Abstract
We report here the characterization of an adapter protein identified in a yeast 2-hybrid screen with the use of Bcr-Abl as the bait. Grb4 bound to Bcr-Abl in a variety of systems, both in vitro and in vivo, and is an excellent substrate of the Bcr-Abl tyrosine kinase. The association of Grb4 and Bcr-Abl in intact cells was mediated by an src homology (SH)2–mediated phosphotyrosine-dependent interaction as well as an SH3-mediated phosphotyrosine-independent interaction. Grb4 has 68% homology to the adapter protein Nck and has similar but distinct binding specificities in K562 lysates. Subcellular localization studies indicate that Grb4 localizes to both the nucleus and the cytoplasm. Coexpression of kinase-active Bcr-Abl with Grb4 resulted in the translocation of Grb4 from the cytoplasm and the nucleus to the cytoskeleton to colocalize with Bcr-Abl. In addition, expression of Grb4 with kinase-active Bcr-Abl resulted in a redistribution of actin-associated Bcr-Abl. Finally, coexpression of Grb4 and oncogenic v-Abl strongly inhibited v-Abl–induced AP-1 activation. Together, these data indicate that Grb4 in conjunction with Bcr-Abl may be capable of modulating the cytoskeletal structure and negatively interfering with the signaling of oncogenic Abl kinases. Grb4 may therefore play a role in the molecular pathogenesis of chronic myelogenous leukemia. (Blood. 2000;96:618-624)
Adaptor proteins are an emerging class of proteins that contain functional src homology (SH) domains, the SH2 and SH3 domains, and lack intrinsic enzymatic function.1They play a critical role in the formation of multimeric protein complexes and connect signaling molecules to upstream and downstream signaling events.1 Grb2, one of the first adapter proteins studied, exists in a complex with a second protein, Son of Sevenless (SOS), which catalyzes Ras GTP/GDP exchange.2 Grb2 is known to bind to Bcr-Abl, the chimeric oncogene of chronic myeloid leukemia, at an autophosphorylation site in Bcr and has been suggested as being involved in Bcr-Abl–mediated oncogenesis.3,4 Bcr-Abl was also shown to bind to other adapter molecules such as Grb10, Shc, Crk, and CrkL that were implicated in Bcr-Abl–mediated transformation.5-7 Screening for adapter molecules interacting with Bcr-Abl thus seems an efficient method of identifying molecules important in Bcr-Abl–induced transformation.
In order to identify adapter molecules that interact with Bcr-Abl in a phosphotyrosine-dependent manner, we established a modified version of the yeast 2-hybrid screen using the DNA binding domain of the LexA transcription factor fused to the bait.8 The LexA protein has the inherent ability to dimerize and so allowed the dimerizaton of Bcr-Abl and, consequently, its autophosphorylation. Using this system, we identified several known and novel interactions of proteins with Bcr-Abl.5
We report here the identification of an adapter protein, Grb4, that shares 68% homology with Nck, an adapter protein with an SH3-SH3-SH3-SH2 configuration. Nck links receptor tyrosine kinases, such as the epidermal growth factor (EGF) and platelet-derived growth factor (PDGF) receptors, to downstream signaling pathways9 and has been implicated in SOS-activated Ras signaling, the p21 Cdc42/Rac–activated kinase cascade, Rho, and human Wiskott-Aldrich syndrome protein–mediated actin cytoskeleton changes.10-12 During the preparation of this manuscript, 3 groups independently reported the cloning and identification of Grb4 (also known as Nck-β/Nck-2). The first group screened a human complementary DNA (cDNA) library with a partial mouse Nck cDNA13; the second identified Grb4 as a protein interacting with the LIM-only protein PINCH14; and the third performed polymerase chain reaction with the use of anchor primers and Grb4-specific primers on a human-brain–ready cDNA library.15 We identified Grb4 as 2 C-terminal fragments of the protein that interacted with Bcr-Abl in a yeast 2-hybrid screen. Bcr-Abl and Grb4 interacted with each other both in vitro and in vivo, and the interaction involved both a phosphotyrosine-dependent and a phosphotyrosine-independent interaction. The nuclear localization of Grb4, its translocation from the nucleus to the cytoskeleton to colocalize with wild-type (WT) Bcr-Abl, and its inhibition of AP-1 induction by oncogenic Abl kinases suggest a role for this adapter protein in the negative modulation of pro-mitogenic nuclear signals elicited by oncogenic Abl tyrosine kinases.
Materials and methods
Yeast 2-hybrid system
Cell culture and DNA transfection
We maintained 293 and Cos7 cells in Dulbecco modified Eagle medium (DMEM) (Gibco-BRL, Karlsruhe, Germany) containing 10% fetal calf serum (FCS). K562, Jurkat, Mo7e, and Mo7e/p210 cells were maintained in RPMI 1640 (Gibco-BRL) with 10% FCS (Seromed, Berlin, Germany). Ba/F3 cells were grown in media supplemented with 1.5 ng/mL of murine recombinant interleukin-3 (R & D Systems DPC Bierman, Wiesbaden, Germany). Mo7e cells were grown in media supplemented with granulocyte-macrophage colony–stimulating factor (R & D Systems DPC Bierman). Transfections were performed with the DOTAP transfection reagent (Boehringer, Mannheim, Germany).
Glutathione-S-transferase fusion proteins and binding assays
The Grb4 cDNA was cloned in frame into the vector pGE × 2TK to make glutathione-S-transferase (GST) fusion proteins.16Different Bcr-Abl proteins (aa1-63, aa1-242, aa1-509 representing varying lengths of Bcr) were in vitro translated in the presence of S35-labeled methionine with the use of the transcription and translation (TNT) system (Promega, Madison, WI) as described previously.5 Then, 5 μg of GST-fusion proteins were added and incubated for 1 hour at 4°C. Protein complexes were collected on glutathione agarose beads (Pharmacia, Freiburg, Germany), washed thoroughly with lysis buffer, and subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). In vitro translated proteins were visualized by autoradiography.
Immunoprecipitation and immunoblotting
Immunoprecipitations and immunoblotting were done as previously described.17 Briefly, 1 × 107 cells were solubilized in lysis buffer containing 10 mmol/L Tris-HCl (pH 7.4), 5 mmol/L EDTA, 130 mmol/L NaCl, 1% Triton, 1 mmol/L phenylmethylsulfonyl fluoride, 1 mmol/L Na3VO4, and 10 mg/mL each of phenantroline, aprotinin, leupeptin, and pepstatin. After clarification by centrifugation, antibody-protein complexes were brought down with 30 μL of protein A-Sepharose (Pharmacia). Xpress-tagged Grb4 was precipitated with an anti-Xpress antibody (Invitrogen, Groningen, The Netherlands), enhanced yellow flourescent protein (EYFP)–tagged Grb4 and EYFP-tagged Nck were detected with an anti–enhanced green fluorescent protein (EGFP) antibody (Clontech), and endogenous Grb4 was precipitated and detected with a polyclonal rabbit serum raised against a GST-Grb4 fusion protein. Bcr-Abl and Abl were detected with antibody 8E9 (Pharmingen, Hamburg, Germany), and tyrosine phosphorylation was detected with the monoclonal antiphosphotyrosine antibody 4G10 (Upstate Biotech, Lake Placid, NY), as previously described.17 18 Bands were visualized with the use of the ECL system (Amersham, Braunschweig, Germany).
Far Western analysis
Far Western analysis was performed according to the method previously described.19 Radiolabeled Grb4 was used as a probe on blotted precipitates of K562 lysates, and dried membranes were analyzed by autoradiography.
c-Jun, Elk1, CREB, and AP-1 activation reporter assays
PathDetect In Vivo Signal Transduction Pathway Trans-Reporting Systems (Stratagene, Heidelberg, Germany) were used according to the manufacturer's directions. Then, 293 cells were transfected with a fusion transcription factor encoding the DNA activation domain of Jun, Elk, or CREB and the DNA binding domain of GAL4 and the luciferase GAL4 reporter construct together with 0.5 μg of Grb4 and 100 ng of β-galactosidase cDNA to normalize for transfection efficiency. Cells were cultured for another 3 days in DMEM 10% FCS and then lysed in luciferase lysis buffer (Stratagene). AP-1 activation was examined with the use of a luciferase reporter gene driven by a basic promoter element (TATA box) joined to 7 tandem repeats of the AP-1 binding element. Then, 1 μg of the AP-1/luciferase expression construct and 1 of 3 μg of Grb4 cDNA were co-transfected with 1 μg of pCDNA3.1/v-Abl or v-Abl kinase defective together with varying amounts of empty-vector DNA so that in each experiment equal amounts of DNA were used. Cells were cultured for 3 days in DMEM 10% FCS and then lysed in luciferase lysis buffer. Luciferase activity was measured by means of a luciferase assay kit (Promega, Mannheim, Germany) and a luminometer (Berthold, Pforzheim, Germany).
Intracellular localization of Grb4-EyFP and Bcr-Abl-ECFP
The EGFP cDNA20 was modified with the use of the QuikChange site-directed mutagenesis kit (Stratagene) to change its emission to make 2 different forms of the protein, EYFP (yellow) and ECFP (cyan), that emit at different wavelengths.21 The fusion proteins were expressed in Cos7 cells cultured in DMEM 10% FCS and plated on gelatin-coated Permanox chamber slides (Nunc, Wiesbaden-Biebrich, Germany). At 72 hours following transfection, cells were fixed with 3.7% paraformaldehyde for 15 minutes. Cells were then washed twice with phosphate-buffered saline, overlayed with mounting medium (Molecular Probes, Leiden, The Netherlands), covered with a cover slide, and visualized by means of fluorescence microscopy (Zeiss Axioskop, Oberkochen, Germany) and an imaging system from TILL Photonics (Munich, Germany).
Results
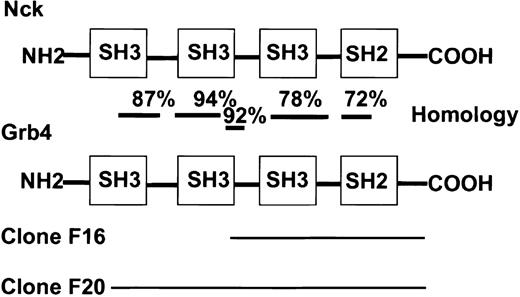
Cloning of 2 C-terminal fragments of the adapter protein Grb4
A cDNA encoding Bcr-AblΔSal (which deletes the actin-binding domain to avoid selection of actin-encoding clones) was fused to the DNA-binding domain of the transcription factor LexA in the bait vector.8 This allowed dimerization and subsequent autophosphorylation of Bcr-Abl in the yeast 2-hybrid system.5 A K562 cDNA library (Clontech) was screened with Bcr-Abl in this modified yeast 2-hybrid system. Among adapter proteins already known to bind to Bcr-Abl, such as Grb2, Crk, or Grb10, the C-terminal end of a novel adapter protein, Grb4, was identified. A portion of the murine Grb4 sequence was already in GenBank (accession no. I13161) and was referred to as Grb4; therefore, the original nomenclature was adhered to.22 The cDNA lacked the very 5′ end of the gene and was referred to as Grb4/F16. Grb4/F16 (aa 154-380) possessed a portion of an SH3 domain, a middle SH3 domain, and a single SH2 domain at the C-terminal end (Figure1). Grb4/F16 interacted with Bcr-Abl in a phosphotyrosine-dependent manner in yeast; ie, Grb4/F16 interacted with WT Bcr-Abl but not with kinase defective (KD) Bcr-Abl (Table 1). A second cDNA clone, Grb4/F20 (aa 32-380), that possessed additional 5′ sequences was identified (Figure 1). Unlike Grb4/F16, Grb4/F20, by virtue of its N-terminal SH3 sequences, interacted with Bcr-Abl in a phosphotyrosine-independent manner in yeast (Table 1). The interactions of Grb4/F16 and Grb4/F20 with Bcr-Abl were specific as the control protein lamin showed no binding to either construct (Table 1).
Homology of Grb4 and Nck.
Two interacting clones of Grb4 were identified in the yeast screen: F16 (aa 154-380) and F20 (aa 32-380). Bars indicate the level of homology in different regions at the amino acid level.
Homology of Grb4 and Nck.
Two interacting clones of Grb4 were identified in the yeast screen: F16 (aa 154-380) and F20 (aa 32-380). Bars indicate the level of homology in different regions at the amino acid level.
Grb4 and Bcr-Abl interaction in yeast
| Constructs . | −TL* . | −THL† . |
|---|---|---|
| F16/Grb4 + WT Bcr-Abl | + | + |
| F16/Grb4 + KD Bcr-Abl | + | − |
| F20/Grb4 + WT Bcr-Abl | + | + |
| F20/Grb4 + KD Bcr-Abl | + | + |
| F16/Grb4 + lamin (control) | + | − |
| F20/Grb4 + lamin (control) | + | − |
| Constructs . | −TL* . | −THL† . |
|---|---|---|
| F16/Grb4 + WT Bcr-Abl | + | + |
| F16/Grb4 + KD Bcr-Abl | + | − |
| F20/Grb4 + WT Bcr-Abl | + | + |
| F20/Grb4 + KD Bcr-Abl | + | + |
| F16/Grb4 + lamin (control) | + | − |
| F20/Grb4 + lamin (control) | + | − |
Interaction of the 2 clones Grb4 F16 and F20 with WT Bcr-Abl, KD Bcr-Abl, or (as a control) lamin in the yeast 2-hybrid screen.
Growth under tryptophan and leucine selection and reflects the transfection efficiency.
Growth under additional histidine selection and reflects the specificity of interaction.
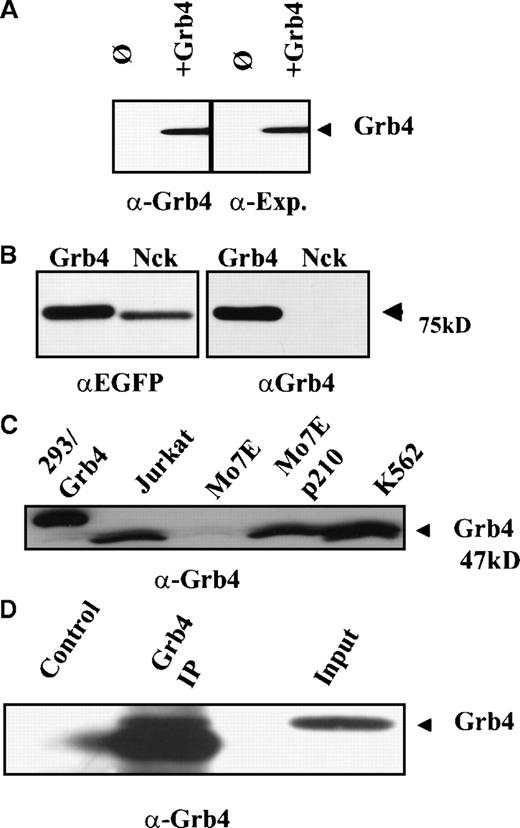
A polyclonal rabbit antibody raised to Grb4/F16 could specifically detect tagged Grb4 on Western analysis, and sensitivity was comparable to the Xpress antibody that recognized the tagged sequence (Figure 2A). This antibody showed minimal cross-reactivity with Nck (Figure 2B, right panel). Grb4 is expressed as a 47-kd protein in all cell lines tested (Figure 2C) as well as in Bcr-Abl–positive patient samples (data not shown). The double bands may represent different phosphorylated forms of Grb4, as it was reported by Chen et al that the slower migrating forms were no longer detectable following alkaline phosphatase treatment of the cells.13 The polyclonal serum could also immunoprecipitate Grb4 albeit with low efficiency (Figure2D).
Grb4 encodes for a 47-kd protein.
(A) We lysed 1 × 107 293 cells overexpressing Xpress-tagged Grb4 and performed an anti-Xpress immunoprecipitation. Bound proteins were resolved with the use of SDS-PAGE, and an anti-Grb4 Western blot was performed with the use of a polyclonal rabbit antiserum (left panel) or an anti-Xpress antibody (right panel). Blots were visualized by means of the ECL detection system. (B) N-terminal EYFP fusion constructs of Grb4 and Nck were transiently expressed in Cos7 cells. After 3 days, 1 × 107 cells were lysed and separated by SDS-PAGE and probed with an anti-EGFP antibody (Clontech) (left panel) and the anti-Grb4 serum (right panel). Bands were visualized by means of the ECL system. (C) Then, 1 × 107 293 (overexpressing Xpress-tagged Grb4), Jurkat, Mo7e, Mo7e expressing p210/Bcr-Abl, and K562 cells were lysed and resolved on SDS-PAGE, and Western blotting was performed with the Grb4 antibody. Native Grb4 migrated at 47 kd and is indicated by the arrow. Tagged Grb4 migrates slightly more slowly (lane 1). (D) Then, 2 × 107 293 cells overexpressing Grb4 were lysed, and an immunoprecipitation (Grb4 Ip) followed by immunoblotting was performed with the use of the rabbit polyclonal Grb4 antibody or a control rabbit antibody.
Grb4 encodes for a 47-kd protein.
(A) We lysed 1 × 107 293 cells overexpressing Xpress-tagged Grb4 and performed an anti-Xpress immunoprecipitation. Bound proteins were resolved with the use of SDS-PAGE, and an anti-Grb4 Western blot was performed with the use of a polyclonal rabbit antiserum (left panel) or an anti-Xpress antibody (right panel). Blots were visualized by means of the ECL detection system. (B) N-terminal EYFP fusion constructs of Grb4 and Nck were transiently expressed in Cos7 cells. After 3 days, 1 × 107 cells were lysed and separated by SDS-PAGE and probed with an anti-EGFP antibody (Clontech) (left panel) and the anti-Grb4 serum (right panel). Bands were visualized by means of the ECL system. (C) Then, 1 × 107 293 (overexpressing Xpress-tagged Grb4), Jurkat, Mo7e, Mo7e expressing p210/Bcr-Abl, and K562 cells were lysed and resolved on SDS-PAGE, and Western blotting was performed with the Grb4 antibody. Native Grb4 migrated at 47 kd and is indicated by the arrow. Tagged Grb4 migrates slightly more slowly (lane 1). (D) Then, 2 × 107 293 cells overexpressing Grb4 were lysed, and an immunoprecipitation (Grb4 Ip) followed by immunoblotting was performed with the use of the rabbit polyclonal Grb4 antibody or a control rabbit antibody.
Similar but distinct binding patterns of Nck and Grb4
The high homology between Grb4 and Nck prompted us to examine the pattern of proteins joined in a complex to each molecule. Far Western analysis was performed with the use of metabolically labeled GST-Grb4 on the bound fraction of K562 lysates precipitated by a panel of GST fusion proteins, including Grb4, Nck, Grb2, Crk, and CrkL (Figure 3). The quantity of each GST-fusion protein was determined by coomassie staining of the gel and found to be equal (data not shown). The binding patterns of Grb4 and Nck in K562 lysates were mostly similar although significant differences could be observed. Solid arrows indicate proteins bound strongly by Grb4 and either weakly or not at all by Gst-Nck. Open arrows indicate proteins that bind to both Grb4 and Nck. The solid circle indicates the migration of GST-Nck, which leads to a faster migration of a band also seen with Grb4. Significantly, Grb4 bound strongly to Bcr-Abl when compared with the binding by Nck or Grb2. Thus, despite their high overall similarity, there are differences in both the specificity and the strength with which Grb4 and Nck join in a complex to proteins in K562 lysates.
Far Western analysis using metabolically labeled S35-GST-Grb4.
We precipitated 5 mg of K562 RIPA protein extract at 4°C overnight with 10 μg of GST or equimolar amounts of various GST fusion proteins as indicated. Bound proteins were resolved on SDS-PAGE before being blotted, blocked, and renatured at room temperature. The membrane was probed with S35-GST-Grb4 and analyzed by autoradiography. Solid arrows indicate proteins bound strongly by GST-Grb4 and either weakly or not at all by Gst-Nck; open arrows indicate proteins that bind both to Gst-Grb4 and to Nck but show marginally stronger binding to Grb4. The solid circle indicates GST-Nck.
Far Western analysis using metabolically labeled S35-GST-Grb4.
We precipitated 5 mg of K562 RIPA protein extract at 4°C overnight with 10 μg of GST or equimolar amounts of various GST fusion proteins as indicated. Bound proteins were resolved on SDS-PAGE before being blotted, blocked, and renatured at room temperature. The membrane was probed with S35-GST-Grb4 and analyzed by autoradiography. Solid arrows indicate proteins bound strongly by GST-Grb4 and either weakly or not at all by Gst-Nck; open arrows indicate proteins that bind both to Gst-Grb4 and to Nck but show marginally stronger binding to Grb4. The solid circle indicates GST-Nck.
In vitro association of Grb4/F16 and Bcr-Abl
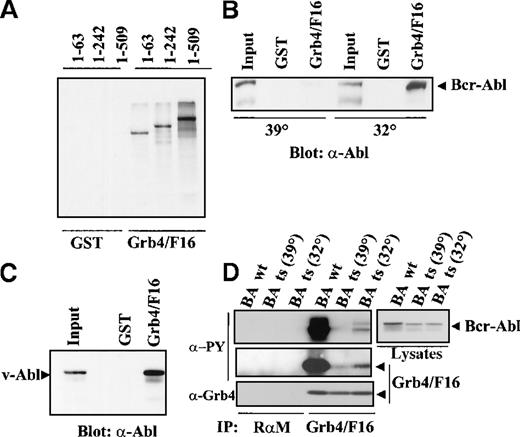
In vitro–translated S35-labeled Bcr-Abl was tested for interaction with Grb4/F16 (Figure 4A). Three different Bcr-Abl constructs were used with internal deletions of Bcr, namely, 1-63 Bcr-Abl, 1-242 Bcr-Abl, and 1-509 Bcr-Abl, the numbers representing different lengths of amino acids in Bcr.5 It was observed that Grb4/F16 bound to all mutants including 1-63 Bcr-Abl. This indicated that binding occurs at an autophosphorylation site within Abl as the first 63 amino acids of Bcr have no known major autophosphorylation site. This distinguishes Grb4 from Grb2, which binds to Tyr 177 in the Bcr part of Bcr-Abl and from other known SH2 adapter proteins, which all bind to phosphorylation sites in the Bcr part of Bcr-Abl.3 4
Complex formation of Grb/F16 and Bcr-Abl.
(A) Grb4/F16 and Bcr-Abl form a complex in vitro. Bcr-Abl mutants with internal deletions of Bcr resulting in varying lengths of Bcr as indicated were in vitro translated and S35 labeled. We allowed 50 μL of the translation mix to autophosphorylate in vitro and then incubated it with 5 μg purified GST or GST-Grb4/F16. Bound fractions were separated by SDS-PAGE, and Bcr-Abl mutants were visualized by autoradiography. (B) The association of Grb4/F16 and Bcr-Abl is kinase-dependent. We transiently transfected 1 × 106 293 cells with a ts mutant of Bcr-Abl and grew these at either 32° (permissive temperature) or 39° C (restrictive temperature) for 8 hours before harvest. Cell lysates were incubated with 5 μg of GST or GST-Grb4/F16, and bound fractions were resolved on SDS-PAGE. Western blotting was performed with the anti-Abl antibody, 8E9. (C) Grb4/F16 binds to v-Abl. We transiently transfected 1 × 106 293 cells with v-Abl. Cells were harvested 48 hours after transfection, and lysates were incubated with 5 μg of GST or GST-Grb4/F16. Bound fractions were run out on SDS-PAGE. Western blotting was performed with the anti-Abl antibody 8E9. (D) Grb4/F16 and Bcr-Abl coimmunoprecipitate in vivo. In this experiment, 1 × 106 293 cells were co-transfected with either WT Bcr-Abl or the ts variant and Grb4/F16. Cells transfected with the ts mutant were shifted to the permissive temperature, 32°, or the restrictive temperature, 39°, for 8 hours before harvest. A coimmunoprecipitation experiment (IP) was performed on cell lysates with the use of an anti-Xpress antibody to detect tagged Grb4 (lanes 5-7) or a rabbit antimouse (RαM) antibody as control (lanes 1-4). Bound fractions were resolved on SDS-PAGE and immunoblotted with a phosphotyrosine antibody (δ-PY) (4G10) (upper 2 left panels), an anti-Xpress antibody to detect Grb4 (lowest left panel), or an anti-Abl antibody (8E9) (right panel).
Complex formation of Grb/F16 and Bcr-Abl.
(A) Grb4/F16 and Bcr-Abl form a complex in vitro. Bcr-Abl mutants with internal deletions of Bcr resulting in varying lengths of Bcr as indicated were in vitro translated and S35 labeled. We allowed 50 μL of the translation mix to autophosphorylate in vitro and then incubated it with 5 μg purified GST or GST-Grb4/F16. Bound fractions were separated by SDS-PAGE, and Bcr-Abl mutants were visualized by autoradiography. (B) The association of Grb4/F16 and Bcr-Abl is kinase-dependent. We transiently transfected 1 × 106 293 cells with a ts mutant of Bcr-Abl and grew these at either 32° (permissive temperature) or 39° C (restrictive temperature) for 8 hours before harvest. Cell lysates were incubated with 5 μg of GST or GST-Grb4/F16, and bound fractions were resolved on SDS-PAGE. Western blotting was performed with the anti-Abl antibody, 8E9. (C) Grb4/F16 binds to v-Abl. We transiently transfected 1 × 106 293 cells with v-Abl. Cells were harvested 48 hours after transfection, and lysates were incubated with 5 μg of GST or GST-Grb4/F16. Bound fractions were run out on SDS-PAGE. Western blotting was performed with the anti-Abl antibody 8E9. (D) Grb4/F16 and Bcr-Abl coimmunoprecipitate in vivo. In this experiment, 1 × 106 293 cells were co-transfected with either WT Bcr-Abl or the ts variant and Grb4/F16. Cells transfected with the ts mutant were shifted to the permissive temperature, 32°, or the restrictive temperature, 39°, for 8 hours before harvest. A coimmunoprecipitation experiment (IP) was performed on cell lysates with the use of an anti-Xpress antibody to detect tagged Grb4 (lanes 5-7) or a rabbit antimouse (RαM) antibody as control (lanes 1-4). Bound fractions were resolved on SDS-PAGE and immunoblotted with a phosphotyrosine antibody (δ-PY) (4G10) (upper 2 left panels), an anti-Xpress antibody to detect Grb4 (lowest left panel), or an anti-Abl antibody (8E9) (right panel).
The interaction of Grb4/F16 and Bcr-Abl is phosphotyrosine dependent
To confirm the phosphotyrosine-dependent interaction of Grb4/F16 and Bcr-Abl observed in yeast, a GST-binding experiment was performed in 293 cells transfected with a temperature-sensitive (ts) Bcr-Abl mutant. GST-Grb4/F16 bound to the kinase-active Bcr-Abl at the permissive (32°) but not at the restrictive temperature (39°) (Figure 4B). GST-Grb4/F16 also bound v-Abl in a pull-down experiment using 293 cells transiently expressing v-Abl (Figure 4C) while a Grb4 SH2 mutant failed to bind to v-Abl in 293 cells (unpublished data). This further strengthens the hypothesis that Grb4 binds the Abl part of Bcr-Abl.
To determine if Grb4 and Bcr-Abl interacted with each other in the context of intact cells, coimmunoprecipitation experiments were performed in 293 cells transiently transfected with Bcr-Abl and Xpress-tagged Grb4/F16. The Xpress antibody was used to immunoprecipitate Grb4/F16. Grb4/F16 and Bcr-Abl showed a strong association with each other in these cells (Figure 4D, lane 4, uppermost and lowest left panels). This interaction was dependent on the kinase activity of Bcr-Abl and occurred with WT Bcr-Abl or ts Bcr-Abl at the permissive temperature for the Bcr-Abl kinase (32°) but not (or only minimally) at the restrictive temperature (39°) (Figure 4D, lanes 4, 5, and 6, upper and lowest left panels). Additionally, Grb4/F16 was phosphorylated efficiently when coexpressed with kinase-active Bcr-Abl (Figure 4D, middle left panel, lane 4). This is most likely a direct effect of an active Bcr-Abl kinase since Grb4/F16 phosphorylation occurred with ts Bcr-Abl only at the permissive temperature (32°) (Figure 4D, middle left panel; compare lanes 5 and 6). Thus, Grb4/F16 forms a complex only with kinase-active Bcr-Abl and is an efficient substrate of the Bcr-Abl kinase.
Two types of interaction are responsible for the formation of the Grb4/Bcr-Abl complex in vivo
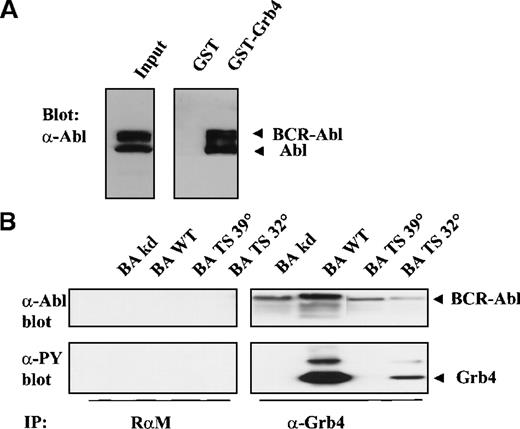
The association of Bcr-Abl with full-length Grb4, unlike that between Grb4/F16 and Bcr-Abl, was a constitutive interaction that occurred irrespective of the activity of the Bcr-Abl kinase (Figure5B, upper panel, lanes 5-8). Interaction of full-length Grb4 with KD Bcr-Abl (Figure 5B, upper panel, lane 5) and with ts Bcr-Abl at the restrictive temperature (39°) (Figure 5B, upper panel, lane 7) indicated that an additional non–phosphotyrosine-dependent interaction was occurring between the 2 proteins. This phosphotyrosine-independent interaction with Bcr-Abl is due to sequences in the first N-terminal SH3 domains, as Grb4/F16, which lacks these SH3 sequences, showed no phosphotyrosine-independent binding (Figure 1). The most likely target of this interaction is a proline-rich stretch in the C-terminal end of Abl.23Phosphorylation of Grb4 by Bcr-Abl remained, however, a process dependent on the activity of the Bcr-Abl kinase (Figure 5B, lower panel, lanes 5-8). In keeping with these data, it was observed that Grb4 bound strongly to both Bcr-Abl and Abl in K562 cells in a GST pull-down experiment, K562 cells constitute a cell line derived from a patient with chronic myeloid leukemia in blast crisis (Figure 5A, right panel). However, a coimmunoprecipitation experiment with endogenous protein in these cells could not be done because the Grb4 antibody precipitates the Grb4 protein very poorly (Figure 2D).
Full-length Grb4 interacts with Bcv-Abl.
(A) Grb4 interacts with Bcr-Abl and Abl in a chronic myeloid leukemia cell line. Cell lysates from 3 × 107 K562 cells were incubated with 5 μg of GST or GST-Grb4 (full length), and the bound fraction was resolved on SDS-PAGE and then blotted with an anti-Abl antibody (8E9). Bands indicated as Bcr-Abl and Abl correspond to 210 kd and 145 kd, respectively. (B) Full-length Grb4 interacts with Bcr-Abl in vivo. Then, 1 × 106 293 cells were transfected with KD Bcr-Abl (BA KD), WT Bcr-Abl (BA WT), or ts Bcr-Abl (BA TS) together with Grb4. Cells transfected with the ts mutant were shifted to 32° or 39° 8 hours before harvest. A coimmunoprecipitation experiment was performed with the use of a rabbit antimouse antibody (RαM) as control (left panels) or an anti-Xpress antibody to detect tagged Grb4 (right panels). Bound fractions were resolved on SDS-PAGE and immunoblotted with an anti-Abl antibody (8E9) (upper panels) or a phosphotyrosine antibody (4G10) (lower panels). In the upper panel, lanes 1 to 4 and 7 to 8 were exposed 3 times longer than lanes 5 and 6. Levels of Grb4 immunoprecipitated in all lanes were equal (not shown).
Full-length Grb4 interacts with Bcv-Abl.
(A) Grb4 interacts with Bcr-Abl and Abl in a chronic myeloid leukemia cell line. Cell lysates from 3 × 107 K562 cells were incubated with 5 μg of GST or GST-Grb4 (full length), and the bound fraction was resolved on SDS-PAGE and then blotted with an anti-Abl antibody (8E9). Bands indicated as Bcr-Abl and Abl correspond to 210 kd and 145 kd, respectively. (B) Full-length Grb4 interacts with Bcr-Abl in vivo. Then, 1 × 106 293 cells were transfected with KD Bcr-Abl (BA KD), WT Bcr-Abl (BA WT), or ts Bcr-Abl (BA TS) together with Grb4. Cells transfected with the ts mutant were shifted to 32° or 39° 8 hours before harvest. A coimmunoprecipitation experiment was performed with the use of a rabbit antimouse antibody (RαM) as control (left panels) or an anti-Xpress antibody to detect tagged Grb4 (right panels). Bound fractions were resolved on SDS-PAGE and immunoblotted with an anti-Abl antibody (8E9) (upper panels) or a phosphotyrosine antibody (4G10) (lower panels). In the upper panel, lanes 1 to 4 and 7 to 8 were exposed 3 times longer than lanes 5 and 6. Levels of Grb4 immunoprecipitated in all lanes were equal (not shown).
v-Abl induced–AP-1 activation is suppressed by Grb4
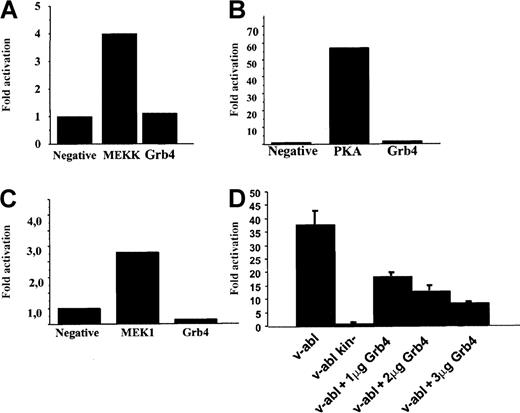
Nck and Bcr-Abl were shown to activate the JNK signaling pathway in a promoter activation assay.24-26 Braverman and Quilliam,15 when reporting the identification of Grb4, described enhanced Elk1 activation when Grb4 was coexpressed with v-Abl. In addition, Abl kinases have been reported to activate the CREB signaling pathway,27 and there are reports of cross-talk between the Jun/Fos/AP-1 pathways and the CREB signaling pathway.28 Therefore, we wished to test the ability of Grb4 to activate the JNK, CREB, and Elk1 signaling pathway. Using a transactivating fusion transcription factor containing the DNA-activation domain of a pathway-specific transcription factor fused to the DNA-binding domain of GAL4, we assessed the ability of Grb4 to activate either the JNK, the Elk-1, or the CREB signaling pathway by means of a luciferase-based reporter assay (see “Materials and methods” for details). Using this assay, we were unable to see any activation by Grb4 itself of the JNK, the Elk1, or the CREB signaling pathways (Figure 6A-C). However, it has been shown that Bcr-Abl itself can activate AP-1 at a low level and that v-Abl is a strong activator of AP-1–dependent transcription.26 29 Therefore, we tested whether coexpression of Grb4 and v-Abl interferes with activation of AP-1. We were able to demonstrate that v-Abl–induced activation of AP-1 was inhibited by Grb4 in a concentration-dependent manner in 293 cells (Figure 6D). The level of v-Abl expression in every set of transfected cells was determined and found to be equal (data not shown). This inhibitory effect of Grb4 on v-Abl–induced AP-1 activation was detectable in several additional cell types (Cos1 and NIH 3T3; data not shown). Thus, Grb4 is an inhibitor of an oncogenic Abl-induced pro-mitogenic pathway.
Grb4 expression does not activate the JNK, Elk1, or the CREB signaling pathway but inhibits v-Abl–induced AP-1 activation.
(A-C) JNK, Elk1, and CREB pathway. We transiently transfected 1 × 105 293 cells with a plasmid expressing Grb4, a beta-galactosidase construct to normalize for transfection efficiency, and a construct for the expression of a fusion transcription factor encoding the DNA-activation domain of Jun, Elk, or CREB and the DNA-binding domain of GAL4. The luciferase gene was under the control of the GAL4 promoter. As a positive control, mitogen-activated protein kinases (MEKK in panel A, MEK1 in panel C) and protein kinase A (PKA in panel B) were used. Results are representative of at least 3 independent experiments. (D) AP-1 activation. We transiently transfected 293 cells with 1 μg of a plasmid expressing v-abl or kinase-inactive v-abl together with increasing amounts of a plasmid expressing Grb4 as indicated, beta-galactosidase, and a reporter construct with 7 AP-1 sites upstream of the luciferase gene. In each reaction, equal amounts of DNA were transfected. Results are the mean and SD determined from an experiment done in triplicate reproduced at least 3 times.
Grb4 expression does not activate the JNK, Elk1, or the CREB signaling pathway but inhibits v-Abl–induced AP-1 activation.
(A-C) JNK, Elk1, and CREB pathway. We transiently transfected 1 × 105 293 cells with a plasmid expressing Grb4, a beta-galactosidase construct to normalize for transfection efficiency, and a construct for the expression of a fusion transcription factor encoding the DNA-activation domain of Jun, Elk, or CREB and the DNA-binding domain of GAL4. The luciferase gene was under the control of the GAL4 promoter. As a positive control, mitogen-activated protein kinases (MEKK in panel A, MEK1 in panel C) and protein kinase A (PKA in panel B) were used. Results are representative of at least 3 independent experiments. (D) AP-1 activation. We transiently transfected 293 cells with 1 μg of a plasmid expressing v-abl or kinase-inactive v-abl together with increasing amounts of a plasmid expressing Grb4 as indicated, beta-galactosidase, and a reporter construct with 7 AP-1 sites upstream of the luciferase gene. In each reaction, equal amounts of DNA were transfected. Results are the mean and SD determined from an experiment done in triplicate reproduced at least 3 times.
Subcellular localization of Grb4
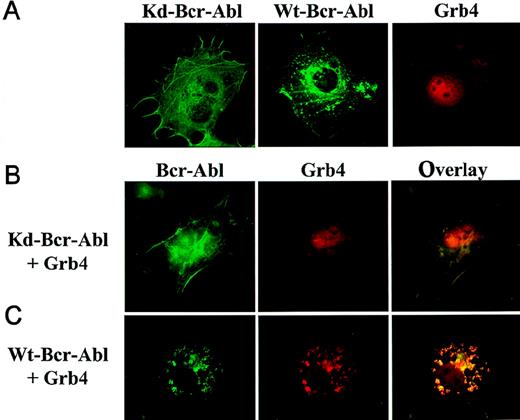
Grb4-EYFP (red) and WT or KD Bcr-Abl-ECFP (green) were created to examine the intracellular localization and colocalization (yellow) of Grb4 and Bcr-Abl simultaneously in the context of intact cells.20,21,30 The detection of these fusion proteins is possible with no or only mild fixation of the cells and for the first time allows Bcr-Abl localization in intact cells. KD Bcr-Abl (Figure7A, left panel) localized to the actin filaments and stress fibers in the cytoskeleton whereas WT Bcr-Abl, in addition, accumulated in juxtanuclear punctate aggregates, which probably contain F-actin (7A, middle panel), confirming previous data by McWhirter et al using immunofluorescence.18 31 Grb4-EYFP showed cytoplasmic and nuclear staining; within the nucleus, Grb4 staining excluded the nucleoli (Figure 7A, right panel). When KD Bcr-Abl and Grb4 were coexpressed, the cytoplasmic fraction of Grb4 showed distinct colocalization with Bcr-Abl in the cytoskeleton while a significant amount of Grb4 remained in the nucleus (7B, middle and right panels). The distribution of KD Bcr-Abl did not change (compare 7A, left panel, and 7B, left panel). In contrast, when Grb4 was coexpressed with WT Bcr-Abl, the distribution of Bcr-Abl itself was significantly altered (compare 7C, left panel, with 7A middle panel): most of the Bcr-Abl appeared in punctate aggregates that were no longer predominantly juxtanuclear, and no WT Bcr-Abl was visible along the stress fibers of the cytoskeleton. In addition, cytoplasmic and nuclear Grb4 colocalized with kinase-active Bcr-Abl in these punctate aggregates, and only a small amount of Grb4 remained in the nucleus (compare 7C, middle panel, and 7B, middle panel and 7A, right panel). This redistribution is specific for Grb4, as expression of EYFP alone shows a diffuse cytoplasmic and nuclear staining that is not altered by the coexpression of Bcr-Abl (data not shown). Thus, coexpression of Grb4 with WT Bcr-Abl leads to translocation of the nuclear pool of Grb4 to colocalize with WT Bcr-Abl in the cytoplasm and to redistribution of actin-bound Bcr-Abl itself, which may reflect reorganization of the cytoskeleton.
Subcellular localization of Bcr-Abl and Grb4.
Cos7 cells transiently transfected with KD or WT Bcr-Abl-ECFP and Grb4-EYFP were visualized by means of fluorescence microscopy. KD Bcr-Abl and WT Bcr-Abl appear in green; Grb4 appears in red; and the colocalization of both proteins appears in yellow. (A) Cells expressing KD Bcr-Abl (left panel), WT Bcr-Abl (middle panel), or Grb4 (right panel). (B) (C) Cells coexpressing KD Bcr-Abl (panel B) or WT Bcr-Abl (panel C) together with Grb4. Left panels show distribution of KD Bcr-Abl and WT Bcr-Abl; middle panels show the distribution of Grb4 with either KD Bcr-Abl or WT Bcr-Abl; and the right panels show merged pictures of the localization of the 2 proteins
Subcellular localization of Bcr-Abl and Grb4.
Cos7 cells transiently transfected with KD or WT Bcr-Abl-ECFP and Grb4-EYFP were visualized by means of fluorescence microscopy. KD Bcr-Abl and WT Bcr-Abl appear in green; Grb4 appears in red; and the colocalization of both proteins appears in yellow. (A) Cells expressing KD Bcr-Abl (left panel), WT Bcr-Abl (middle panel), or Grb4 (right panel). (B) (C) Cells coexpressing KD Bcr-Abl (panel B) or WT Bcr-Abl (panel C) together with Grb4. Left panels show distribution of KD Bcr-Abl and WT Bcr-Abl; middle panels show the distribution of Grb4 with either KD Bcr-Abl or WT Bcr-Abl; and the right panels show merged pictures of the localization of the 2 proteins
Discussion
We describe here the identification of Grb4 as a Bcr-Abl–interacting adapter protein. Grb4 has an overall homology of 68% on the amino acid level to the adapter protein Nck and, like Nck, consists of 3 SH3 domains and 1 SH2 domain. Nck has been described as binding in a phosphotyrosine-independent manner via its SH3 domain to proline-rich sequences in the C-terminal end of Bcr-Abl. The functional consequence of this interaction is unknown.32 Similarily, Grb4 also showed this phosphotyrosine-independent binding to Bcr-Abl via its SH3 domains. However, the SH2 domain of Grb4 mediated an additional phosphotyrosine-dependent interaction with Bcr-Abl in yeast, in vitro, and in vivo. Grb4 interacted with both Bcr-Abl and Abl and is an efficient substrate of the Bcr-Abl kinase.
Binding experiments with Bcr-Abl deletion mutants and v-Abl indicated that the phosphotyrosine-dependent binding to Bcr-Abl occurred at an autophosphorylation site in the Abl portion of Bcr-Abl. A Grb4/F16 mutant in which the FLVR region within the SH2 domain of Grb4 was mutated failed to bind to v-Abl (data not shown). This raises the interesting possibility that Grb4, in contrast to other SH2 adapter proteins (eg, Grb2), is a potential mediator not only of Bcr-Abl but also of c-Abl, v-Abl, or Tel-Abl induced signals.
Far Western analyses on K562 lysates showed that Nck and Grb4 had similar protein-binding specificities although some significant differences could be found. Interestingly, we detected stronger binding of Bcr-Abl to Grb4 than to Nck or to Grb2, the other SH2-mediated binding adapter of Bcr-Abl in this pull-down assay. In addition, we determined that Grb4 and Nck co-precipitated with an individual set of phosphorylated proteins in 293 cells (S.C. and J.D., unpublished results, October 1999).
Stein et al reported that Nck is a central molecule linking EphB1 signaling to JNK.24 Nck also binds NIK, a Ste20-related kinase that activates the SAPK/JNK cascade.33,34 In addition Bcr-Abl has been shown to activate the JNK signaling pathway.25,26 35 To test whether Grb4 represents a potential link between Bcr-Abl and JNK, we examined the effect of Grb4 on JNK activation. We were unable to demonstrate any JNK activation in several cell types tested. In addition, JNK kinase assays showed no significant activation by the expression of Grb4 (unpublished data).
Additionally, we examined AP-1 activation as both Bcr-Abl and v-Abl have been shown to activate this signaling pathway.25,36Interestingly, we could demonstrate that concomitant Grb4 expression consistently inhibited v-Abl–induced AP-1 activation in several cell types tested. These data suggest that overexpression of Grb4 is capable of inhibiting an Abl-induced pro-mitogenic pathway. In line with these data, Chen et al demonstrated that Grb4 expression inhibited EGF- and PDGF-stimulated DNA synthesis.13 Chen et al showed that an SH2-domain mutant of Grb4 did not inhibit EGF- and PDGF-stimulated DNA synthesis, suggesting that this effect is mediated by the SH2 domain of Grb4.13 In contrast, this mutant caused an even more pronounced inhibition of v-Abl–induced AP-1 activation (data not shown), implying that the inhibition of the AP-1 pathway is an SH3-mediated response. Thus, both the SH2 and the SH3 domains of Grb4 seem to be involved in the inhibition of pro-mitogenic signals elicited by growth factors or oncogenic Abl.
These data are somewhat contrary to those of Braverman and Quilliam reporting enhanced activation of the pro-mitogenic Elk1 signaling pathway when Grb4 was expressed with v-Abl.15 However, the increase in activation measured with Grb4 was low, and the authors report that growth of cells in bovine calf serum from various sources can result in significant differences in the magnitude of adapter-protein–induced transcriptional activation.15 We were not able to measure any significant coactivation of the Elk1 reporter used in this study by coexpression of v-Abl and Grb4 in several cell lines. Thus, differences in both studies may be due to the reporter assay and the culture conditions used.15
Cellular localization studies indicate that Grb4, like Nck, localizes to both the nucleus and the cytoplasm.37 Together with KD Bcr-Abl, only the cytoplasmic pool of Grb4 colocalized with KD Bcr-Abl to actin filaments and stress fibers of the cytoskeleton. However, when coexpressed with WT Bcr-Abl, cytoplasmic and nuclear Grb4 colocalized with WT Bcr-Abl and redistributed into punctate aggregates containing F-actin, indicating reorganization of the cytoskeleton.31Thus, Grb4-complex formation with kinase-active Bcr-Abl leads to a redistribution of actin-bound Bcr-Abl. It is possible that tyrosine phosphorylation of Grb4 by Bcr-Abl leads to the observed changes in the cytoskeleton structure by recruiting other integrin- or actin-modulating proteins. Indeed, Tu et al described Grb4 as a molecule interacting with PINCH, a LIM-only protein interacting with integrin-linked kinase (ILK).14 38 It was suggested that through its interaction with PINCH, Grb4 may link growth factor receptors with integrin signaling. Together with our data, this may suggest a role for Grb4 in cytoskeletal and integrin regulation.
The inhibition of v-Abl kinase–induced AP-1 activation by Grb4 expression makes it unlikely that Grb4 expression contributes to a pro-mitogenic signal in the course of chronic myeloid leukemia. However, we obtained experimental evidence that the nuclear pool of Grb4 is mediating the AP-1 inhibition observed since this inhibition could also be detected with AP-1 activation through MEKK and JNK (unpublished data). Therefore, it could be hypothesized that inhibition of AP-1 activation through nuclear Grb4 is blocked by sequestering Grb4 in the cytoplasm by Bcr-Abl or v-Abl, thus leading to a growth advantage of the cell.
Progenitor cells in chronic myeloid leukemia are known to have defects in cell adhesion, and recently accumulated evidence suggests that Bcr-Abl may contribute to transformation not by promoting mitogenic signals but by the inhibition of apoptosis and dysregulation of adhesion signals.36 39 Given that Grb4 influences the distribution of Bcr-Abl to the cytoskeleton and associates with integrin-interacting proteins, it seems reasonable to speculate that Grb4 may link the chimeric oncogene to integrin-signaling pathways, thus playing a role in Bcr-Abl–induced dysregulation of cell adhesion.
Acknowledgments
We thank Petra Seipel for technical assistance and Jean Wang, Donald Kohn, and Michael Hallek for cDNAs and cell lines.
Partially supported by grants to J.D. from the José-Carreras Stiftung and by Sonderforschungsbereich Grant no. 456 to J.D. and C.P. T.J. is supported by a fellowship from the Deutsche José-Carreras Stiftung.
S.C. and T.J. contributed equally to this work.
Reprints:Justus Duyster, Department of Internal Medicine III, Technical University of Munich, Trogerstr 32, D-81675 Munich, Germany; e-mail: justus.duyster@lrz.tum.de.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal