes (Figure).
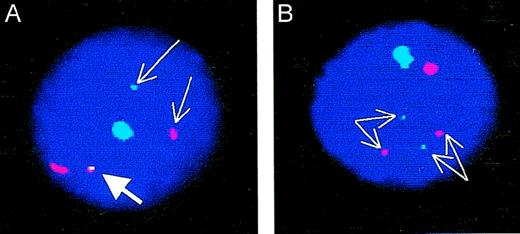
Cohybridization of sex-chromosome specific probes and BCR-ABL fusion probes. (A) BCR-ABL–positive cell male cell. Thin arrows point to the ABL (red) and BCR (green) signals; the thick arrow points to the BCR-ABL fusion. The large signals identify the X (red) and Y (green) chromosomes. (B) BCR-ABL–negative male cell. Thin arrows point to the 2 separate ABL (red) and BCR (green) signals. The large signals identify the X (red) and Y (green) chromosomes. Note that the “conventional” dual-color color FISH probe (without additional red signal on the derivative chromosome 9) was used in these experiments.
Cohybridization of sex-chromosome specific probes and BCR-ABL fusion probes. (A) BCR-ABL–positive cell male cell. Thin arrows point to the ABL (red) and BCR (green) signals; the thick arrow points to the BCR-ABL fusion. The large signals identify the X (red) and Y (green) chromosomes. (B) BCR-ABL–negative male cell. Thin arrows point to the 2 separate ABL (red) and BCR (green) signals. The large signals identify the X (red) and Y (green) chromosomes. Note that the “conventional” dual-color color FISH probe (without additional red signal on the derivative chromosome 9) was used in these experiments.
Patient and donor characteristics in correlation to IP-FISH and RT-PCR for BCR-ABL
| No. of patients . | Time from transplant (y) . | Sex mismatch . | No. of specimens analyzed . | Chimerism, % donor . | IP-FISH positive . | RT-PCR positive . |
|---|---|---|---|---|---|---|
| 11 | 3.5 (0.1-8) | Yes | 24 | 100 | 0/24 | 0 |
| 3 | 3.4 (1.5-12) | Yes | 6 | 90-99.8 | 0/6 | 0 |
| 17 | 2.1 (0.3-10.9) | No | 26 | NA | 1/26 | 0 |
| No. of patients . | Time from transplant (y) . | Sex mismatch . | No. of specimens analyzed . | Chimerism, % donor . | IP-FISH positive . | RT-PCR positive . |
|---|---|---|---|---|---|---|
| 11 | 3.5 (0.1-8) | Yes | 24 | 100 | 0/24 | 0 |
| 3 | 3.4 (1.5-12) | Yes | 6 | 90-99.8 | 0/6 | 0 |
| 17 | 2.1 (0.3-10.9) | No | 26 | NA | 1/26 | 0 |
NA = not applicable.
Assessment of false-positive rates in patients with Ph-negative hematopoietic disorders
| Nuclei analyzed . | Specimens tested . | Positive nuclei (mean) . | % positive . |
|---|---|---|---|
| 100 | 47 | 0.08 ± 0.28 | 0.08 |
| 200 | 37 | 0.19 ± 0.62 | 0.095 |
| 300 | 22 | 0.5 ± 1.01 | 0.17 |
| 400 | 15 | 0.4 ± 1.06 | 0.1 |
| 500 | 15 | 0.53 ± 1.36 | 0.11 |
| Nuclei analyzed . | Specimens tested . | Positive nuclei (mean) . | % positive . |
|---|---|---|---|
| 100 | 47 | 0.08 ± 0.28 | 0.08 |
| 200 | 37 | 0.19 ± 0.62 | 0.095 |
| 300 | 22 | 0.5 ± 1.01 | 0.17 |
| 400 | 15 | 0.4 ± 1.06 | 0.1 |
| 500 | 15 | 0.53 ± 1.36 | 0.11 |
Eleven out of 14 patients with sex-mismatched donors were found to be complete chimeras at a median of 3.5 years after transplantation. In a total of 24 samples, no BCR-ABL–positive interphases were detectable in these patients (Table 1). Three out of 14 patients were mixed chimeras (90%-99.8% donor, total of 6 samples). Cohybridization revealed that the recipient cells were BCR-ABL–negative and thus represented residual normal hematopoiesis. Seventeen patients after sex-matched allografting were followed by FISH for BCR-ABL only, and a total of 26 samples were analyzed at a mean of 2.1 years after tranplantation. Only one sample was positive (6 of 500 cells).
In our series only a single specimen scored positive, with a value just above our cutoff (mean + 3 SD = 4.6 for 500 analyzed nuclei). Moreover, complete donor chimerism was present in approximately 80% of patients after sex-mismatched transplantation, which rules out the presence of any substantial number of Ph-positive cells. In the remaining patients recipient cells were present but they were exclusively BCR-ABL–negative and thus represented residual Ph-negative hematopoiesis that survived the conditioning.
It is not easy to explain the discordance between the findings of Chomel et al and our results. Different FISH systems were used in the 2 studies (Chomel et al used one from Oncor, and we used one from Vysis), but it is not readily obvious why this should result in such a great discrepancy. False-positive rates appear to be similar in both systems and are in the range of 0.1%. But Chomel et al studied only 1000 cells from Ph-negative patients, which might have led to an underestimate of the false-positive rate in their system. Over 30 000 nuclei were analyzed to establish the false-positive rate in our laboratory. In order to resolve this issue it would be crucial to directly compare the performance of both FISH systems in the same specimens. Although our results refer exclusively to patients after allogeneic SCT, it should be noted that Chomel et al also found between 1% and 12% Ph-positive cells in 11 complete cytogenetic responders (CCR) to therapy with IFN-alpha. Although this is certainly less surprising than in allografted patients, it is still in contrast to a previous report that employed the same FISH probe and found a normal pattern in 4 out of 6 specimens from CCR.4
In conclusion, in our hands RT-PCR reliably predicts negative IP-FISH. Conversely, reemergence of IP-FISH positive nuclei likely indicates relapse and warrants close follow-up.
Persistence of BCR-ABL genomic rearrangement in chronic myeloid leukemia patients in complete and sustained cytogenetic remission after interferon- therapy or allogeneic bone marrow transplantation
Poitiers, France
CHU de Poitiers
Poitiers, France
Poitiers, France
Poitiers, France
CHU de Poitiers
Poitiers, France
Poitiers, France
Poitiers, France
CHU de Poitiers
Poitiers, France
Poitiers, France
Poitiers, France
CHU de Poitiers
Poitiers, France
Poitiers, France
Poitiers, France
CHU de Poitiers
Poitiers, France
Poitiers, France
We appreciate the letters by M. Deininger et al and by A. Chase et al regarding our report.1-1 Both letters mainly address the important question of the residual disease after bone marrow transplantation (BMT) for chronic myeloid leukemia (CML) chronic-phase patients. M. Deininger et al reported their own series of patients undergoing transplantation. The residual disease was assessed on 14 patients after sex-mismatched transplantation and on 17 patients after sex-matched transplantation. For that purpose, residual disease was assessed using sex-chromosome–specific probes from Vysis. With these probes, most of their patients were found to be FISH negative with complete donor chimerism in 80% of patients. Three out of 14 patients were mixed chimeras, but cohybridization with BCR-ABL probes was negative. For these 3 cases it is not easy to explain why residual Ph-negative hematopoiesis survived the conditioning regimen while leukemic cells, which are likely to be more resistant, were totally eliminated. In addition we feel that it is not so easy to score nuclei after cohybridization and to distinguish signals based on their different sizes. Different delays from the times of BMT and of FISH analyses could explain the discrepancies between the studies. Hence the FISH analysis was performed at a median of 3.5 years after transplantation in the study by Deininger et al whereas it was done at a median of 2 years afterward in our study. Thus it is possible that total elimination of the leukemic cells would take a more prolonged period of time. But in our study patient 13 was persistently positive at 56 months after transplantation (with BCR-ABL probes, Y probes, and Y amplification by RT-PCR). The other 2 studies used Vysis probes, whereas we used the Oncor system. But we agree with Deininger that these 2 systems should not produce substantial differences. Chase et al have studied the residual disease of 11 consecutive CML patients who underwent BMT with a sex-mismatched donor. The use of X or Y FISH probes showed that 8 patients were negative with no host cells, and 3 were also negative by FISH with the use of BCR-ABL probes from Vysis. But the absence of host cells does not demonstrate that no more leukemic cells are present in the bone marrow, although we agree that residual leukemia blood cells would have the patient's sex chromosome complement. Chase et al referred also to their previous study in which they suggested that, at the DNA level, patients were cured after BMT.1-2 For that purpose they used patients' specific PCR primers for genomic DNA amplification, that is, DNA-PCR. But they also found discrepancies with 3 patients who were DNA-PCR–positive and BCR-ABL- mRNA–negative by RT-PCR. It is obvious, however, that their results challenge our data.
We agree with Deininger et al and Chase et al that BCR-ABL FISH is technically demanding. The analysis of each nucleus requires a sufficient duration of observation under the microscope in order to detect the fusion signal indicating a cell possessing the translocation. We also paid great attention to the preparation of the slides: only high-quality slides were scored. To avoid false positive signals, we performed 2 controls: (1) dilutions of Ph+ cells in Ph− cells analyzed by FISH and Southern-blot that exhibited a very good concordance between the 3 methods even in the low percentage of Ph+ cells and (2) the study of 1000 nuclei from patients with hematologic disorders other than CML showing a false positive rate of 0.1%. This study of 1000 nuclei is sufficient to determine the false positive rate because we believe that an accurate and sufficient duration of observation of each nucleus is more important than the observation of a large number of nuclei. Indeed, the direct observation of fusion signals is sometimes difficult and can be underestimated. Moreover, to avoid false negative results, some nuclei were computerized in order to validate the detection of the fusion signal. The slides were also scored without knowledge of the RT-PCR results, and this was done for the IFN-treated patients, as well as for the BMT- treated patients.
An increased risk of leukemia relapse has been associated with the detection of BCR-ABL mRNA in bone marrow transplant patients, particularly if such detection has been observed repeatedly more than 100 days after transplantation.1-3 Moreover, an increased relapse rate was noticed if BCR-ABL transcripts were detected between 6 and 12 months after transplantation.1-4 Conversely, a period of sustained PCR negativity is generally associated with a high probability of cure or prolonged leukemia-free survival. Hence the risk of relapse must be evaluated at the RNA levels by quantitative RT-PCR rather than at the DNA level with FISH.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal