Abstract
Defects in a triad of organelles (melanosomes, platelet granules, and lysosomes) result in albinism, prolonged bleeding, and lysosome abnormalities in Hermansky-Pudlak syndrome (HPS). Defects in HPS1, a protein of unknown function, and in components of the AP-3 complex cause some, but not all, cases of HPS in humans. There have been 15 inherited models of HPS described in the mouse, underscoring its marked genetic heterogeneity. Here we characterize a new spontaneous mutation in the mouse, cappuccino (cno), that maps to mouse chromosome 5 in a region conserved with human 4p15-p16. Melanosomes ofcno/cno mice are immature and dramatically decreased in number in the eye and skin, resulting in severe oculocutaneous albinism. Platelet dense body contents (adenosine triphosphate, serotonin) are markedly deficient, leading to defective aggregation and prolonged bleeding. Lysosomal enzyme concentrations are significantly elevated in the kidney and liver. Genetic, immunofluorescence microscopy, and lysosomal protein trafficking studies indicate that the AP-3 complex is intact in cno/cno mice. It was concluded that the cappuccino gene encodes a product involved in an AP-3–independent mechanism critical to the biogenesis of lysosome-related organelles.
Introduction
Hermansky-Pudlak syndrome (HPS) is characterized by autosomal recessive inheritance, oculocutaneous albinism, prolonged bleeding (platelet storage pool disease, SPD), and abnormal lysosomal enzyme levels as a result of defects in 3 subcellular organelles—melanosomes, platelet dense bodies, and lysosomes.1 Chediak-Higashi syndrome (CHS) is a related disease with similar defects. A major difference, however, is that giant granules are present in most cells of CHS patients.2In the mouse, 16 models of SPD that map to distinct chromosomal regions have been described. CHS in human beings and the homologous mouse mutation beige (bg) are caused by defects in the lysosomal trafficking regulator gene (LYST/Lyst), encoding a novel protein of unknown function.3,4LYSTcontains motifs found in the yeast Vps15 gene, which functions in intracellular protein trafficking,2 and shares a region of homology with stathmin, suggesting possible involvement in microtubule-mediated lysosomal transport.4The remaining mouse SPD mutations closely resemble HPS.5 6
HPS is clinically heterogeneous in humans. The worldwide incidence of HPS is unknown, but it is very high in some populations owing to founder effects. In Northwestern Puerto Rico, its frequency is approximately 1 in 1800.7 HPS is associated with considerable morbidity and mortality. Oculocutaneous albinism results in nystagmus, decreased visual acuity (approximately 20/200), and skin damage upon exposure to sunlight.8 The bleeding diathesis generally manifests as easy bruising, frequent nose and gum bleeding, and excessive bleeding during menstruation, after surgery, and during childbirth.8 Bleeding can be severe; a recent study documented major bleeding events, some life-threatening, in approximately 40% of HPS patients studied.9
Lysosomal ceroid lipofuscin accumulation accounts for the most serious complications of HPS: bleeding granulomatous colitis and pulmonary fibrotic disease. Other, less common complications include kidney failure and cardiomyopathy.10 Bleeding granulomatous colitis occurs in 15% of HPS cases, and some patients require colostomies.11 Pulmonary fibrosis is progressive and causes death in the fourth to fifth decades in approximately 50% of HPS patients.12 The risk and severity of fibrotic lung disease are highest in the Puerto Rican form of HPS.9Although steroid treatments provide symptomatic relief, no curative therapies are available for these devastating HPS sequelae.
The mouse models of HPS show striking clinical correlations with human HPS.6 All show autosomal recessive inheritance. All have δ-SPD caused by a deficiency of platelet delta granule (dense-body) contents (adenine nucleotides, serotonin), resulting in prolonged bleeding. The gunmetal (gm) mutation differs in that both platelet dense bodies and α-granules are affected (αδ-SPD).13 All of the mutations show varying degrees of coat-color dilution, and lysosomes are defective in all but 3 (subtle gray, sut; cocoa, coa; andgm).6 Three mutations (pallid, pa; muted, mu; mocha, mh) represent a distinct subset in which associated inner ear abnormalities (absent or abnormal otoliths and reduced inner ear pigmentation) lead to head tilting and balance problems.6,14 Inner ear abnormalities are also reported in human HPS.15
Puerto Rican HPS maps to chromosome 10 and is caused by defects in theHPS1 gene.16-18 HPS1 is both a cytosolic and peripheral membrane protein of unknown function.19,20 The mouse homologue of Puerto Rican HPS is the pale ear (ep) mutation located on chromosome (Chr) 19.21,22 The genetic defects in 4 other mouse models of classic HPS have recently been identified. The pearl (pe, Chr 13) and mh (Chr 10) mutations involve defects in the β3A and δ subunits, respectively, of the AP-3 complex, which functions in endosomal-lysosomal protein trafficking.23-25 Defects in the β3A subunit of AP-3 have recently been identified in human HPS.26 The pa (Chr 2) mutation is a defect in a novel protein, pallidin, which binds to syntaxin-13, a t-snare protein that mediates vesicle docking and fusion.27 In ashen (ash), Rab27a, a protein involved in targeting and fusing transport vesicles to their acceptor membrane, is defective.5 Although not considered a classic HPS model because of associated α-granule defects, Rab function is disrupted in gunmetal owing to a mutation within the alpha subunit of Rab geranylgeranyl transferase.28
Many human HPS patients have no defects in the HPS1 gene or in the genes encoding AP-3 complex proteins, indicating that the causes of HPS in humans are heterogeneous, as in the mouse. It is increasingly clear that cloning of the remaining mouse mutations will provide additional candidate genes for human HPS, as well as insights into the mechanisms of organelle biogenesis and protein and vesicle trafficking, and identify potential novel pathways critical to the biogenesis of melanosomes, dense bodies, and lysosomes.
We have identified a new mouse HPS mutation, cappuccino (cno), located on Chr 5. Cappuccino mice have severe HPS with both phenotypic similarities to and distinctions from the other known mouse models. Mapping of multiple candidate genes, immunofluorescence microscopy, and lysosomal protein trafficking studies indicate that the cno gene is not a component of the AP-3 complex. These observations, together with data from the other cloned mouse mutants, implicate defects in multiple pathways in the pathogenesis of HPS.
Materials and methods
Mice
The cappuccino mutation, first identified because it causes severe coat-color dilution, occurred on the C3H/HeJ inbred strain at The Jackson Laboratory. Cappuccino segregates as an autosomal recessive trait. C3H/HeJ +/cno mice are phenotypically indistinguishable from normal control C3H/HeJ +/+ mice. The C3H/HeJ-cno strain was maintained by mating C3H/HeJ +/cno females to C3H/HeJ cno/cno males and intercrossing heterozygotes. Heterozygotes or inbred C3H/HeJ +/+ mice served as normal controls for all studies.
Complete blood counts
Adult whole blood was collected in EDTA-coated Microtainer tubes (Becton Dickinson, Rutherford, NJ). Complete blood counts were determined by means of an Advia 120 Multi-species whole blood analyzer (Bayer, Tarrytown, NY).
Histology
For tissue pathology, mice were anesthetized with avertin and perfused through the left ventricle with 20 mL Bouin's fixative. Tissues (spleen, bone marrow, liver, kidney, and small intestine) were removed and placed in fresh fixative overnight. Lungs were removed prior to perfusion. Paraffin sections (5 μm) were stained with hematoxylin and eosin. Whole eyes were fixed in 0.8% glutaraldehyde/1.2% paraformaldehyde in 0.1 mol/L phosphate buffer, pH 7.2, for 2 hours and embedded in plastic, as described.29 30 Sections (2 μm) were stained with hematoxylin and eosin.
Electron microscopy
To obtain platelet-rich plasma, whole blood was collected in acid-citrate-dextrose (0.13 mol/L citric acid, 0.15 mol/L sodium citrate, 0.1 mol/L dextrose) and centrifuged at 120g for 20 minutes at room temperature. Platelet dense bodies were enumerated in air-dried unstained, unfixed whole platelets as described.14 For ultrastructural studies, platelet-rich plasma was centrifuged at 1300g for 5 minutes, and the pellet was fixed overnight at 4°C in 2% glutaraldehyde/2% paraformaldehyde in 0.1 mol/L cacodylate buffer, pH 7.2. Whole eyes were fixed as described above. Small sections containing the choroid and retina were dissected, and fixation continued for 24 hours. All samples were post-fixed in osmium and routinely processed for transmission electron microscopy.30 A JEOL 100CXII (JEOL, Tokyo, Japan) transmission electron microscope was used for all studies.
Bleeding time, platelet aggregation, and serotonin assays
Bleeding times were determined by the tail-tip method, as described.31 Platelet aggregation in response to high (4 μg/mL) and low (1 μg/mL) collagen was measured by impedance with the use of a whole blood aggregometer (Chronolog, Havertown, PA). Adenosine triphosphate (ATP) release was determined simultaneously by luminescence. Platelet serotonin concentration was determined fluorometrically as described.32
Lysosomal enzyme assays
Tissues were obtained from females to avoid potential testosterone-induction effects. Kidney, liver, and spleen β-glucuronidase and α- and β-galactosidase activities were measured fluorometrically as previously described.33 34Protein concentrations were determined by the method of Bradford (Bio Rad, Hercules, CA).
Fine mapping of cno
An F2 intercross was established with the use of C3H/HeJ cno/cno and Mus musculus castaneus (CAST/Ei) mice. There were 525 homozygous F2cno/cno mice generated (1050 meioses). DNA was prepared from spleens as described35 and from tail tissue by means of a commercial kit (Gentra Systems, Research Triangle Park, NC). Candidate gene complementary DNAs (cDNAs) were mapped by Southern blot analysis. Anonymous DNA microsatellite markers were mapped by polymerase chain reaction (PCR) with the use of primers obtained from Research Genetics (Huntsville, AL).36,37 PCR reactions were carried out in 96-well plates as described.38 Products were resolved and scored on ethidium bromide–stained 1% to 5% NuSieve gels (FMC Bioproducts, Rockland, ME).
Mapping of candidate genes
Candidate genes (Ap3m1, Ap3m2, Ap3s1, Ap3s2, Vps41) were mapped by Southern blot analysis by means of The Jackson Laboratory BSS interspecific backcross [(C57BL/6JEi x SPRET/Ei)F1 x SPRET/Ei] panel.39 Hybridization probes consisted of segments of the open reading frames (ORFs) and untranslated regions (UTRs) and were derived from the inserts of expressed sequence tag (EST) mouse cDNA clones (IMAGE Consortium, Lawrence Livermore National Laboratory, Livermore, CA), as follows: IMAGE 536161 (complete ORF, 5′ UTR and 3′ UTR of Ap3m1), IMAGE 737067 (codons 231 to stop and 3′ UTR of Ap3m2), IMAGE 585789 (codon 10 to stop and 3′ UTR ofAp3s1), IMAGE 438481 (complete ORF, 5′ UTR and 3′ UTR ofAp3s2) and IMAGE 464051 (codons 705 to stop and 3′ UTR ofVps41). Identity of the EST clones was verified by DNA sequencing. Blots were hybridized at 42°C in formamide and washed at 65°C for highest stringency, as previously described.40
Segregation of alleles was followed by the use of an Ap3m1EcoRV restriction fragment length polymorphism (RFLP) (C57BL/6JEi, 9.0 kilobases [kb]; SPRET/Ei, 3.0 kb); an Ap3m2 PstI RFLP (C57BL/6JEi, 7.8 kb; SPRET/Ei, 5.0 kb); an Ap3s2 BamHI RFLP (C57BL/6JEi, 6.3 kb; SPRET/Ei, approximately 20.0 kb); and aVps41 BglII RFLP (C57BL/6JEi, 3.6 kb; SPRET/Ei, 2.3 kb). Southern blotting using the Ap3s1 probe revealed multiple fragments indicating 3 closely related, unlinked genes (see “Results”). The typing data have been placed in the Mouse Genome Database (Accession # J:61187) and can be accessed through the World Wide Web (http://www.jax.org).
Fibroblast culture and immunofluorescence analyses
Skin fibroblasts derived from C3H/HeJ (wild-type control), mocha, and cappuccino mice were prepared and cultured as described previously.20 Cells grown on glass coverslips were fixed and immunostained41 with the use of affinity-purified rabbit antibodies to either the δ42 or β3A43 subunits of the AP-3 complex, followed by Cy3-conjugated donkey anti-rabbit immunoglobulin (Jackson Immunoresearch, West Grove, PA). For antibody internalization experiments, fibroblasts grown on glass coverslips were incubated with the rat monoclonal antibody 1D4B against mouse LAMP-1 (Developmental Studies Hybridoma Bank at the University of Iowa, Iowa City, IA) in culture medium (Dulbecco's modified Eagle's medium, 20% [vol/vol] fetal bovine serum, 25 mmol/L Hepes [pH 7.4], 2 mmol/L glutamine, 0.1 g/L streptomycin, 100 IU/mL penicillin) for 15 minutes at 37°C. Following incubation, cells were washed for 5 minutes in ice-cold phosphate-buffered saline (PBS), fixed in PBS containing 2% formaldehyde for 10 minutes at room temperature, and stained for 30 minutes at room temperature with Cy3-conjugated donkey anti-rat immunoglobulin (Jackson Immunoresearch) diluted in PBS, 0.1% saponin, 0.1% bovine serum albumin. Stained samples were rinsed with PBS and mounted on glass slides with Fluoromount G (Southern Biotechnologies, Birmingham, AL). Fluorescence images were acquired on a Zeiss 410 LSM confocal microscope (Carl Zeiss, Thornwood, NY). Quantitation of antibody internalization was carried out as described previously.26
Results
Melanosomes are deficient and immature in cnomice
The first cappuccino mouse was identified owing to severe reduction in skin and eye pigment compared with its normal littermates (Figure 1A). Pigment reduction in homozygous cno mice rivals that of pa, the mouse HPS mutation with the most severe pigment loss.6 Allelism testing and genetic mapping (see below) confirmed that cnowas distinct from pa and all other known mouse SPD mutations.
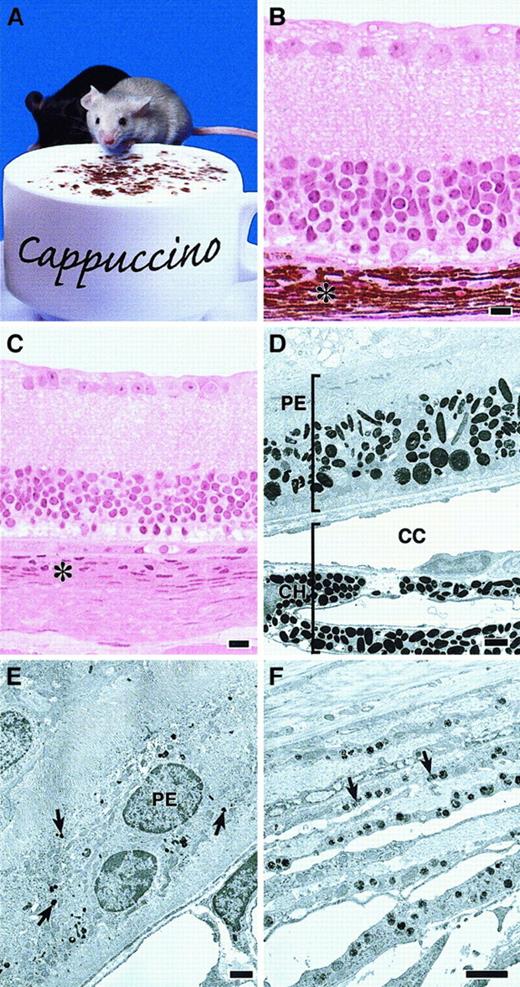
Defective melanosomes in homozygous cappuccino mice.
(A) A cno/cno mouse (foreground) and a normal littermate. Note marked coat-color dilution in the mutant. (B, C) Plastic embedded light microscopy images of normal (panel B) andcno/cno (panel C) eyes showing the profound reduction in the choroid layer (asterisk) characteristic of cno homozygotes. Other layers are normal. (Note: The photoreceptor and outer-segments layers are missing in the C3H/HeJ strain.) (D) Electron photomicrograph of a normal (+/cno) eye with the retinal pigmented epithelial (PE) layer toward the top and the choroid (CH) below. CC, chorio-capillaries. (E) cno/cnoretinal PE layer. Note severe reduction in number and size of the melanosomes (arrows). (F) cno/cno choroid layer at higher magnification showing small, incompletely melanized melanosomes (arrows). Bars, 10 μm (B-C), 1 μm (D-F).
Defective melanosomes in homozygous cappuccino mice.
(A) A cno/cno mouse (foreground) and a normal littermate. Note marked coat-color dilution in the mutant. (B, C) Plastic embedded light microscopy images of normal (panel B) andcno/cno (panel C) eyes showing the profound reduction in the choroid layer (asterisk) characteristic of cno homozygotes. Other layers are normal. (Note: The photoreceptor and outer-segments layers are missing in the C3H/HeJ strain.) (D) Electron photomicrograph of a normal (+/cno) eye with the retinal pigmented epithelial (PE) layer toward the top and the choroid (CH) below. CC, chorio-capillaries. (E) cno/cnoretinal PE layer. Note severe reduction in number and size of the melanosomes (arrows). (F) cno/cno choroid layer at higher magnification showing small, incompletely melanized melanosomes (arrows). Bars, 10 μm (B-C), 1 μm (D-F).
Melanosomes in cno/cno eyes are strikingly reduced in number throughout the retinal pigmented epithelium and choroid layers (Figure1B-F). The melanosomes present are significantly smaller and less melanized than normal. We conclude that cno causes severe oculocutaneous albinism.
Platelets are defective in cno/cnomice
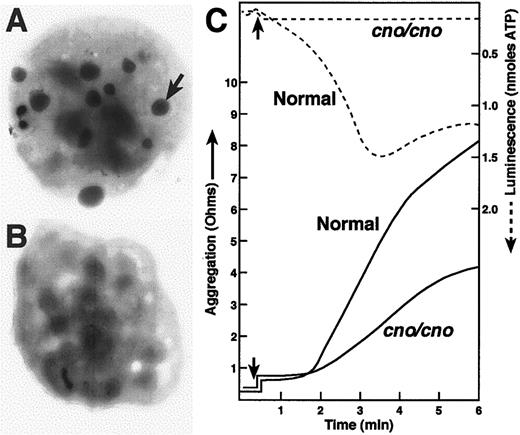
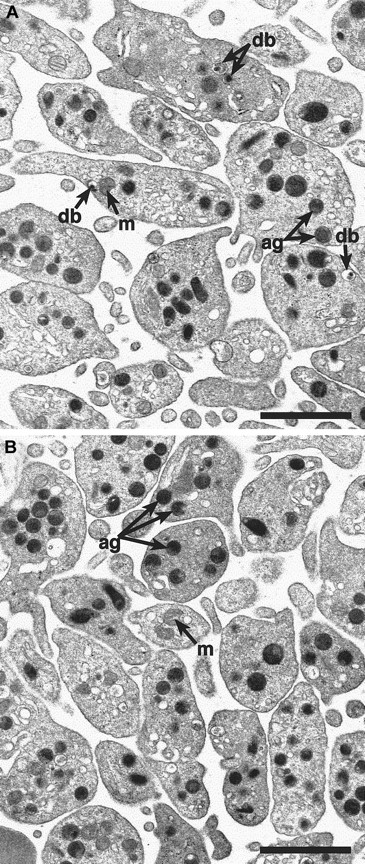
To determine if cno/cno mice have platelet SPD, we performed complete blood counts and measured bleeding times on adult mice. Despite normal blood cell counts, including platelets (Table1), bleeding times are markedly elevated, suggesting a qualitative platelet disorder (Table2). Electron microscopical examination of air-dried whole platelets reveals a dramatic decrease in visible dense bodies (Figure 2 A-B; Table 2); loss of dense body electron density suggests a low Ca++content.6 Similarly, platelet serotonin levels are decreased to approximately 3% of normal (Table 2). Collagen-induced platelet aggregation is significantly reduced, and ATP release is undetectable (Figure 2C). Hence, a profound deficiency of dense body contents characterizes homozygous cno platelets. Ultrastructurally, these platelets contain abundant α-granules whose size and number appear normal (Figure 3). The data are consistent with δ-SPD in cno/cnomice.
Hematologic parameters in adult cno/cno mice
| Genotype . | No. . | WBC (×103/μL) . | RBCs (×106/μL) . | HGB (g/dL) . | HCT (%) . | PLT (×103/μL) . | MPV (fL) . |
|---|---|---|---|---|---|---|---|
| Normal | 3 | 5.4 ± 0.7 | 9.1 ± 0.2 | 15.4 ± 0.4 | 43.4 ± 1.1 | 1030 ± 95 | 5.0 ± 0.1 |
| cno/cno | 4 | 7.5 ± 2.2 | 9.3 ± 0.3 | 15.4 ± 0.6 | 43.5 ± 2.1 | 1116 ± 108 | 5.0 ± 0.3 |
| Genotype . | No. . | WBC (×103/μL) . | RBCs (×106/μL) . | HGB (g/dL) . | HCT (%) . | PLT (×103/μL) . | MPV (fL) . |
|---|---|---|---|---|---|---|---|
| Normal | 3 | 5.4 ± 0.7 | 9.1 ± 0.2 | 15.4 ± 0.4 | 43.4 ± 1.1 | 1030 ± 95 | 5.0 ± 0.1 |
| cno/cno | 4 | 7.5 ± 2.2 | 9.3 ± 0.3 | 15.4 ± 0.6 | 43.5 ± 2.1 | 1116 ± 108 | 5.0 ± 0.3 |
All values are mean ± SD. There were no significant differences.
WBC indicates white blood cells; RBCs, red blood cells; HGB, hemoglobin; HCT, hematocrit; PLT, platelet, MPV, mean platelet volume.
Analyses of platelet structure and function in adultcno/cno mice
| Genotype . | Bleeding time (min) . | No. dense bodies per platelet* . | Platelet serotonin (μg/109 platelets) . |
|---|---|---|---|
| Normal | 1.5 ± 0.6 (7) | 12.0 ± 1.4 | 5.92 ± 0.84 (5) |
| cno/cno | 15.9 ± 2.9 (7)† | 2.0 ± 0.5† | 0.20 ± 0.03 (5)† |
| Genotype . | Bleeding time (min) . | No. dense bodies per platelet* . | Platelet serotonin (μg/109 platelets) . |
|---|---|---|---|
| Normal | 1.5 ± 0.6 (7) | 12.0 ± 1.4 | 5.92 ± 0.84 (5) |
| cno/cno | 15.9 ± 2.9 (7)† | 2.0 ± 0.5† | 0.20 ± 0.03 (5)† |
All values are mean ± SD; number in parentheses indicates sample number.
56 platelets analyzed from 3 cno/cno mice and 34 from 2 normal mice.
P < .001 by 2-tailed Student t test.
Severe λ-SPD, resulting in prolonged bleeding, in cappuccino mice.
(A) (B) Electron micrographs of air-dried, unstained normal (panel A) and mutant (panel B) platelets. The arrow illustrates one of the many dense bodies visible in the normal platelet and lacking in thecno/cno platelet shown. Most cno/cno platelets have no visible dense bodies, but a small number have 1 or 2. (C) Platelet aggregation (solid lines) was determined in whole blood by the impedance method in response to collagen (4 μg/mL). Note the reduction of cno/cno platelet aggregation. Thecno/cno platelet-aggregation response to low collagen (1 μg/mL) was likewise reduced (not shown). Release of ATP, determined simultaneously by luminescence (dashed lines), is undetectable incno/cno. Arrows indicate the time of collagen addition.
Severe λ-SPD, resulting in prolonged bleeding, in cappuccino mice.
(A) (B) Electron micrographs of air-dried, unstained normal (panel A) and mutant (panel B) platelets. The arrow illustrates one of the many dense bodies visible in the normal platelet and lacking in thecno/cno platelet shown. Most cno/cno platelets have no visible dense bodies, but a small number have 1 or 2. (C) Platelet aggregation (solid lines) was determined in whole blood by the impedance method in response to collagen (4 μg/mL). Note the reduction of cno/cno platelet aggregation. Thecno/cno platelet-aggregation response to low collagen (1 μg/mL) was likewise reduced (not shown). Release of ATP, determined simultaneously by luminescence (dashed lines), is undetectable incno/cno. Arrows indicate the time of collagen addition.
Alpha-granules appear normal in
cno/cno platelets. Transmission electron microscopy reveals a qualitatively normal structure and number of α-granules in cno/cno platelets. Representative photomicrographs showing normal (panel A) and cno/cno (B panel) platelets. m indicates mitochondria; db, dense bodies; ag, α-granules. Bar, 1 μm.
Alpha-granules appear normal in
cno/cno platelets. Transmission electron microscopy reveals a qualitatively normal structure and number of α-granules in cno/cno platelets. Representative photomicrographs showing normal (panel A) and cno/cno (B panel) platelets. m indicates mitochondria; db, dense bodies; ag, α-granules. Bar, 1 μm.
Abnormal lysosomal enzyme concentrations in cno/cnokidney and liver
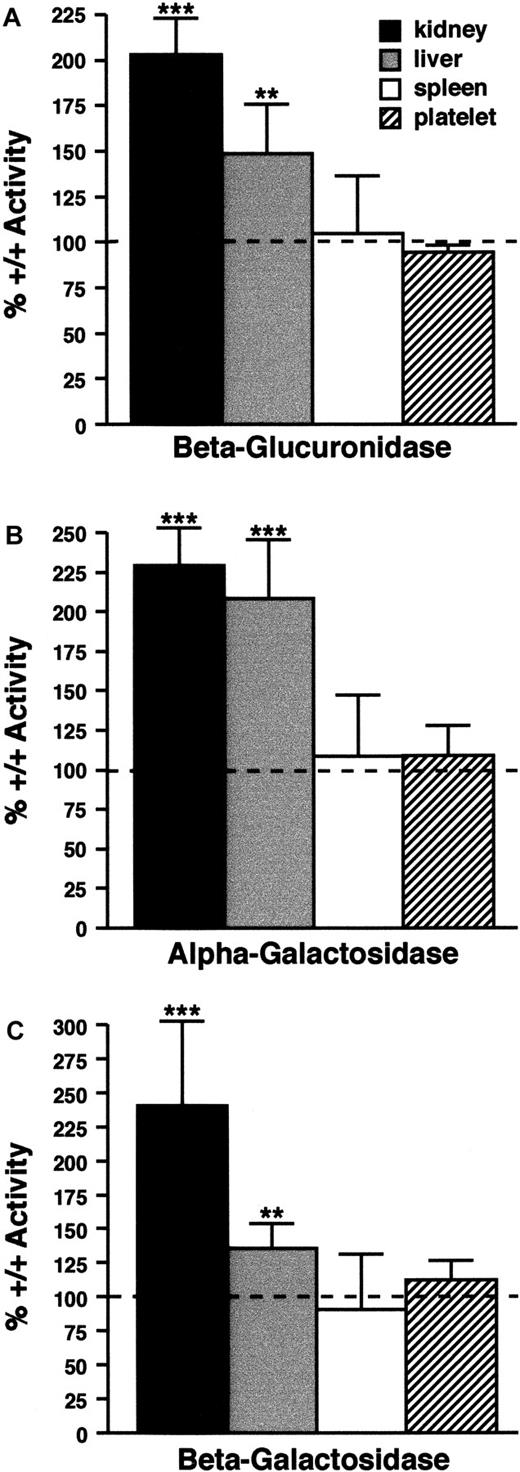
In cno/cno kidneys, β-glucuronidase, α-galactosidase, and β-galactosidase activities are increased 2.0 to 2.5 times (Figure 4). In this respect, cno resembles the majority of mouse SPD mutations, which cause a lysosomal secretion defect leading to enzyme accumulation in the kidney. However, cno differs in that it elevates enzyme levels in the liver (Figure 4). Abnormal liver lysosomal enzyme secretion has not been described in any other mouse SPD model. The cno/cno lysosome defect is not a generalized phenomenon, however, as spleen and platelet enzyme levels are normal (Figure 4). We conclude that cno/cno mice show the triad of organelle defects characteristic of HPS.
Lysosomal enzyme levels.
Lysosomal enzyme levels are abnormal in some cno/cnotissues. (A) (B) (C) β-glucuronidase (panel A), α-galactosidase (panel B), and β-galactosidase (panel C) activity in kidneys, liver, spleen, and platelets from cno/cno mice. Results are expressed as the percentage of wild-type activity (100%), indicated by the dashed line in each panel. Enzyme levels are significantly increased in cno/cno kidney and liver, but not spleen or platelets. **P < .01; ***P < .001; n = 5 for all determinations.
Lysosomal enzyme levels.
Lysosomal enzyme levels are abnormal in some cno/cnotissues. (A) (B) (C) β-glucuronidase (panel A), α-galactosidase (panel B), and β-galactosidase (panel C) activity in kidneys, liver, spleen, and platelets from cno/cno mice. Results are expressed as the percentage of wild-type activity (100%), indicated by the dashed line in each panel. Enzyme levels are significantly increased in cno/cno kidney and liver, but not spleen or platelets. **P < .01; ***P < .001; n = 5 for all determinations.
The cno mutation maps to Chr 5
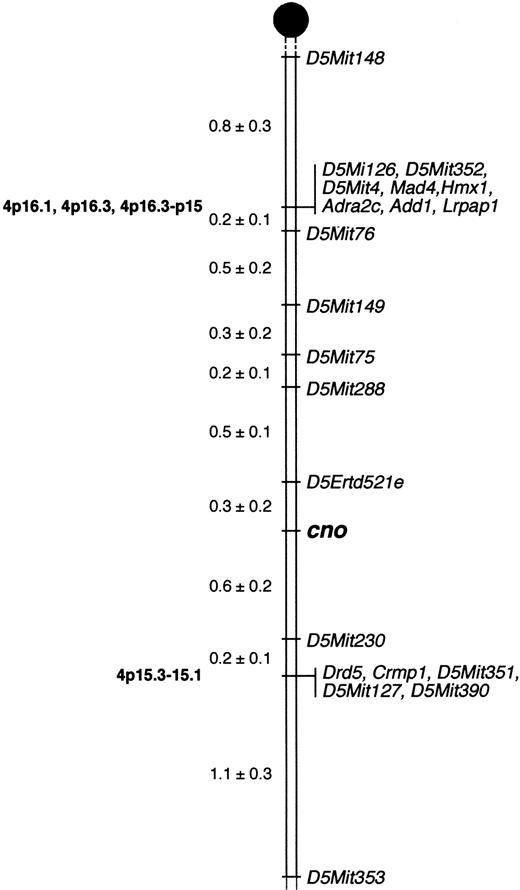
We mapped cno to Chr 5 (Figure5) by analyzing 1050 meioses obtained from 525 F2 intercross mice (see “Materials and methods”). Three recombinations occurred betweencno and the closest proximal marker, D5Ertd521e, corresponding to a linkage distance of 0.3 cM. Six recombinants were seen with the closest distal marker, D5Mit230, a linkage distance of 0.6 cM. Hence, the chromosomal interval containingcno is within 0.9 cM. These results will facilitate positional cloning of the gene. (Note: A linkage distance of 0.9 cM corresponds to a physical distance of 1.8 × 106base pairs, on the basis of estimates that 1 cM = 2 MB.44) No other known mouse HPS mutations map to this region of Chr 5, confirming that cno is a novel gene.
Localization of cappuccino.
Cappuccino localizes to mouse Chr 5. Mapping of anonymous DNA markers (Mit and Erdt) and multiple candidate genes relative to cnoin 525 offspring from an F2 intercross between C3H/HeJcno/cno and CAST/Ei narrowed the critical interval containing cno to a 0.9-cM region on proximal Chr 5. The percentage of recombination (± SE) between adjacent loci is given to the left. Missing typings were inferred from surrounding data where assignment was unambiguous. Vertical lines indicate nonrecombinant markers and genes. Locations of corresponding human genes, where known, are indicated on the left in bold. Gene names and references are available from The Jackson Laboratory Mapping Resource throughhttp://www.jax.org/resources/documents/cmdata.
Localization of cappuccino.
Cappuccino localizes to mouse Chr 5. Mapping of anonymous DNA markers (Mit and Erdt) and multiple candidate genes relative to cnoin 525 offspring from an F2 intercross between C3H/HeJcno/cno and CAST/Ei narrowed the critical interval containing cno to a 0.9-cM region on proximal Chr 5. The percentage of recombination (± SE) between adjacent loci is given to the left. Missing typings were inferred from surrounding data where assignment was unambiguous. Vertical lines indicate nonrecombinant markers and genes. Locations of corresponding human genes, where known, are indicated on the left in bold. Gene names and references are available from The Jackson Laboratory Mapping Resource throughhttp://www.jax.org/resources/documents/cmdata.
Analysis of candidate genes
Pearl23,45 and mocha24 mice have defects in components of the AP-3 complex. AP-3 is a heterotetrameric complex consisting of 4 ubiquitous subunits (δ, β3A, μ3A, and ς3A or ς3B). We mapped the mouse genes encoding ς3B (Ap3s2), μ3B (Ap3m2), μ3A (Ap3m1), ς3A (Ap3s1), and the Vps41 gene, which encodes a vesicular transport protein in yeast, on The Jackson Laboratory BSS mapping panel39 to identify potential candidate genes forcno. Figure 6 shows the map positions for these loci.
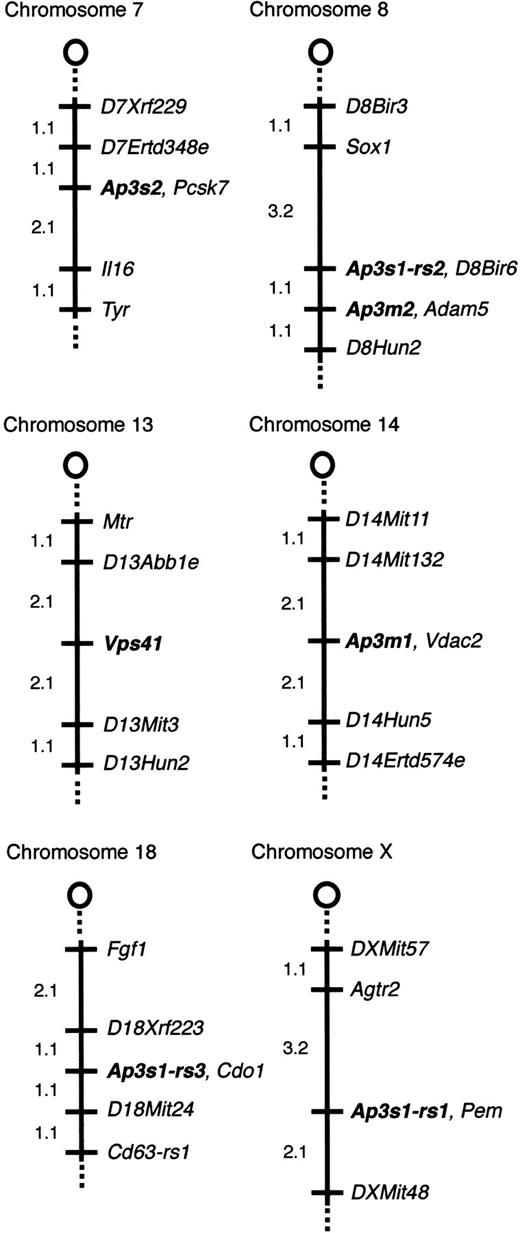
Chromosomal locations of
Ap3s2, Ap3m2,Vps41, Ap3m1, andAp3s1-related sequences. Thecno gene does not colocalize with AP-3 complex genes or the mouse homolog of Vps41. The relevant portion of each chromosome is depicted with the centromere toward the top. Loci are listed in order from the centromere. Loci mapping to the same position are listed in alphabetical order. The percentage of recombination between adjacent loci is given to the left of the figure. Missing typings were inferred from surrounding data where assignment was unambiguous. The panel data and references for mapping the other loci are available from The Jackson Laboratory Mapping Resource throughhttp://www.jax.org/resources/documents/cmdata.
Chromosomal locations of
Ap3s2, Ap3m2,Vps41, Ap3m1, andAp3s1-related sequences. Thecno gene does not colocalize with AP-3 complex genes or the mouse homolog of Vps41. The relevant portion of each chromosome is depicted with the centromere toward the top. Loci are listed in order from the centromere. Loci mapping to the same position are listed in alphabetical order. The percentage of recombination between adjacent loci is given to the left of the figure. Missing typings were inferred from surrounding data where assignment was unambiguous. The panel data and references for mapping the other loci are available from The Jackson Laboratory Mapping Resource throughhttp://www.jax.org/resources/documents/cmdata.
Ap3s2 is nonrecombinant with Pcsk3, placingAp3s2 on Chr 7, approximately 39 cM distal to the centromere, a region that shows conserved homology with human 15q26.46 The mouse mutation taupe (tp) is located on Chr 7 at approximately 42 cM.39 Taupe homozygotes have diluted coat color47 but normal bleeding times.6 The mouse SPD mutation reduced pigmentation (rp) is located on Chr 7 significantly proximal toAp3s2 at approximately 2 cM, and ruby-eye 2 (ru2) is at approximately 24 cM.46
Ap3m2 is nonrecombinant with Adam5 on Chr 8, approximately 8 cM distal to the centromere, a region conserved with human chromosome 8p12-p11. No mouse SPD mutations localize to Chr 8.
Ap3m1 maps to proximal (approximately 2 cM) Chr 14, a region conserved with human 10q24. No SPD mutations have been mapped in this region.
Multiple Ap3s1-related fragments were detected by Southern blotting of genomic DNA. Analysis of the BSS mapping panel revealed 3 independently segregating, closely related sequences.Ap3s1-rs1 (related sequence 1) localizes to the X chromosome. Ap3s1-rs2 maps to Chr 8, approximately 1 cM proximal to Ap3m2, and Ap3s1-rs3 maps to Chr 18. Notably, PCR amplification of genomic DNA using mouseAp3s1-specific primers produces multiple products (not shown), again suggesting the presence of highly related sequences.
Vps41 maps to Chr 13, approximately 8 cM distal to the centromere, a region conserved with human 7q13-p13. Several mouse mutants map to proximal Chr 13, including satin (sa, 17 cM) and the SPD mutations muted (mu) and sandy (sdy) (at 21 and 22 cM, respectively).
Together, these data exclude these 7 genes as candidate genes forcno and suggest that additional genes encoding sigma-related isoforms of the AP-3 complex may exist.
The distribution of AP-3 components and LAMP-1 antibody internalization are normal in cno/cno fibroblasts
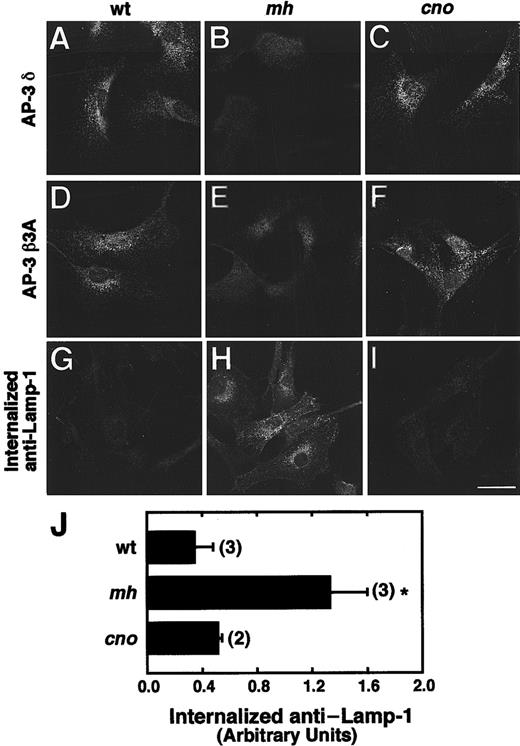
The cellular distribution of AP-3 subunits and the internalization of anti–LAMP-1 in wild-type, mocha, and cappuccino fibroblasts were examined. The distribution of the δ and β3A AP-3 subunits is normal in cno/cno (Figure 7C,F), and as previously described,24 both subunits are markedly diminished in mh/mh (Figure 7B,E) fibroblasts compared with wild-type (Figure 7A,D).
The distribution of AP-3 proteins and trafficking of LAMP-1 in cno/cno fibroblasts.
The distribution of AP-3 proteins and trafficking of LAMP-1 are normal in cno/cno fibroblasts. (A-F) Immunofluorescence staining of fibroblasts derived from wild-type C3H/HeJ (wt; panels A and D), mocha (mh; panels B and E) and cappuccino (cno; panels C and F) mice, with the use of antibodies to the δ (panels A-C) and β3A (panels D-F) subunits of the AP-3 complex. Note the characteristic punctate pattern of AP-3 in the cytoplasm of wt andcno/cno fibroblasts and its apparent absence frommh/mh fibroblasts. (G-I) Fluorescence microscopy detection of anti–LAMP-1 monoclonal antibody internalized by wt (panel G),mh/mh (panel H) and cno/cno (panel I) fibroblasts for 15 minutes at 37°C. Bar, 20 μm. (J) Quantitation of antibody internalization experiments. The amounts of internalized antibody in samples, prepared as in panels G through I, were estimated by image analysis of 5 randomly selected fields, as described.26Background-corrected values are expressed in arbitrary units of fluorescence per cell (mean ± SD of the number of experiments indicated in parentheses). *P < .01.
The distribution of AP-3 proteins and trafficking of LAMP-1 in cno/cno fibroblasts.
The distribution of AP-3 proteins and trafficking of LAMP-1 are normal in cno/cno fibroblasts. (A-F) Immunofluorescence staining of fibroblasts derived from wild-type C3H/HeJ (wt; panels A and D), mocha (mh; panels B and E) and cappuccino (cno; panels C and F) mice, with the use of antibodies to the δ (panels A-C) and β3A (panels D-F) subunits of the AP-3 complex. Note the characteristic punctate pattern of AP-3 in the cytoplasm of wt andcno/cno fibroblasts and its apparent absence frommh/mh fibroblasts. (G-I) Fluorescence microscopy detection of anti–LAMP-1 monoclonal antibody internalized by wt (panel G),mh/mh (panel H) and cno/cno (panel I) fibroblasts for 15 minutes at 37°C. Bar, 20 μm. (J) Quantitation of antibody internalization experiments. The amounts of internalized antibody in samples, prepared as in panels G through I, were estimated by image analysis of 5 randomly selected fields, as described.26Background-corrected values are expressed in arbitrary units of fluorescence per cell (mean ± SD of the number of experiments indicated in parentheses). *P < .01.
The surface expression and internalization of lysosomal membrane proteins (eg, CD63, LAMP-1, LAMP-2) is increased in fibroblasts from human HPS patients with defects in the AP-3 complex, indicating that AP-3 is involved in the intracellular trafficking of lysosomal membrane proteins.26 Consistent with these findings, LAMP-1 internalization is increased in mh/mh fibroblasts (Figure7H) compared with wild-type (Figure 7G). Notably, LAMP-1 internalization in cno/cno fibroblasts (Figure 7I) is normal. Quantitative analyses confirmed the fluorescence microscopy data (Figure 7J). Together, the data indicate that the cnogene functions in a pathway critical to organelle biogenesis that is independent of the AP-3 complex.
No gross pathology is evident in mutant tissues at 10 months of age
Considerable morbidity is associated with human HPS owing to the development of rapidly progressing fibrotic restrictive lung disease in the fourth to fifth decades of life.7,9,48 In a recent analysis of lung pathology in mouse models of SPD, alterations in the lung architecture were described.49 Lesions were noted at 22 months of age and characterized as enlarged airspaces, resulting in a “honeycomb” histological appearance. With the exception of one mutant, light ear (le), the severity of the lung lesions inversely correlated with life-span. We examined several tissues (see “Materials and methods”) from 10-month-old cno/cno mice. No gross abnormalities were detected at the light microscopy level in any of the tissues surveyed, including the lung.
Behavioral abnormalities suggest otolith defects in cnomice
Abnormal postural and balance reflexes can be discerned in approximately 75% of cno homozygotes. A broad range of severity is seen. Some mice show abnormal head tilting and balance defects that are apparent only upon handling and close observation. Others show extreme leaning to one side and are unable to stand upright. These observations suggest that cno causes inner ear defects similar to mu, mh, and pa.
Discussion
The autosomal recessive mutation cappuccino was first identified in an affected mouse because of the severe coat-color dilution. Many pigment-dilution mutations in the mouse are models of human platelet SPD, a heterogeneous group of diseases that includes HPS and CHS.6 Here, we demonstrate that cappuccino homozygotes show the classic HPS phenotype: severe oculocutaneous albinism, markedly prolonged bleeding time, and tissue-specific abnormal lysosomal enzyme secretion. Immunofluorescence staining with LAMP-1 revealed no evidence of giant granules in cno/cnofibroblasts (data not shown), as is seen in CHS.
In homozygous cno mice, melanosomes are immature and dramatically decreased in number; platelet dense bodies are deficient in ATP and serotonin; and lysosomal enzyme concentrations are elevated in kidney and liver, but normal in spleen and platelets. Abnormal postural reflexes observed in cno homozygotes indicate that the cno mutation belongs to the subset of mouse SPD mutations in which otoliths are abnormal, bringing the number in this subfamily to 4. Although human HPS is associated with fibrotic lung disease and granulomatous colitis,15 no tissue pathology is evident in cno/cno mice at 10 months of age. To determine whether cno/cno mice develop abnormal lung architecture with age, as occurs in several mouse HPS models by 22 months,49will require further analyses once more aged mice become available.
Genetic mapping narrowed the location of cno to a 0.9-cM interval on Chr 5 flanked proximally by D5Ertd521e and distally by D5Mit230, confirming that cno is distinct from the other known mouse SPD mutations. Multiple genes known to localize to proximal Chr 5 were mapped relative to cno; none colocalize with cno. This analysis eliminates several potential cno candidate genes, such as alpha adducin (Add1), which is ubiquitously expressed and present in platelets.50 In addition, we mapped several genes encoding components of the AP-3 complex and the mouse homolog ofVps41, proteins known to function in intracellular trafficking. None of these genes mapped to Chr 5, eliminating them as candidate genes for cno.
Mapping of the gene encoding the sigma 3A subunit of AP-3 (Ap3s1) indicated the presence of 3 highly homologous genes on mouse chromosomes X, 8, and 18. At present, it is not possible to assign Ap3s1 unequivocally to any of these map positions. Additional analyses will be required to isolate and characterize theAp3s1-related sequences. These data suggest that additional isoforms related to the sigma subunit of the AP-3 complex may exist.
All 4 subunits of the AP-3 complex are missing in mocha homozygotes, presumably because the loss of the δ subunit destabilizes the entire complex.24 In pearl mice, in which the β3A subunit is defective, all other AP-3 subunits are decreased in all tissues examined except the brain, reflecting the existence of the neuronal-specific β3B subunit.51 Here, we examined the status of β3A and δ subunits of AP-3 in cno/cnofibroblasts by immunofluorescence. No differences in protein levels or intracellular distribution were observed compared with wild-type.
Human patients deficient in HPS1 protein (HPS type1) show normal trafficking of lysosomal integral membrane proteins, such as CD63 and LAMP-1.20 Type 2 HPS patients, however, in whom the β3A subunit of the AP-3 complex is defective, have increased trafficking of lysosomal proteins through the plasma membrane.26Consistent with this observation, plasma membrane trafficking of lysosomal proteins is increased in mocha and pearl mice. All other mouse SPD mutants tested (pallid, cocoa, muted, sandy, reduced pigmentation, ruby eye, ruby eye 2, and light ear) show normal trafficking.20 In this study, we found that trafficking of lysosomal membrane proteins in cno/cno mice is normal. Together, the genetic mapping and functional analyses provide compelling evidence that the cno defect does not involve AP-3 complex genes or affect their function.
No evidence of a stable association between the HPS1 protein and AP-3 proteins has been detected.20 Because HPS-1 and HPS-2 patients show normal and abnormal lysosomal membrane protein trafficking, respectively, it appears that HPS can occur in humans and in mice through defects in distinct pathways that do not alter protein trafficking in lysosomes (and, presumably, platelet dense bodies and melanosomes). An example is the pallid mouse, in which the syntaxin 13–binding protein, pallidin, is defective, suggesting that vesicle docking and fusion events downstream of AP-3–dependent processes in organelle biogenesis are disrupted.27 Hence, the potential for identifying other genetic causes of HPS in humans and for elucidating fundamental biological mechanisms by cloning cnoand the remaining mouse SPD genes is apparent.
In summary, we have characterized a new mouse model of HPS,cno (cappuccino). Its phenotype is similar to other severe HPS mouse mutations in that bleeding is prolonged owing to a marked deficiency of platelet dense body contents, and severe oculocutaneous albinism occurs as a result of quantitatively and qualitatively abnormal melanosome formation. Lysosomal enzyme concentrations are elevated in the kidney, as occurs in all but 3 of the other mouse HPS mutants. Unique to homozygous cno mice, however, liver lysosomal enzyme concentrations are also increased. The explanation for this observation is presently unknown. Extensive linkage analyses have eliminated many potential candidate genes, including genes encoding AP-3 complex subunits, and narrowed the cno critical interval to a 0.9-cM region of Chr 5. The AP-3 complex is intact and lysosomal membrane protein trafficking is normal in cno/cnofibroblasts. The data indicate that the cno gene product causes HPS symptoms via an AP-3–independent pathway. Taken together, studies of mouse HPS mutations to date make it clear that the development of organelles—with the correct complement of proteins and targeted to the correct location in the cell—is extremely complex, involving multiple genes and interacting gene pathways.
Acknowledgments
We thank Jane E. Barker and Lucy B. Rowe for critical review of the manuscript; Mary E. Barter for assistance with mapping figures; Jennifer L. Smith for graphics; and Marie Perry and Kim Gray for identifying the first cappuccino mutants in the Jax Research Systems' C3H/HeJ breeding colony.
Supported by National Institutes of Health grants HL55321 (L.L.P.), RR01183 (E.M.E.), HL31698 (R.T.S.), and EY12104 (R.T.S.); The March of Dimes (L.L.P.) and American Heart Association (L.L.P.); and The National Cancer Institute CA34196 (The Jackson Laboratory).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Luanne L. Peters, The Jackson Laboratory, 600 Main St, Bar Harbor, ME 04609; e-mail: luanne@aretha.jax.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal