Abstract
A 38-kd protein that associates with F-actin structures in activated platelets and endothelial cells was purified, cloned, and characterized. The protein contains an N-terminal PDZ motif, a large intervening sequence, and a C-terminal LIM domain and was identified as the human homolog of rat CLP36. The study showed that CLP36 associates with actin filaments and stress fibers that are formed during shape change and spreading of platelets and during migration and contraction of endothelial cells. CLP36 binds to α-actinin-1 as shown by coimmunoprecipitation, pull-down experiments, yeast 2-hybrid analysis, and blot overlay assays and colocalizes with α-actinin-1 along endothelial actin stress fibers. In contrast to α-actinin-1, CLP36 was absent from focal adhesions in both activated platelets and endothelial cells. The N-terminal part of CLP36 containing the PDZ domain and the intervening region, but not the LIM domain, targeted enhanced green fluorescent protein fusion proteins to stress fibers in endothelial cells. Yeast 2-hybrid analysis demonstrated that the intervening sequence, but not the PDZ or the LIM domain of CLP36, binds to the spectrinlike repeats 2 and 3 of α-actinin-1. The study further shows that CLP36 binds to α-actinin in resting platelets and translocates as a CLP36/α-actinin complex to the newly formed actin cytoskeleton in activated platelets. The results indicate that CLP36 binds via α-actinin-1 to actin filaments and stress fibers in activated human platelets and endothelial cells. The study suggests that CLP36 may direct α-actinin-1 to specific actin structures and at this position might modulate the function of α-actinin-1.
Introduction
The actin cytoskeleton is a complex protein network that not only provides cellular structure but is also fundamental for cellular dynamics, such as shape change, migration, spreading, and contraction of nonmotile cells.1,2 Endothelial cells exhibit on activation by inflammatory, atherogenic, or hemostatic stimuli a rapid change of their actin cytoskeleton to rounded contracted cells with stress fibers.3-5 Stress fibers have been recognized as the contractile organelle of nonmuscle cells bearing structural and functional resemblance to the myofibrils of skeletal muscle.6 They are not only found in vitro in cultured cells but are also an in vivo phenomenon in arterial vascular endothelial cells,7,8 where they are prominent in regions of high-shear stress and above early atherosclerotic lesions.9 10
Blood platelets show within seconds of activation a dramatic reorganization of the cytoskeleton such as the formation of new actin filaments and filament bundles, contractile actomyosin rings, and stress fiber–like structures. This rapid reorganization of the cytoskeleton underlies and leads to platelet shape change and spreading.11,12 It is becoming clear that the cytoskeletal remodeling on cell activation is mediated by high-affinity interactions between specific proteins. On stimulation of platelets, proteins that interact directly with F-actin, such as myosin and actinin, rapidly translocate to the F-actin–rich cytoskeleton.13 Also the small GTP-binding proteins Rho, Rac, and CDC42 that regulate cytoskeletal structures and are activated on platelet stimulation associate with the actin cytoskeleton in stimulated platelets.14-17 We have observed previously that proteins of 38-kd molecular mass translocated to the cytoskeleton in activated platelets.18 Here we report the purification and cloning of a 38-kd protein that was identified as the human homolog of rat CLP3619 and almost identical to hCLIM1 isolated from a human adenocarcinoma complementary DNA (cDNA) library.20
CLP36 contains a N-terminal PDZ domain, a large intervening sequence, and a C-terminal LIM domain. The PDZ and LIM domains are modular protein interaction motifs21,22 and mediate protein association with the cytoskeleton and with proteins involved in signal transduction.23-28 We found that CLP36 associates with actin filaments and stress fibers in activated human platelets and endothelial cells. This association is mediated through its constitutive binding to α-actinin-1, which involves the intervening region of CLP36 and the spectrinlike repeats 2 and 3 of α-actinin-1. The results were presented in part previously at the 7th Erfurt conference on platelets and have been published in abstract form.29
Materials and methods
Cell culture
Human erythroleukemia (HEL) cells for cDNA cloning were grown in RPMI 1640 medium (Biochrom KG, Berlin, Germany) containing 10% fetal calf serum, 200 mmol/L glutamine, and penicillin/streptomycin (10 000 U/mL) to a density of 40 000 cells/mL medium. Human umbilical arterial and venous endothelial cells were obtained and cultured as described.4
Purification of CLP36 from platelets
Platelets from human volunteers were treated with acetylsalicylic acid and isolated as described.30 Proteins of 38 kd have previously been found to translocate to the cytoskeleton of activated platelets.18 We purified one platelet protein of 38-kd molecular mass with an isoelectric pH (pI) of 6.5 by preparative isoelectric focusing, using the Rotofor-chamber (BioRad Laboratories, Hercules, CA) and subsequent 2D-polyacrylamide gel electrophoresis. After partial gel digestion of the protein with endoproteinase Lys-C from Lysobacter enzymogenes (Boehringer Mannheim, Germany), the resulting peptides were separated by reversed-phase high-pressure liquid chromatography and microsequenced. Seven peptides were obtained and used to search the expressed sequence tag (EST) database from The Institute for Genomic Research (TIGR), resulting in a complete match with THC 94 235 (Tentative Human Consensus Sequence).
cDNA cloning
Total RNA was isolated from HEL cells, using RNeasy (Qiagen, Hilden, Germany) and purified through oligo(dT)-cellulose (Sigma, Deisenhofen, Germany). First-strand cDNAs were synthesized with oligo(dT)-priming and Moloney murine leukemia virus (M-MLV) reverse transcriptase (GIBCO BRL, Karlsruhe, Germany). To amplify the clp36 cDNA, polymerase chain reaction (PCR) was performed with specific primers, based on the open reading frame (ORF) in THC 94 235 (sense 5′-ACCACCCAGCAGATAGAC-3′ and antisense 5′-AACCAAAGTAAGCAGAGAAC-3′). The PCR product of 1.1 kilobase (kb; expected length 1.093) was electro-eluted from the gel at a constant voltage of 2 V/cm for 3 hours, purified by phenol/chloroform extraction, reamplified by PCR, and extracted from low-melt agarose (QIAquick Gel Extraction Kit, Qiagen). The clp36 cDNA was blunt-end inserted into the HincII site of the pCR-Script Amp SK+vector (Stratagene, La Jolla, CA) and cloned in Escherichia coli XL1-blue (Stratagene). With the use of the Sac I and Hind III site, the clp36 cDNA was subcloned into the pQE-32 vector. For synthesis of glutathione-S-transferase (GST)-tagged LIM motif of CLP36 (amino acid residues (aa) 257-329; GST-LIM), the lim cDNA was subcloned into the pGEX-5X-2 (Pharmacia, Freiburg, Germany) vector by digestion with Xho I and Eco RI.
clp36 cDNA was also obtained from human umbilical venous endothelial cells (HUVECs). RNA was isolated from HUVECs using PureScript (Biozym, Oldendorf, Germany), and cDNA was synthesized using the CapFinder PCR cDNA Synthesis Kit (Clontech). Endothelial clp36 cDNA was obtained by PCR using the clp36 specific primers.
DNA sequencing
The clp36 cDNA from endothelial cells and HEL cells, the various His-tag and enhanced green fluorescent protein (EGFP)-fusion constructs from clp36, and the various clp36 constructs for the yeast 2-hybrid were sequenced on both strands by MediGenomix (München, Germany) and MWG (Ebersberg, Germany).
Expression of recombinant CLP36
The His-tagged CLP36 and His-tagged LIM of CLP36 were expressed in E coli XL1-blue and purified nondenaturating by Ni-NTA affinity chromatography according to instructions of the manufacturer (Quiagen). GST-LIM of CLP36 was expressed in E coli NM 522 and purified with glutathione agarose beads (Sigma).
Antibody production
Two potentially immunogenic peptides of CLP36 were chosen according to their hydrophilicity and sequence difference to homologous proteins. Peptide 14 (EDQIYCEKHARER; CLP36 [302-314]) and Peptide 17 (ESEEKGDPNKPSGFRS, CLP36 [224-239]) were synthesized and coupled through an N-terminal cysteine to KLH (Pierce, Rockford, IL). Antibodies to recombinant His-CLP36 and the 2 peptides of CLP36 were raised in rabbits (pab production, Herbertshausen). Antisera were purified using protein A columns (Protein A-Sepharose CL-4B, Sigma).
Immunoblots
Proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, blotted, and detected by enhanced chemoluminescence as described.30 The dilutions of the rabbit anti-CLP36 immunoglobulin and the monoclonal mouse anti–α-actinin immunoglobulin M (IgM) (BM-75.2; Sigma) were 1:20 000 and 1:5000, respectively. The dilutions of the horseradish-peroxidase-linked secondary antimouse and antirabbit antibodies (Amersham) were both 1:10 000.
Expression of CLP36 in platelets was estimated by loading a SDS gel with increasing amounts of GST-LIM and platelet proteins of a defined cell number. The amount of loaded GST-LIM molecules was calculated by measuring absorption at 280 nm, using the extinction coefficient of GST (ε = 40 920 M−1 cm−1). The proteins were blotted, and CLP36 and GST-LIM were immunodetected with anti–14-peptide antibody and quantified densitometrically. CLP36 in the platelet samples and GST-LIM were quantified densitometrically, and the amount of platelet CLP36 was calculated based on the GST-LIM standard curve.
Immunoprecipitation
Suspensions of washed platelets were adjusted to a concentration of 1 × 109 cells/mL. Unstimulated platelets, platelets preincubated with cytochalasin D (2 μmol/L) for 2 minutes, or BAPTA-AM (60 μmol/L) and EGTA (5 mmol/L) for 30 minutes, or platelets activated by 1 U/mL thrombin (Boehringer Mannheim) for 1 minute were lysed in an equal volume of 2 × lysis buffer (“RIPA”; pH 7.4, 2% Triton X-100, 2 mmol/L EGTA, 100 mmol/L HEPES, 150 mmol/L NaCl, 2 mmol/L sodium orthovanadate, 1 tablet protease inhibitor cocktail/5 mL [No. 1 836 153, Boehringer Mannheim]) for 20 minutes on ice. The lysates were clarified by centrifugation at 15 600g for 10 minutes at 4°C. The supernatant was precleared by incubation with 5 μg/mL preimmune serum and Protein A Sepharose CL-4B for 1 hour, followed by centrifugation at 15 600g for 10 minutes. The supernatant was incubated for 1 hour at 4°C with 15 μg anti-CLP36 antibody and, subsequently, with Protein A Sepharose (0.75 mg in 10 μL RIPA) for 1 hour. All steps were performed at 4°C. After centrifugation, the immunoprecipitates were washed 3 times with 1.5 mL of ice-cold lysis buffer, and resuspended in 2 × sample buffer (125 mmol/L Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 3.1% dithiothreitol, 0.01 mg/mL bromphenol blue) and treated for 5 minutes at 95°C before gel separation.
To determine the binding domains of CLP36 to α-actinin, platelet lysates were incubated with the His-tagged fusion proteins His-CLP36, His-CLP36ΔLIM (aa 1-257) or His-LIM (aa 258-329) (1 hour, 10 μg each). The his-tag fusion proteins and associated proteins were subsequently pulled down with anti–RGS-[His]4 antibody (5 μg; Qiagen). The precipitates were analyzed on silver-stained protein SDS gels for the presence of α-actinin.
Blot overlay assay
Platelet proteins resolved by SDS-polyacrylamide gel electrophoresis (PAGE) were blotted onto nitrocellulose membranes (Amersham). Membranes were then blocked overnight at room temperature in triethanolamine-buffered saline (TBS), 0.05% Tween-20, 3% nonfat dry milk (Bio-Rad). Subsequent incubations with biotinylated His-tagged CLP36 (100 ng/mL; biotinylation with N-+-Biotinyl-6-amino-capronsäure-N-succinimidylester) were performed for 4 hours at room temperature in incubation buffer (TBS, 0.05% Tween-20, 0.5% nonfat dry milk [Biorad]). The blots were washed 3 times for 10 minutes with incubation buffer, incubated with Extravidin-peroxidase (Sigma) for 1 hour at room temperature, and washed 3 times with TBS, 0.05% Tween-20. Bands were detected by enhanced chemiluminescence (Amersham).
Isolation of cytoskeletal and membrane skeletal fractions of platelets
Cytoskeletal fractions were prepared similarly to the method described previously,17 31 using a 2 × Triton lysis buffer (pH 7.3) containing 2% Triton X-100, 0.1 mol/L Tris-HCl, 0.2 mol/L NaCl, 4 mmol/L MgCl2, 20 mmol/L EGTA, 200 μmol/L Pefablock-SC (Boehringer Mannheim), 20 μg/mL leupeptin, and 200 μg/mL aprotinin.
Platelet spreading
Platelet spreading was carried out as described32with some modifications. Round glass coverslips (Eppendorf) were coated for 1 hour with 100 μL poly-l-lysine (0.1 mg/mL) (Sigma), washed twice with phosphate buffered saline (PBS), air dried, and put into 24-well culture dishes. Aliquots (0.2 mL) of washed platelet suspensions or, in some experiments, platelet-rich plasma were adjusted to 3 × 107 cells/mL and pipetted on the coverslips, which were centrifuged for 5 minutes at 250g at room temperature. Platelets were allowed to spread by incubation at 37°C for 5 to 10 minutes. Platelets not attached were removed by rinsing the coverslips with PBS.
Immunofluorescence microscopy
Spread platelets or endothelial cells (subconfluent and confluent), grown on coverslips, were fixed with fixation buffer (60 mmol/L Pipes, pH 6.1, 25 mmol/L Hepes, 10 mmol/L EGTA, 3 mmol/L MgCl2) containing 3.7% formaldehyde (Sigma) for 10 minutes at room temperature or overnight at 4°C. Coverslips were rinsed briefly with PBS containing 1 mmol/L MgCl2 and 1 mmol/L CaCl2, bathed in permeabilization buffer (0.2% Triton X-100 in PBS) for 5 minutes at room temperature and washed 3 times with PBS. Cells were incubated with the primary antibodies or rhodamine-phalloidin (Molecular Probes, Leiden, The Netherlands) in a moist chamber at room temperature for 1 hour. F-actin was stained with rhodamine-phalloidin (50-fold dilution). For primary antibodies we used: The polyclonal anti-CLP36 immunoglobulin (25 μL of 30 μg/mL corresponding to 0.75 μg) or anti–17-peptide immunoglobulin, the monoclonal anti–α-actinin antibody (clone BM-75.2; Sigma) and the monoclonal antivinculin antibody (ICN, Aurora, OH), diluted as recommended. Preimmune serum immunoglobulin (25 μL of 30 μg/mL) or incubation of permeabilized cells with His-CLP36 (16 μg) for 1 hour before or 12 hours after addition of anti-CLP36 antibody (0.75 μg) served as control for the specific staining of CLP36.
The coverslips were washed 3 times and incubated in a solution containing the secondary antibodies fluorescein (FITC)-conjugated goat antirabbit IgG (Dianova, Hamburg, Germany) or Alexa 568 goat-anti mouse IgG dye (sulfonated rhodamine derivative, Molecular Probes), all diluted 1:200. After 3 washing steps, the cells were mounted in Immuno Floure Mounting Medium (ICN). An inverted Leica microscope equipped with the TCS confocal system and an Ar/Kr laser was used to obtain images through × 63 and × 100 objectives (type UV 1,32NA oil PL APO). Stacks of images were analyzed using the LeicaNT software. Cells in Figure 3Ai-Ciii, Figure 4, and Figure 6 were viewed on a Leica fluorescence microscope RBM 3.
Cloning of EGFP-clp36 constructs
Sequences of clp36 (nt 1-987), clp36ΔLIM (nt 1-771), and clp36LIM (nt 772-987) were cloned in pβactin-EGFP developed in the laboratory of Dr A. Matus (Friedrich Miescher-Institute, Basel). pβactin-EGFP is identical to the vector described by Ludin et al,33 except that EGFP (Clontech) was used instead of GFP. The HindIII site and the ClaI site in the polylinker at the 5′-end of the EGFP sequence were used in cloning. The amino terminal primers contained the Kozak consensus sequence GCC GCC AGC CAT GA after the HindIII site for efficient expression. cDNA sequences for the construction of gene fusion were obtained by PCR for clp36 on a pCR-Script Amp SK+ vector containing clp36 cDNA from HEL cells (see above). The EGFP fusion plasmids were transfected in HUVECs by electroporation.
Electroporation of HUVECs
Cells (2 × 106) were treated with trypsin/EDTA, spun down, washed with PBS, and resuspended in 1 mL cold PBS. Cells were incubated with 20 μg EGFP fusion plasmid for 5 minutes at 0°C and electroporated at 240 V and 960 μF, using the BioRad gene pulser II. Transfected cells were incubated at 0°C for 7 minutes, resuspended in 10 mL pre-warmed RPMI 1640, and plated onto collagen-coated coverslips (see below). After an overnight period of gene expression, cells were fixed and mounted as described (see below) and viewed on a Leica fluorescence microscope RBM 3.
Yeast 2-hybrid interactions
To define the interacting domains between CLP36 and α-actinin-1, the Gal4-based Matchmaker 2-Hybrid System 2 (Clontech) was used. Nonmuscle α-actinin-1 cDNA was obtained from endothelial cell cDNA, using the primers (based on the α-actinin-1 sequence) 5′-ATGGACCATTATGATTCTCAGC-3′ (forward) and 5′-TTAGAGGTCACTCTCGCCG-3′ (backward). The PCR-product was used to reamplify the spectrinlike repeat 1, spectrinlike repeats 1-2, spectrinlike repeats 2-3, spectrinlike repeats 1-3 of α-actinin-1, and full-length α-actinin-1. These constructs were subcloned into pACT-2 (“prey”), using the EcoRI and XhoI cloning sites. CLP36, CLP36 (aa 1-82), CLP36 (aa 1-104), CLP36 (aa 1-127), CLP36 (aa 81-256), CLP36 (aa 1-256), and CLP36 (aa 257-329) were amplified from the CLP36 cDNA of HEL cells and subcloned into pAS2-1 (“bait”), using the NdeI and PstI cloning sites. Bait and prey plasmids were cotransformed into electrocompetent Saccharomyces cerevisiae Y190 (Clontech) (pulse 1, 5 kV, 25 μF, 200 Ω) and plated onto synthetic dropout agar (SD medium) containing histidine but no tryptophan and leucin to select for clones carrying both plasmids. Protein expression of CLP36 constructs was confirmed by Western blot analysis. To screen for 2-hybrid interactions, the β-galactosidase activity was determined by qualitative blue/white screening using the Colony-Lift Filter Assay as recommended by the manufacturer (Clontech). The LacZ reporter gene in yeast strain Y190 is under the control of the GAL1 upstream activating sequence and, therefore, inducible by a positive 2-hybrid interaction. To assay yeast transformants for LacZ reporter gene expression, colonies were plated on SD medium lacking tryptophan and leucine.
To verify positive and negative interactions from the β-galactosidase assay, yeast transformants were also tested for HIS3 reporter gene expression by growth on selective SD Leu−Trp−His− agar plates. 3- Aminopteridin (30 mmol/L) was added to the medium to suppress constitutive leaky expression of the HIS3 gene. Clones that turned out to be positive in the β-galactosidase assay were also able to grow on HIS− selective SD agar. The growth of colonies that were negative for β-galactosidase gene expression was completely suppressed (not shown). Therefore, both reporter genes LacZ and HIS3 signaled the same positive and negative 2-hybrid interactions.
Results
Protein purification and cloning of CLP36
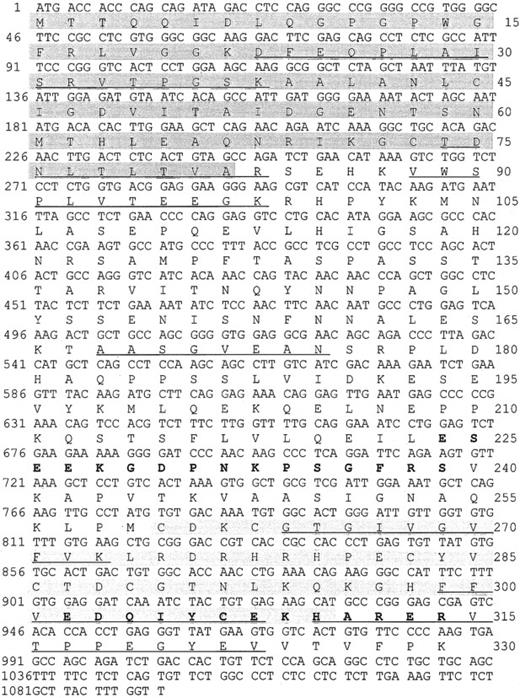
We purified a 38-kd protein from human platelets and obtained 7 peptides. Their sequences mapped completely with a tentative human consensus sequence present in the EST database. On the basis of this sequence information, specific primers were designed, and the cDNA of the 38-kd protein was obtained by PCR using cDNA from HEL cells. The cDNA contains a 987-bp ORF encoding a protein of 329 amino acids with a calculated molecular mass of 36 072 Da, and a pI of 6.56. Within the deduced amino acid sequence of the ORF, all 7 peptides, which had been obtained by microsequencing of purified P38, were identified (underlined in Figure 1). The protein shows 88% sequence identities with the C-terminal LIM-domain protein of 36 kd from both rat and mouse (Genbank/EMBL accession no. U 23769 and AF 053367). The protein was, therefore, denoted human CLP36. Rat CLP36 was previously cloned from rat hepatocytes by differential screening of a substractive (normoxic minus hypoxic) cDNA library.19 Clp36 was also sequenced after PCR amplification of HUVEC cDNA and found to be identical with that from HEL cells. Furthermore, alignment of the clp36 nucleotide sequence with the sequence data in EST databases (dbEST and preblast_est), which were obtained from many human tissues and cells, showed complete identity to more than a hundred different EST sequences. This is important, because an almost identical human clp36 sequence has been described recently and has been designated as hCLIM1.20 This gene has 3 different nucleotides in positions 60, 61, and 63 that could not be found in the available EST-sequence databases. The latter 2 nucleotide exchanges lead to a glycine-to-arginine exchange in position 21 of hCLIM1.20 Glycine in position 21 is within a conserved domain of the PDZ motif and also present in the other proteins of the PDZ/LIM-domain proteins (see below). One possible reason for the sequence difference between clp36 and hCLIM1 is that hCLIM1, which had been isolated from a human adenocarcinoma cDNA library,20is a mutated form of clp36.
Nucleotide sequence and deduced amino acid sequence of the human clp36 cDNA from HEL cells.
An ORF within clp36 cDNA consisting of 987 base pair (bp) codes for a protein of 329 amino acid residues. Numbering of nucleotides and amino acid residues is at the left and right margins, respectively. Underlined in the amino acid sequence are 7 peptide sequences obtained by microsequencing of purified human platelet CLP36. Pale shading indicates the PDZ domain, whereas gray shading indicates the LIM domain. The 2 peptides that were selected to generate antipeptide antibodies are displayed in bold.
Nucleotide sequence and deduced amino acid sequence of the human clp36 cDNA from HEL cells.
An ORF within clp36 cDNA consisting of 987 base pair (bp) codes for a protein of 329 amino acid residues. Numbering of nucleotides and amino acid residues is at the left and right margins, respectively. Underlined in the amino acid sequence are 7 peptide sequences obtained by microsequencing of purified human platelet CLP36. Pale shading indicates the PDZ domain, whereas gray shading indicates the LIM domain. The 2 peptides that were selected to generate antipeptide antibodies are displayed in bold.
Human CLP36 contains a C-terminal LIM domain of 51 amino acid residues displaying the known LIM-consensus motif and an N-terminal PDZ domain of 81 amino acid residues (shaded in Figure 1). CLP36 belongs to a new group of LIM proteins that contain an N-terminal PDZ domain and 1 or 3 C-terminal LIM domains. Human CLP36 shows a 47% overall identity to the human actinin-associated LIM protein (hALP) in striated muscle that binds to α-actinin-227 and a 43% identity to RIL, a protein that has been implicated in the phenotypic reversion of transformed rat fibroblasts34 and binds to a specific protein tyrosine phosphatase.35 To the same family of proteins belong Enigma,36 Enigma-Homolog,37and Cypher38 that contain one N-terminal PDZ domain and 3 C-terminal LIM domains. The homology of the PDZ domain of CLP36 to the PDZ domains of ALP (55% amino acid identity) and RIL (66%) and of the LIM domain of CLP36 to the LIM domain of ALP (67%) and RIL (57%) is higher compared with the PDZ and LIM domains of Enigma, Enigma-Homolog, and Cypher (data not shown).
Immunoprecipitation and immunoblotting of CLP36
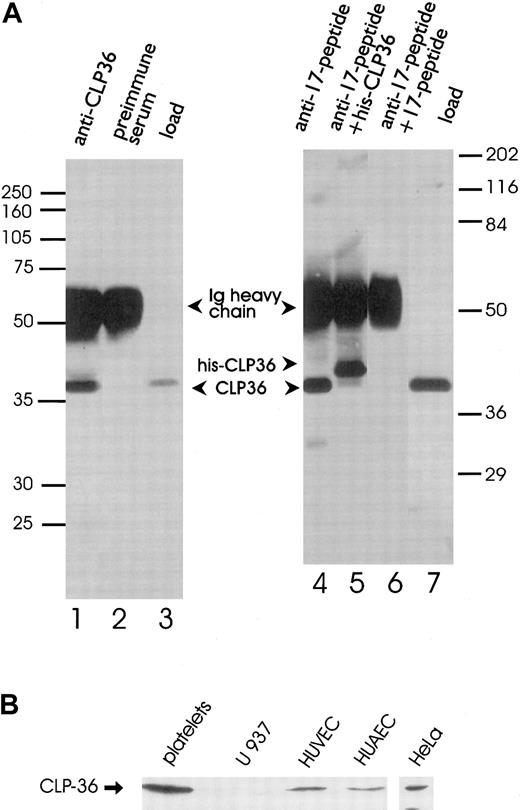
Polyclonal antibodies directed against the recombinant His-CLP36 and 2 specific peptides of CLP36 (aa 302-314, designated as “14-peptide,” and aa 224-239, designated as “17-peptide,”; bold in Figure 1) were used for immunoblot analysis. All 3 antibodies recognized specifically one band of about 38 kd in human platelets. The anti-CLP36 and anti–peptide-17 antibodies were able to specifically immunoprecipitate CLP36 from platelet lysate. Preimmune serum or anti–17-peptide antiserum preadsorbed with the 17-peptide did not immunoprecipitate platelet CLP36. Also, recombinant His-CLP36 added to the platelet lysate effectively competed with endogenous CLP36 for immunoprecipitation (Figure 2A). We found an expression of 105 molecules CLP36 per platelet, corresponding to about 0.3% of the total protein content in platelets (for the measurement and calculation see “Materials and methods”).
Immunoprecipitation and immunoblot analysis of CLP36.
(A) Specific immunoprecipitation of platelet CLP36 by 2 different anti-CLP36 antibodies. Platelet lysates (1 mL) were incubated with anti-CLP36 Ig (lane 1), preimmune serum Ig (lane 2), or 15 μg of anti–17-peptide-antibody (lanes 4-6). The addition of 50 μg 17-peptide (lane 6) or 50 μg His-CLP36 (lane 5) to the platelet extract blocked immunoprecipitation of endogenous CLP36. Load (lanes 3 and 7) indicates aliquots of the platelet lysate before immunoprecipitation. Immunoprecipitates were separated by SDS-polyacrylamide gels and immunoblotted with anti-CLP36 Ig. Molecular mass values are given as kd; marker on the left: Rainbow full range (Amersham); marker on the right: Broad range (Biorad). (B) Expression of endogenous CLP36 in various human cell types. Homogenates (250 μg protein) of platelets, the monocytic cell line U937, human umbilical venous endothelial cells (HUVEC), human umbilical arterial endothelial cells (HUAEC), and HeLa cells were separated by SDS-PAGE, blotted, and probed with anti-CLP36 antibody.
Immunoprecipitation and immunoblot analysis of CLP36.
(A) Specific immunoprecipitation of platelet CLP36 by 2 different anti-CLP36 antibodies. Platelet lysates (1 mL) were incubated with anti-CLP36 Ig (lane 1), preimmune serum Ig (lane 2), or 15 μg of anti–17-peptide-antibody (lanes 4-6). The addition of 50 μg 17-peptide (lane 6) or 50 μg His-CLP36 (lane 5) to the platelet extract blocked immunoprecipitation of endogenous CLP36. Load (lanes 3 and 7) indicates aliquots of the platelet lysate before immunoprecipitation. Immunoprecipitates were separated by SDS-polyacrylamide gels and immunoblotted with anti-CLP36 Ig. Molecular mass values are given as kd; marker on the left: Rainbow full range (Amersham); marker on the right: Broad range (Biorad). (B) Expression of endogenous CLP36 in various human cell types. Homogenates (250 μg protein) of platelets, the monocytic cell line U937, human umbilical venous endothelial cells (HUVEC), human umbilical arterial endothelial cells (HUAEC), and HeLa cells were separated by SDS-PAGE, blotted, and probed with anti-CLP36 antibody.
The analysis of expression of CLP36 in other human cell types by Western blot analysis revealed the presence of CLP36 in venous and arterial human umbilical endothelial cells and HeLa cells, but not in the monocytic cell line U937 (Figure 2B). The presence of CLP36 messenger RNA (mRNA) in endothelial cells was confirmed by performing reverse transcriptase-polymerase chain reaction (RT-PCR) and cDNA sequencing.
Immunocytochemical localization of CLP36 in resting and activated platelets
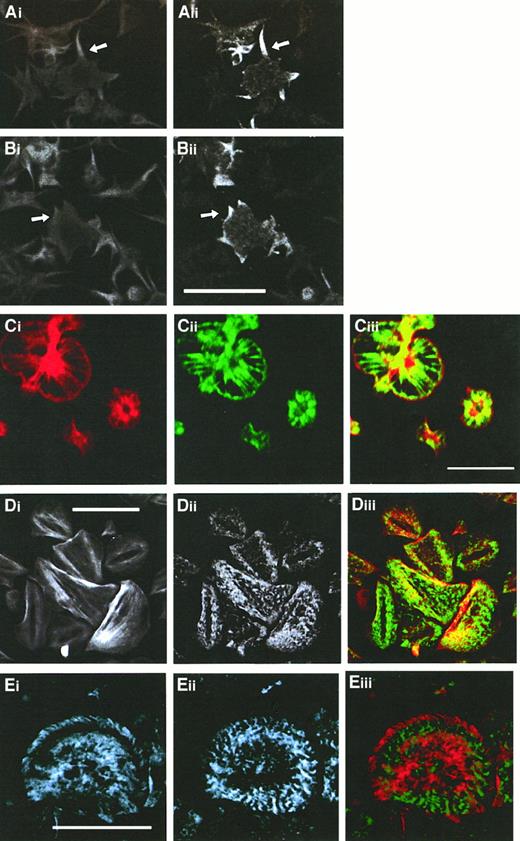
Immunofluorescence studies using either anti-CLP36 or anti–17-peptide antiserum were performed on spread platelets that allow, because of their larger size, a higher resolution of cytoskeletal structures. Adherent platelets show various stages of activation from shape change characterized by an irregular cell surface and extrusion of pseudopodia to spreading with the extension of veils between the radially outgrowing filopodia.32,39 These processes are associated with the formation of new actin structures: microfilament bundles in the pseudopodia (Figure3Ai), lamellipodia at the edge of the extending veil (Figure 3Bi), microfilaments radiating outward from the platelet center, and a thin filament ring at the rim in the fully spread platelet (Figure 3Ci). After completion of the spreading phase, platelets exhibit further distinct actin patterns: Parallel cables of actin filaments resembling stress fibers traversing the platelet or microfilament bundles arranged in a triangular pattern or concentric rings with a center free of filamentous actin are seen (Figure 3Di). In this late stage, platelets can also exhibit a vortexlike actin filament arrangement (Figure 3Eii).40 Double labeling of CLP36 and F-actin in spread human platelets revealed a colocalization (Figure 3). CLP36 was associated with microfilaments in long pseudopodia (Figure3Ai-ii) and short filopods and lamellipods (Figure 3Bi-ii). In the fully spread platelet, CLP36 was highly concentrated along the radially outgrowing actin filaments without being present in the F-actin–rich center—the region where the secretory granules are concentrated. CLP36 could also be found at the rim of the spread platelet, on the inner side of the cortical actin belt (Figure 3Ci-iii). In late stages of spreading, CLP36 is seen along actin stress fibers in a dotted regular pattern along parallel actin bundles (Figure 3Di-ii), although the overlay (Figure 3Diii) showed only in part a colocalization because of the different and inhomogeneous intensities of the F-actin and CLP36 signals.
Comparison of the subcellular localization of CLP36 and F-actin during various phases of platelet spreading.
Colocalization of F-actin (Ai,Bi, Ci,Di) and CLP36 (Aii-Dii) in early stages (Ai-Bii), and subsequent stages (Ci-ii) of platelet spreading, and after completion of spreading (Di-ii) was revealed by double immunofluorescence staining (merge Ciii,Diii). CLP36 staining of filopods (arrow in Aii) and lamellipods (arrow in Bii) formed during the early phase of spreading is homogenous and accumulates in patches (Cii) or dotted pattern (Dii,Eii) on F-actin–based structures in the late stages of platelet spreading. Note that CLP36 locates to the inner side of the cortical actin-filament belt during the spreading process (Ciii). Double immunofluorescent staining of the focal contact protein vinculin (Ei) and CLP36 (Eii), merge (Eiii); vinculin accumulates in the late stage of spread platelets in patches along circles or spirals at the microfilament tips.40 Bar, 10 μm.
Comparison of the subcellular localization of CLP36 and F-actin during various phases of platelet spreading.
Colocalization of F-actin (Ai,Bi, Ci,Di) and CLP36 (Aii-Dii) in early stages (Ai-Bii), and subsequent stages (Ci-ii) of platelet spreading, and after completion of spreading (Di-ii) was revealed by double immunofluorescence staining (merge Ciii,Diii). CLP36 staining of filopods (arrow in Aii) and lamellipods (arrow in Bii) formed during the early phase of spreading is homogenous and accumulates in patches (Cii) or dotted pattern (Dii,Eii) on F-actin–based structures in the late stages of platelet spreading. Note that CLP36 locates to the inner side of the cortical actin-filament belt during the spreading process (Ciii). Double immunofluorescent staining of the focal contact protein vinculin (Ei) and CLP36 (Eii), merge (Eiii); vinculin accumulates in the late stage of spread platelets in patches along circles or spirals at the microfilament tips.40 Bar, 10 μm.
Immunofluorescence microscopy in resting and activated endothelial cells
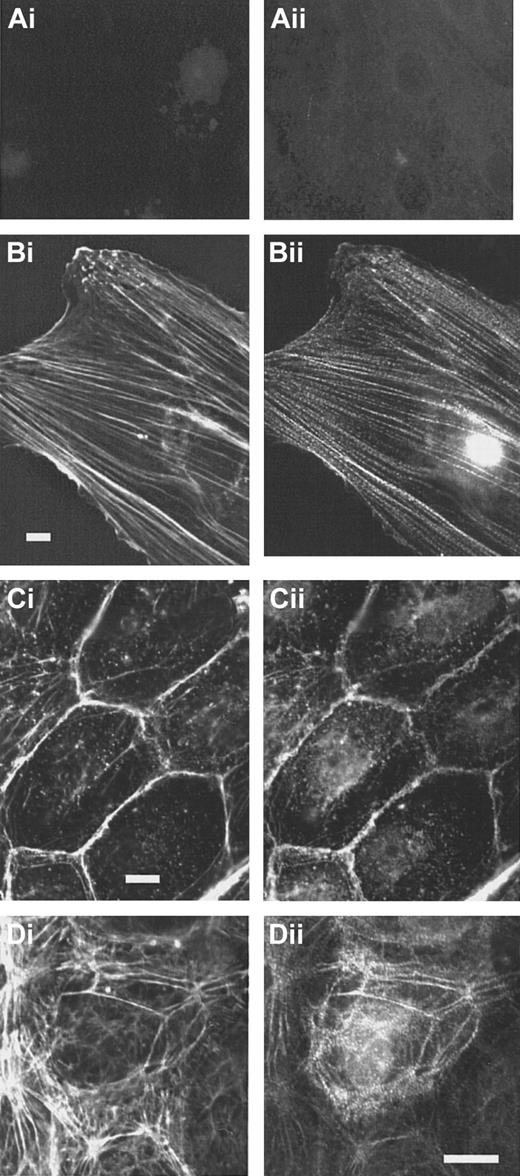
In subconfluent endothelial cells, stress fibers are abundant. In contrast to the controls (Figure4Ai-ii), specific staining of the cells with the anti-CLP36 immunoglobulin revealed that CLP36 was localized on stress fibers showing a dotted staining pattern (Figure 4Bi-ii). Like in platelets, CLP36 was absent from focal adhesions (visualized by staining with antivinculin antibodies) that are prominent in subconfluent endothelial cells (Figure 5Ci-ii).
CLP36 associates with actin stress fibers in endothelial cells.
Double immunofluorescence staining of CLP36 (Bii,Cii,Dii) and F-actin (Bi,Ci,Di). Controls (Ai-ii) for the specific staining of CLP36 were incubated with preimmune serum IgG (Ai) or with His-CLP36 for 1 hour before addition of anti-CLP36 antibody (Aii). Shown are subconfluent migrating endothelial cells (Ai-Bii), resting confluent endothelial cells (Ci-ii), and confluent endothelial cells exposed to thrombin (1 U/mL) for 2 minutes (Di-ii).
CLP36 associates with actin stress fibers in endothelial cells.
Double immunofluorescence staining of CLP36 (Bii,Cii,Dii) and F-actin (Bi,Ci,Di). Controls (Ai-ii) for the specific staining of CLP36 were incubated with preimmune serum IgG (Ai) or with His-CLP36 for 1 hour before addition of anti-CLP36 antibody (Aii). Shown are subconfluent migrating endothelial cells (Ai-Bii), resting confluent endothelial cells (Ci-ii), and confluent endothelial cells exposed to thrombin (1 U/mL) for 2 minutes (Di-ii).
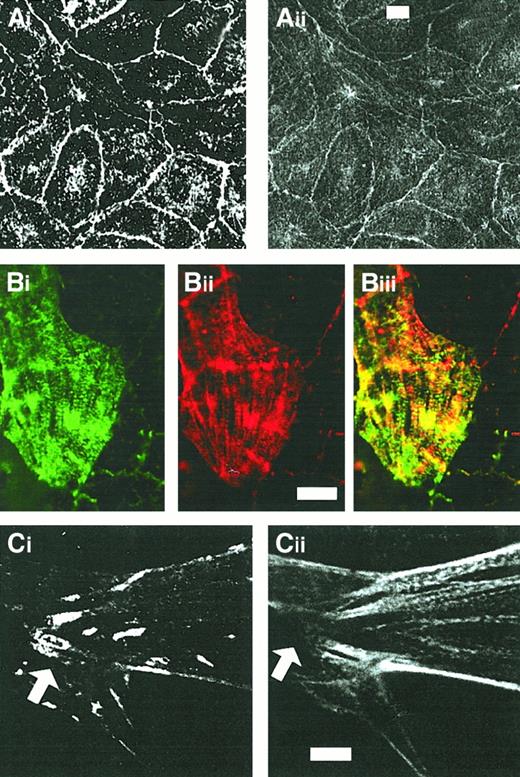
CLP36 colocalizes with α-actinin but not with vinculin in human endothelial cells.
Shown are double-immunofluorescence micrographs of resting confluent cells (Ai-ii) and migrating cells (Bi-Ci). Note the colocalization of CLP36 (Aii,Bi,Cii) and α-actinin (Ai,Bii) along stress fibers (Biii) and the absence of CLP36 from focal adhesions (arrow in Cii) stained with antivinculin antibody (Ci). Bars, 5 μm (Aii,Bii,Cii).
CLP36 colocalizes with α-actinin but not with vinculin in human endothelial cells.
Shown are double-immunofluorescence micrographs of resting confluent cells (Ai-ii) and migrating cells (Bi-Ci). Note the colocalization of CLP36 (Aii,Bi,Cii) and α-actinin (Ai,Bii) along stress fibers (Biii) and the absence of CLP36 from focal adhesions (arrow in Cii) stained with antivinculin antibody (Ci). Bars, 5 μm (Aii,Bii,Cii).
In resting confluent endothelial cells, CLP36 colocalized with the peripheral actin filaments beneath the plasma membrane (Figure 4Ci-ii). Confluent endothelial cells activated with thrombin for various times exhibit a dramatic reorganization of their actin cytoskeleton. CLP36 translocated to the newly formed actin structures after thrombin activation (Figure 4Di-ii).
The periodic dotted staining pattern of CLP36 on actin stress fibers in subconfluent and thrombin-stimulated endothelial cells resembled the decoration of stress fibers with anti–α-actinin antibodies in other types of cells.6 41 We performed, therefore, double immunofluorescence staining of subconfluent endothelial cells with anti–α-actinin and anti-CLP36 antibodies. α-Actinin colocalized with CLP36 on actin stress fibers (Figure5Bi-iii). In contrast to CLP36, anti–α-actinin staining was also prominent on lamellipods and focal contacts (not shown). In confluent endothelial cells, CLP36 colocalized with α-actinin on the plasma membrane at cell-cell contacts (Figure5Ai-ii).
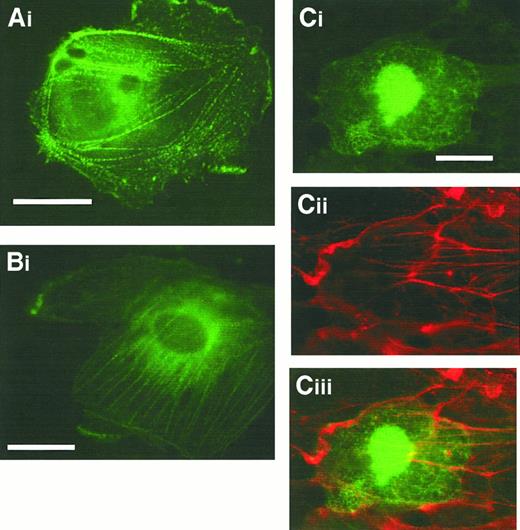
Targeting of heterologously expressed CLP36 to actin stress fibers
To examine more directly the localization of CLP36 in vivo and to identify the domain of CLP36 responsible for its targeting to the actin cytoskeleton, we transfected endothelial cells with CLP36 gene constructs that result in a C-terminal fusion protein with EGFP. Transfection of the vector, which only contained EGFP, served as control. Examination of fixed migrating endothelial cells showed that CLP36 and CLP36 containing the PDZ domain and the large intervening sequence but lacking the LIM domain were able to target EGFP to actin stress fibers in a dotted pattern—similar to that seen with indirect immunofluorescence (Figure 6A-B). The EGFP-CLP36 fusion protein did not target to focal adhesion, confirming our indirect immunofluorescence studies. These proteins also localized to cortical actin filaments. EGFP fused to the LIM domain did not bind to the actin cytoskeleton (Figure 6Ci-ii). It was found in the cytoplasm and concentrated in the nucleus. This nuclear trapping may be due to the small size of the fusion protein (35 kd), letting it pass the nuclear pores. It was observed also with EGFP alone and is, therefore, not specific.
The N-terminal part of CLP36 containing the PDZ domain and the intervening region but not the LIM domain targets CLP36 to stress fibers.
Fixed migrating human endothelial cells expressing CLP36-EGFP (Ai), CLP36ΔLIM-EGFP (Bi), and CLP36LIM-EGFP (Ci), (Cii) F-actin staining of the cell shown in Ci, (Ciii) overlay. Bars, 20 μm.
The N-terminal part of CLP36 containing the PDZ domain and the intervening region but not the LIM domain targets CLP36 to stress fibers.
Fixed migrating human endothelial cells expressing CLP36-EGFP (Ai), CLP36ΔLIM-EGFP (Bi), and CLP36LIM-EGFP (Ci), (Cii) F-actin staining of the cell shown in Ci, (Ciii) overlay. Bars, 20 μm.
CLP36 binds to α-actinin-1
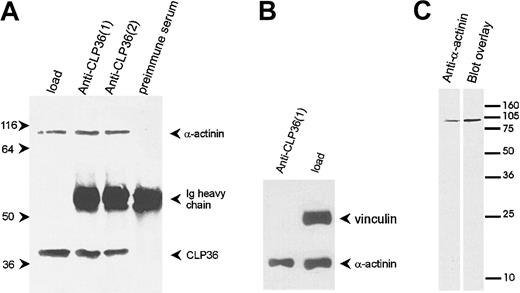
To determine whether the colocalization of CLP36 and α-actinin in endothelial cells was due to a direct interaction of the 2 proteins, CLP36 was immunoprecipitated from platelet lysates (Figure7A). Western blot analysis showed that α-actinin specifically coimmunoprecipitated with CLP36. Vinculin, which binds also to α-actinin42 but is not expected to bind to CLP36 (see above), was not coimmunoprecipitated with CLP36 (Figure 7B). The interaction between CLP36 and α-actinin was also detected by the blot overlay assay. Biotinylated CLP36 bound to a single platelet protein that co-migrated with platelet α-actinin (Figure 7C).
CLP36 associates in vivo with α-actinin in platelets.
(A) Coimmunoprecipitation of α-actinin in anti-CLP36 immunoprecipitates. Anti-CLP36 antibodies from 2 different rabbits (designated 1 and 2) were used. (B) No coimmunoprecipitation of CLP36 with the focal contact protein vinculin. The anti-CLP36 immunoprecipitates were resolved by SDS-PAGE, blotted, and probed with anti-CLP36, anti–α-actinin, or antivinculin antibody. Load indicates platelet lysate before immunoprecipitation corresponding to 7% of lysate used for immunoprecipitation. The experiment is representative for 10 independent experiments. (C) Interaction between CLP36 and platelet α-actinin as detected by the blot overlay assay. Platelet proteins were blotted, and the blot was probed with anti–α-actinin antibody (left) or with biotinylated-CLP36 (right).
CLP36 associates in vivo with α-actinin in platelets.
(A) Coimmunoprecipitation of α-actinin in anti-CLP36 immunoprecipitates. Anti-CLP36 antibodies from 2 different rabbits (designated 1 and 2) were used. (B) No coimmunoprecipitation of CLP36 with the focal contact protein vinculin. The anti-CLP36 immunoprecipitates were resolved by SDS-PAGE, blotted, and probed with anti-CLP36, anti–α-actinin, or antivinculin antibody. Load indicates platelet lysate before immunoprecipitation corresponding to 7% of lysate used for immunoprecipitation. The experiment is representative for 10 independent experiments. (C) Interaction between CLP36 and platelet α-actinin as detected by the blot overlay assay. Platelet proteins were blotted, and the blot was probed with anti–α-actinin antibody (left) or with biotinylated-CLP36 (right).
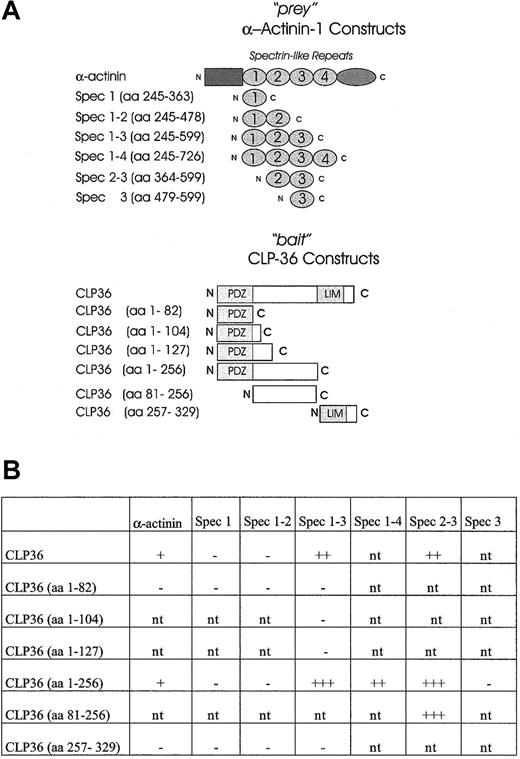
To establish a direct interaction of CLP36 with α-actinin-1 and to map the domains that mediate the interaction of CLP36 with α-actinin-1, the yeast 2-hybrid system was used. It has been shown previously that spectrinlike repeats of skeletal muscle α-actinin-2 mediated the interaction with ALP.27 Therefore, constructs containing spectrinlike repeats of endothelial α-actinin-1 (“prey” vector) were created and tested for an interaction with various constructs of CLP36 (“bait” vector) (Figure8A). As expected, CLP36 interacted directly with α-actinin-1 in the yeast 2-hybrid system. The interacting regions could be limited to the large intervening sequence of CLP36 and the spectrinlike repeats 2-3 of endothelial α-actinin-1 (Figure 8B). The interaction in the yeast 2-hybrid system was strongest with CLP36 (aa 1-256) or CLP36 (aa 81-256) as bait and the spectrinlike repeats 2-3 of α-actinin-1 as prey. The 3 CLP36 constructs containing either the PDZ domain alone or the PDZ domain plus small parts of the intervening sequence of CLP36 did not interact with α-actinin-1 or the spectrinlike repeats 1-3 of α-actinin-1. Also no interaction of the LIM domain of CLP36 and α-actinin-1 or various spectrinlike repeats was detectable. These data indicate that in the yeast 2-hybrid system the intervening sequence, but not the PDZ or the LIM domain, mediates the binding of CLP36 to α-actinin-1.
Mapping of the interacting domains of CLP36 and α-actinin-1 by use of the yeast 2-hybrid system.
(A) CLP36 and α-actinin-1 and their respective domains were constructed and cloned in “bait” and “prey” plasmids. (B) CLP36 (1-256) and CLP36 (81-256) interact strongly with a construct consisting of spectrinlike repeats 2-3 of α-actinin-1. β-Galactosidase activity of each transformant was visualized. The + indicates slow appearance (within 3-4 hours) of light blue colonies, +++ indicates rapid appearance (within 30-60 minutes) of dark blue colonies in the X-Gal assay on Trp−Leu−drop-out medium. The − indicates colonies that are white in the X-Gal screening assay; nt, interaction not tested.
Mapping of the interacting domains of CLP36 and α-actinin-1 by use of the yeast 2-hybrid system.
(A) CLP36 and α-actinin-1 and their respective domains were constructed and cloned in “bait” and “prey” plasmids. (B) CLP36 (1-256) and CLP36 (81-256) interact strongly with a construct consisting of spectrinlike repeats 2-3 of α-actinin-1. β-Galactosidase activity of each transformant was visualized. The + indicates slow appearance (within 3-4 hours) of light blue colonies, +++ indicates rapid appearance (within 30-60 minutes) of dark blue colonies in the X-Gal assay on Trp−Leu−drop-out medium. The − indicates colonies that are white in the X-Gal screening assay; nt, interaction not tested.
To further test whether the LIM domain may be involved in α-actinin binding in intact cells, platelet lysates were incubated with the His-tagged fusion proteins His-CLP36, His-CLP36ΔLIM (aa 1-257), or His-LIM (aa 258-329). The His-tagged fusion proteins were subsequently immunoprecipitated with anti–His-tag antibody. We found that α-actinin was specifically coprecipitated with both the full-length His-CLP36 and the His-CLP36ΔLIM, whereas α-actinin could not be detected in His-LIM immunoprecipitates (data not shown), therefore excluding a role of the LIM domain in the binding of CLP36 to α-actinin.
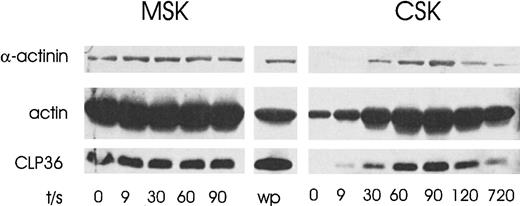
Coordinate and reversible translocation of CLP36 and α-actinin-1 to the cytoskeleton of activated platelets
On platelet aggregation, there is a rapid increase in actin polymerization of 70% to 80% of total platelet actin. Under these conditions, several regulatory proteins translocate to the F-actin–rich cytoskeleton.13 17 To determine whether CLP36 redistributes to the cytoskeleton, platelets were aggregated by stimulation of the thrombin receptor. We found a rapid increase of CLP36 in the cytoskeleton that was reversible and correlated with an increase of α-actinin (Figure 9). CLP36 and α-actinin were present in the membrane skeleton, but no significant changes of these proteins on stimulation could be observed. Given the finding that CLP36 binds to α-actinin-1 in resting platelets, these results indicate that both proteins might translocate as a complex from the cytosol to the F-actin–rich cytoskeleton.
Coordinate and reversible translocation of CLP36 and α-actinin-1 to the cytoskeleton of activated platelets.
Platelets were activated for various times with the thrombin-receptor activating peptide TRAP (20 μmol/L). The proteins in the cytoskeleton (CSK) and the membrane cytoskeleton (MSK) were separated by SDS-PAGE and immunoblotted using anti-CLP36, antiactin, and anti–actinin-1 antibodies; wp indicates whole platelet proteins.
Coordinate and reversible translocation of CLP36 and α-actinin-1 to the cytoskeleton of activated platelets.
Platelets were activated for various times with the thrombin-receptor activating peptide TRAP (20 μmol/L). The proteins in the cytoskeleton (CSK) and the membrane cytoskeleton (MSK) were separated by SDS-PAGE and immunoblotted using anti-CLP36, antiactin, and anti–actinin-1 antibodies; wp indicates whole platelet proteins.
Discussion
Human CLP36 was purified from platelets and cloned from HEL cells. We found by PCR amplification/cDNA sequencing and Western blot analysis that CLP36 was also expressed in endothelial cells and HeLa cells. CLP36 could not be detected in various human monocytic cell lines. Analysis of the cDNA database revealed that the EST tags, which matched completely the CLP36 cDNA, were derived from many tissues (eg, lung, liver, pancreas, spleen, colon, thyroid, placenta, and retina) and cell types (endothelial cells, senescent fibroblasts, Jurkat T cells, and colon carcinoma cells). Similarly, Northern blot analysis, using hCLIM1 as probe, has revealed a wide human tissue distribution with strong expression in heart and skeletal muscle; moderate expression in spleen, small intestine, colon, placenta, and lung; and weak expression levels in liver, thymus, kidney, prostate, and pancreas.20
Immunofluorescence studies showed that CLP36 translocated to newly formed actin filaments and stress fibers, but not to focal adhesions, in both activated platelets and endothelial cells. CLP36 did not interact directly with F-actin in F-actin cosedimentation assays (data not shown). Instead we found by coimmunoprecipitation, yeast 2-hybrid analysis and blot overlay assay that CLP36 bound to α-actinin-1, an actin-binding and cross-linking protein. CLP36 showed a constitutive binding to α-actinin in resting platelets and a coordinate increase of CLP36 and α-actinin in the F-actin–rich cytoskeleton on platelet aggregation. We conclude that CLP36 and α-actinin-1 are incorporated as a complex to newly formed actin filaments and stress fibers.
The nonmuscle α-actinin isoforms present in platelets and endothelial cells contain EF-hand regions that bind Ca++. In vitro, Ca++ decreases the affinity of α-actinin binding to actin.43-46 We found that lowering or increasing the cytosolic Ca++ level, or cell activation, did not change the amount of α-actinin-1 that coimmunoprecipitated with CLP36 (data not shown). These results indicate that the binding of the 2 proteins is not modulated by cytosolic Ca++ or other signal-transduction pathways in intact platelets. Furthermore, depolymerization of F-actin by treating platelets with cytochalasins did not change the amount of α-actinin-1 coimmunoprecipitated with CLP36, indicating that the binding of CLP36 to α-actinin-1 is constitutive and independent of the α-actinin-1/F-actin interaction.
Yeast 2-hybrid analysis indicated that the intervening region of CLP36, but not its PDZ or LIM domain, bound to the spectrinlike repeats 2 and 3 within the rod domain of α-actinin-1. This is in contrast to the yeast 2-hybrid study of the related protein ALP that has been shown to bind through its PDZ domain to the spectrinlike repeats of muscle actinin-2.27 Differences of the sequence of the PDZ domains of CLP36 and ALP (which share only a 55% homology) or of the sequence of α-actinin-1 and α-actinin-2, or methodological differences of the yeast 2-hybrid system used could underlie these different results. Our results appear also not to be in agreement with a yeast 2-hybrid investigation just published, showing that hCLIM1, which represents probably a mutated CLP36 (as discussed in the “Results” section), binds through its LIM domain to the EF-hand region α-actinin-2.47 A possible reason for this apparent discrepancy might lay in the different C-terminal Ca++-binding EF-hand regions of α-actinin-1 and α-actinin-2. Muscle α-actinin-2 in contrast to nonmuscle α-actinin-1 is known to bind to actin in a Ca++-insensitive manner.43-46 In further experiments to address the question whether the LIM domain mediates the binding of CLP36 to α-actinin-1 in vivo, no evidence to support such an interaction was obtained. First, the addition of the His-tagged CLP36-LIM domain to platelet lysates did not pull down α-actinin. Second, our heterologous expression studies in endothelial cells using various EGFP/CLP36 fusion proteins showed that not the LIM domain but the N-terminal part of CLP36 containing the PDZ domain and intervening region targeted EGFP to stress fibers.
CLP36 belongs to a new structurally and functionally distinct group of proteins containing a C-terminal PDZ and one or more N-terminal LIM domains. These proteins might link cell signaling to actin dynamics, because, as far as it has been elucidated, they bind to the actin cytoskeleton as well as to signaling enzymes: ALP and Cypher expressed in striated muscle bind through their PDZ domain to α-actinin-2,27,38 and Enigma binds through its PDZ domain to tropomyosin.48 However, their LIM domains appear to link these proteins to specific protein kinases and phosphatases: RIL binds through its LIM domain to the protein tyrosine phosphatase PTP-BL,35 and LIM domains mediate, at least in vitro, the binding of Enigma to receptor tyrosine kinases,49 and of Enigma-homolog and Cypher to protein kinase C.37,38 ALP and Cypher are both specifically expressed in skeletal and cardiac muscle and colocalize with α-actinin-2 to Z lines in myofibers of striated muscle.27,38 In contrast, CLP36, in addition to a high expression in heart and muscle cells, is present in many other tissues and cells (present study).19 20
The binding of CLP36 to α-actinin-1 and the colocalization of both proteins imply a role of CLP36 in modulating the function of α-actinin. α-Actinin is a rod-shaped protein of 100 kd, which forms homodimers.50 We found that CLP36 binds to the spectrinlike repeats 2-3, which form symmetric antiparallel dimers within the actinin rod.51 The globular N-terminal head of actinin contains the actin-binding domain,52 whereas the C-terminal tail binds to β-subunits of integrins.53 The latter interaction is expected to target α-actinin-1 to adhesion plaques. It is remarkable that CLP36 was absent from focal adhesions in both activated platelets and endothelial cells. By binding to the spectrin repeats of α-actinin-1, CLP36 might prevent the binding of α-actinin-1 to integrin subunits and to focal adhesions and direct the molecule to actin filaments and stress fibers. At this position, CLP36 could modulate the function of α-actinin-1, such as increasing its actin cross-linking and bundling activities.
Acknowledgments
We greatly appreciate the excellent technical assistance of U. Wielert and C. Meister. Some of the experiments are part of the thesis of K.B. at the University of Munich.
Supported by the Deutsche Forschungsgemeinschaft (Si 274; GRK 438) and the August-Lenz-Stiftung.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Note added in proof
A very recent study reports that CLP36 binds via its PDZ-domain to actinin-1 and actinin-4 in colon epithelial cells.54
Author notes
Wolfgang Siess, Institut für Prophylaxe und Epidemiologie der Kreislaufkrankheiten, Universität München, Pettenkoferstr. 9, D 80336 München, Germany; e-mail: wolfgang.siess@klp.med.uni-muenchen.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal