In 1957, Charlotte Friend described a novel retroviral disease in mice characterized by splenic enlargement, erythroleukemia, and death.1 This rapidly progressive disease, now known as Friend disease, has provided a powerful tool for the study of multistage carcinogenesis. Transformed murine erythroleukemia (MEL) cells, isolated from Friend virus–infected mice, have also provided a versatile model system for the study of erythroblast differentiation and erythropoietin (Epo)-induced signal transduction in vitro. Over the years, Friend disease has provided major insights into the molecular evolution of leukemia, the normal mechanisms of Epo receptor (EpoR) activation and, most recently, the molecular mechanisms of leukemia resistance. In this review, we summarize the molecular insights gained from the study of Friend disease. We focus on recent advances, with a special emphasis on studies of the Friend virus susceptibility locus,Fv2. 2
Multistage Friend disease
Following inoculation of a susceptible mouse with Friend virus complex, Friend disease evolves in 2 stages.3,4 In the first stage, the mouse develops massive splenic enlargement due to the polyclonal proliferation of splenic erythroblasts. These erythroblasts are not frankly transformed, however, and their transfer to another animal does not result in tumors.5 The primary mitogenic event underlying this polyclonal expansion is constitutive activation of the EpoR.4,6 During this early stage of disease, there is a marked expansion of erythroid progenitor cells (burst-forming units–erythroid [BFU-E] and colony-forming units–erythroid [CFU-E]).7 8
The second stage of Friend disease is marked by the emergence of fully transformed cells in the spleen and, eventually, the blood, bone marrow, and liver.5 These transformed cells form spleen colonies in genetically anemic Sl/Sldmice, are tumorigenic in vivo, and give rise to stable cell lines in vitro.5,9,10 Mice infected with the polycythemia-inducing strain of Friend virus (see below) often die during the first stage of disease because of splenic rupture. Mice that survive the first stage, however, will develop leukemia.11 Most emerging leukemic clones either fail to express p53, or they express a mutant form of p53 that is inactive, demonstrating that loss of p53 function is a common event in disease progression.12-14 In addition, anets family transcriptional activator, Sfpi1/PU.1 (spleen focus-forming virus [SFFV] proviral integration 1), is activated in 70% to 95% of Friend virus–induced erythroleukemias.15,16 The high incidence of p53 inactivation and Sfpi1/PU.1 activation suggests that these events are critical to the evolution of erythroleukemia. Isolated MEL cells have the characteristic appearance of immature erythroblasts. These cells remain transformed and Epo-independent and demonstrate constitutive activation of the EpoR.17 Molecular events underlying the first and second stages of Friend disease have been reviewed.4,18 19
Friend virus complex consists of 2 viruses, a replication-competent Friend helper virus (F-MuLV) and a replication-defective SFFV. There are at least 2 naturally occurring variants of SFFV, called SFFVP (polycythemia strain) and SFFVA (anemia strain). Mice infected with the viral complex of SFFVP and F-MuLV (also known as FVP) become polycythemic, while mice infected with SFFVA and F-MuLV (FVA) develop anemia as a result of volume expansion.20 In contrast to other acutely oncogenic retroviruses, SFFV does not carry a constitutively active, mutant form of a host gene. Instead, SFFV encodes a truncated form of a retroviral envelope protein, gp55, which can bind and activate the EpoR.6,21 Indeed, virally encoded gp55 can replace Epo in supporting the formation of erythroid colonies in vitro, but only in the presence of the EpoR.22
The polycythemia- and anemia-inducing strains of Friend virus exhibit differential transforming effects in vitro.23 Both SFFVP and SFFVA induce BFU-E in methylcellulose in the absence of exogenous Epo. However, in contrast to SFFVP, SFFVA-induced bursts require Epo to undergo terminal differentiation. FVA-infected erythroblasts proliferate in vivo despite low levels of endogenous Epo.24 Withdrawal of Epo from FVA-infected erythroblasts at the CFU-E stage results in rapid commitment to undergo apoptosis.24 Thus, while FVA-infected erythroblasts can proliferate in the absence of Epo, Epo is required for their survival and terminal differentiation. Erythroblasts isolated from the spleens of FVA-infected mice (FVA cells) provide an excellent model system for the study of Epo-dependent terminal differentiation in vitro.25
The molecular interaction between the EpoR and gp55
Early evidence from several investigators, including Hankins, Ruscetti, and Kabat, demonstrated that SFFV is the causative agent in the first stage of Friend disease. In fact, the gp55 oncoprotein alone was shown to induce BFU-E in vitro.26 Moreover, gp55P (from SFFVP) supports the growth of Epo-dependent cell lines in vitro, suggesting that gp55Pmight activate the cell surface EpoR directly.27
The next clue to the function of the gp55 envelope arose from the cloning of the EpoR itself. The murine EpoR was expression-cloned from Epo-independent MEL cells expressing cell surface Epo-binding proteins.28 The EpoR polypeptide was shown to be the first member of the cytokine receptor superfamily, which includes the receptors for interleukin (IL)-2, IL-3, granulocyte-macrophage colony-stimulating factor, and several other hematopoietic cytokines.29,30 Initially, the EpoR was heterologously expressed in an IL-3–dependent, pro-B lymphocyte line, Ba/F3. Ba/F3-EpoR cells could be grown in either Epo or IL-3, demonstrating that the EpoR activates the appropriate signal transduction pathways in these cells. Next, Ba/F3-EpoR cells were infected with a retrovirus encoding gp55P. The EpoR and gp55P were shown to form a physical and functional complex in these cells, resulting in their transformation to growth factor independence. Expression of gp55 in parental Ba/F3 cells, in the absence of the EpoR, failed to transform the cells, demonstrating that both proteins are required for Epo-independent growth.6
The nature of the molecular interaction between gp55P and the EpoR is now understood in considerable detail. Gp55Pbinds to the EpoR both at the cell surface31 and within the endoplasmic reticulum32 of Friend virus–infected erythroblasts. Binding of gp55P results in the stabilization and constitutive activation of the EpoR.32Gp55P selectively activates the murine EpoR but not the human EpoR, despite the high (92%) amino acid identity between these polypeptides.33,34 Gp55P does not activate other members of the cytokine receptor superfamily. The critical contact between gp55P and the murine EpoR occurs within the transmembrane region of these cell surface proteins,35-37and a productive interaction between the EpoR and gp55Poccurs when both proteins are expressed at the cell surface.35 37-39 The gp55P-EpoR interaction cannot be transmitted by cell-to-cell contact or by the administration of soluble gp55P to EpoR-expressing cells (A.D., unpublished observations, September 1999).
The gp55 envelope proteins of SFFVA and SFFVPdiffer in a small number of amino acid residues, primarily in the transmembrane region, and these differences appear to account for the phenotypic variation of the disease.35,36,40Gp55A does not support the growth of Epo-dependent cell lines, which raises the question of whether it functionally interacts with the EpoR. Gp55 from Rauscher leukemia virus (R-gp55; which is closely related to gp55A) physically associates with the EpoR.34 However, R-gp55 does not detectably interact with the EpoR at the cell surface.41 Part of the explanation for these differences may be that gp55A and R-gp55, when compared with gp55P, are expressed at low levels on the cell surface.42 Recently, it was shown that gp55A induces the formation of CFU-E from fetal liver cells without exogenous Epo.22 Neither gp55A nor gp55P were able to induce CFU-E formation from fetal liver cells that lacked the EpoR. Thus, the EpoR is required for gp55A-induced erythroid colony formation. Whether this reflects a requirement for a direct interaction between gp55A and the EpoR remains to be determined.
The specific interaction between gp55P and the EpoR underscores the importance of the EpoR in the evolution of erythroleukemia. The EpoR can be constitutively activated by other molecular mechanisms, also resulting in the development of erythroleukemia. For instance, the EpoR can be activated by point mutations34,43 or by erythroblast autocrine secretion of Epo.44 Indeed, a constitutively active form of the EpoR can result in induction of Friend-like disease.45 Other molecular events resulting in up-regulation of EpoR activity, such as insertional activation of EpoR expression,46,47 truncation of the EpoR carboxy terminus,48,49 or cellular loss of the negative regulatory phosphatase, Shp-1,50-53 may also contribute to the evolution of erythroleukemia.
Structure of the native cell surface EpoR
Although the EpoR polypeptide has been molecular cloned,28 there are still questions about the structure and composition of the cell surface EpoR complex. Several studies indicate that the EpoR is a simple monomeric cytokine receptor capable of homodimerization in response to Epo binding. A soluble secreted form of the EpoR is sufficient for high-affinity Epo binding, supporting a model of a single receptor subunit.54 The crystal structure confirms the homodimeric structure of the Epo-bound, activated EpoR.55,56 Additional studies suggest a more complex membrane-bound EpoR conformation.57,58 Small synthetic peptides are capable of activating EpoR dimerization, providing a possible avenue for Epo-mimetic drug design.59 60
Other studies suggest that the native cell surface EpoR complex may contain additional subunits or accessory proteins. First, the EpoR displays at least 2 different affinities for Epo, raising the possibility of distinct receptor classes.61,62 Second, cross-linking studies with 125I-Epo reveal multiple classes of cell surface EpoR, suggesting the presence of additional receptor subunits.31,63-65 In one study, the EpoR was cross-linked to a cell surface tyrosine kinase protein.66 In addition, the EpoR interacts with a cytoplasmic tyrosine kinase, Jak2.67 Third, forced expression of the EpoR complementary DNA in some lymphoid cell lines, such as CTLL68,69 and HT-2,70 confers Epo binding but not Epo-induced mitogenesis. These cells lack a required factor in the trans-acting environment.70 One study suggests that the Kit receptor tyrosine kinase activates the EpoR directly, thereby accounting for the synergistic activities of Epo and Kit ligand (Mgf), also known as SCF, in erythropoiesis.71 Emerging data discussed below suggest that a truncated form of the Stk receptor tyrosine kinase, encoded by the Fv2 Friend virus susceptibility locus, may also function as a subunit of the EpoR complex.2
Host genes regulating susceptibility to Friend virus
A number of host genes have been identified that affect the susceptibility of mice to Friend virus–induced erythroleukemia (Table1). These genes can be divided into several categories based on their proposed mechanism of resistance.72 The first group consists of genes that dominantly interfere with the infection of target cells by the retrovirus. Fv4 encodes a protein related to the MuLV Env protein, which blocks ecotropic retroviral receptors.73Fv1 encodes a Gag-related protein, which interferes with the retroviral life cycle.74 The second group consists of genes that alter the immune response to Friend virus infection.Fv3 affects the susceptibility of mice to Friend virus–induced immunosuppression.75 TwoH2-linked loci, Rfv1 and Rfv2, and one non–H2-linked locus, Rfv3, affect the recovery from Friend virus infection.76 The third group consists of genes that affect the potential of infected target cells to proliferate and differentiate. Fv5 determines whether FVP will cause anemia or polycythemia in certain strains of mice.77Mutations in Kit and Mgf either diminish the number of available target cells or their potential to proliferate.78 79 These genes are required for normal erythropoiesis, emphasizing the importance of pre-existing erythropoietic pathways for the development of Friend disease.
Host genes that affect susceptibility to Friend virus–induced disease
| . | Gene . | Function . |
|---|---|---|
| Group 1: interference with retroviral infection | Fv4 | Blocks retroviral cell surface receptors |
| Fv1 | Interferes with retroviral life cycle | |
| Group 2: altered immune response | Fv3 | Susceptibility to Friend virus-induced immunosuppression |
| Rfv1, Rfv2 | H2-linked determinants of immune response | |
| Rfv3 | Non–H2-linked determinant of immune response | |
| Group 3: regulators of erythroid cell proliferation and differentiation | Fv5 | Determines anemia vs polycythemia |
| Kit, Mgf | Required for normal erythropoiesis | |
| Fv2 | Determines proliferative response to gp55 |
| . | Gene . | Function . |
|---|---|---|
| Group 1: interference with retroviral infection | Fv4 | Blocks retroviral cell surface receptors |
| Fv1 | Interferes with retroviral life cycle | |
| Group 2: altered immune response | Fv3 | Susceptibility to Friend virus-induced immunosuppression |
| Rfv1, Rfv2 | H2-linked determinants of immune response | |
| Rfv3 | Non–H2-linked determinant of immune response | |
| Group 3: regulators of erythroid cell proliferation and differentiation | Fv5 | Determines anemia vs polycythemia |
| Kit, Mgf | Required for normal erythropoiesis | |
| Fv2 | Determines proliferative response to gp55 |
Fv2 is another host factor that affects the proliferation of SFFV-infected erythroblasts.80 Susceptibility at theFv2 locus, which is dominant, is present in most inbred strains of mice. The exception is C57BL/6 and related strains, which are Fv2-resistant (Fv2rr). Fv2 does not interfere with retroviral entry into cells or the retroviral life cycle.81 Instead, Fv2 determines the potential of SFFV-infected erythroblasts to proliferate in response to gp55.Fv2 acts in a cell autonomous manner.82Indirect evidence suggests that Fv2 and gp55 are functionally interrelated. Fv2-mediated resistance can be circumvented by deletions within the ecotropic domain of gp55, but the same mutants are less active than wild-type gp55 inFv2-sensitive (Fv2ss) mice.83
Fv2 encodes a truncated form of the Stk receptor tyrosine kinase: implications for the mechanism of EpoR signaling
Recently it has been shown that Fv2 is Ron, a member of the Met subfamily of receptor tyrosine kinases.2Ron encodes the Stk receptor tyrosine kinase, and a naturally expressed truncated form of Stk (SF-Stk) confers susceptibility to Friend virus–induced erythroleukemia. The evidence can be summarized as follows. First, the expression of SF-Stk correlates with susceptibility to Friend disease. SF-Stk is the major expressed form of Stk in Fv2ss erythroid cells; however, SF-Stk is expressed at very low levels or is absent inFv2rr erythroid cells. The 5′ end of the SF-Stk transcript has been mapped to intron 10 ofRon.84 The presumptive basis for the difference in expression is a 3-nucleotide deletion in C57BL/6 DNA, which mutates a GATA binding site and causes a 6-fold difference in promoter activity. Second, targeted disruption of Stk inFv2ss mice causes resistance to Friend disease. Third, enforced expression of SF-Stk in Fv2rrmice confers susceptibility to Friend disease. These data strongly suggest that SF-Stk confers susceptibility to gp55-mediated erythroblastosis and that C57BL/6 mice are resistant because they fail to express the protein.
The molecular mechanism by which SF-Stk confers susceptibility to Friend disease is not known. Possibly, full-length Stk would have the same effect as SF-Stk if expressed at an equivalent level. Alternatively, there may a unique requirement for SF-Stk. The predicted protein encoded by the SF-Stk transcripts lacks the extracellular domain of Stk but retains the transmembrane and kinase domains.84 By analogy to other receptor tyrosine kinases, loss of the extracellular domain may contribute to constitutive activation.85 SF-Stk is closely related to the avian oncogene v-sea, which causes erythroblastosis and anemia in chickens.86 The v-sea oncogene is a transmembrane protein, which has an extracellular domain related to an avian retroviral envelope and an intracellular domain derived from c-sea. One possible mechanism is that gp55 and SF-Stk together form the functional equivalent of v-sea. Crosstalk could occur between gp55, SF-Stk, and components of the EpoR complex (eg, Jak2), resulting in activation of downstream signaling cascades. In support of this model, SF-Stk and the EpoR have been shown to physically interact in COS cells.87 Based on differences in molecular weight, Stk does not appear to be one of the proteins previously identified by 125I-Epo cross-linking to the EpoR.31 63-65 A second possibility is that SF-Stk functions independently of the gp55-EpoR complex to enhance erythroid cell survival or proliferation.
The lack of an erythroid defect in (Fv2rr) C57BL/6 mice suggests that SF-Stk does not have a role in normal erythropoiesis. Still, the role of full-length Stk remains to be determined. Stk is the receptor for macrophage-stimulating protein (also known as hepatocyte growth factor–like protein or Hgfl).88 Targeted disruption of the first exon of Stk in mice causes increased nitric oxide production from peritoneal macrophages in response to interferon-γ.89Under normal conditions, these mice are viable and the mutation has no effect on erythropoiesis. In contrast, deletion of a large part of the Stk gene is associated with early embryonic lethality due to a failure of implantation.90 One explanation for these findings may be that the exon 1 mutation of Stk is a hypomorphic allele rather than a null allele of Stk. Studies in mice that are chimeric for Stk-deficient cells should resolve whether Stk has a role in normal erythropoiesis.
The Met subfamily of receptor tyrosine kinases includes Met, Ron, Stk (the murine homolog of Ron), and v-sea. These receptors have a distinctive 2-tyrosine docking site in the C-terminal tail that mediates interactions with multiple SH2-containing signal transducers.91 Ligand binding elicits a complex biologic response that includes cell growth, dissociation, motility, and polarization.92 Recent evidence suggests that the Met subfamily may function as second subunits or coreceptors for the cytokine receptor family. Stk has been shown to colocalize with and activate the IL-3 receptor common β-chain, another member of the cytokine receptor family.93 Hgf, the ligand for Met, has a synergistic effect with Epo on Stat5 activation and erythroid burst formation, suggesting that Met can productively interact with the EpoR.94 Finally, it is intriguing that another member of the Met family, v-sea, causes erythroblastosis and anemia in chickens.86 Studies on potential interactions between v-sea and the avian EpoR await cloning of the avian EpoR.
Based on the likely physical and functional interaction between the EpoR and Stk,87 several questions emerge. For instance, it will be interesting to determine (1) whether activation of the EpoR results in activation of Stk kinase activity or phosphorylation of the multifunctional docking site; (2) whether activation of Stk by Hgfl results in EpoR phosphorylation or activation of Jak2; (3) whether EpoR and Stk can be cross-linked at the cell surface or whether previously identified Epo/EpoR cross-linked complexes62 contain Stk; and (4) whether specific activation or phosphorylation events correlate with the progression of Friend disease.
A role for SF-Stk in the evolution of Friend disease?
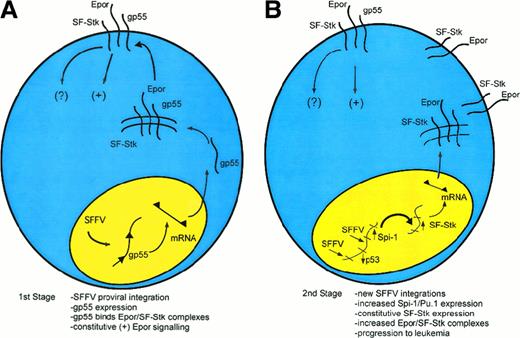
The molecular events underlying the first and second stages of Friend disease are now understood in considerable detail (Figure1). In the first stage of Friend disease, constitutive activation of the EpoR by gp55 causes uncontrolled, polyclonal proliferation of infected erythroblasts (Figure 1A). Susceptibility to gp55-induced erythroblastosis depends on expression of SF-Stk. Recent studies suggest that the mechanism may involve a direct interaction between the EpoR and SF-Stk.87 In the second stage of Friend disease, activation of Sfpi1/PU.1 and loss of p53 contribute to erythroleukemic transformation (Figure 1B and Table2). The transformation-specific targets of Sfpi1/PU.1 are not known, although one potentially relevant target is Fli1.95 Another potential target is SF-Stk. SF-Stk expression is down-regulated during terminal differentiation of FVA-infected, primary erythroblasts (P.N., unpublished observations, December 1998). Potentially, Sfpi1/PU.1 could activate SF-Stk through Ets sites in the SF-Stk promoter.2Constitutive SF-Stk expression, in turn, could contribute to the immortalization of infected erythroblasts. Consistent with this idea, SF-Stk is highly expressed in MEL cells but not most other hematopoietic cell lines. Further, enforced expression of full-length Stk causes ligand-dependent apoptosis of MEL cells but not Ba/F3 cells, indicating that Stk signaling has a direct effect on MEL cell survival.96
A speculative model of multistage Friend disease resulting from an interaction of the EpoR and SF-Stk.
(A) In the first stage of the disease, an SFFV provirus integrates into the host genome and encodes the oncogenic envelope protein, gp55. Then, gp55 forms a high-molecular-weight complex with the EpoR and perhaps SF-Stk, both at the cell surface and within an intracellular compartment. This binding interaction leads to the constitutive activation of the EpoR. SF-Stk may contribute to this mechanism in one or more ways. For instance, SF-Stk may increase the affinity of EpoR-gp55 interaction, increase the expression of cell surface EpoR, act as a coreceptor that activates additional downstream signaling events, or activate unique downstream signaling events that are required for erythroblast transformation. (B) In the second stage of the disease, proviral integrations cause up-regulation of Sfpi1/PU.1 expression and inactivation of wild-type p53 as well as other events (not shown).101 These mutations confer a selective advantage. Although this remains speculative, Sfpi1/PU.1 may activate SF-Stk as well as other target genes, resulting in progression toward erythroleukemia.
A speculative model of multistage Friend disease resulting from an interaction of the EpoR and SF-Stk.
(A) In the first stage of the disease, an SFFV provirus integrates into the host genome and encodes the oncogenic envelope protein, gp55. Then, gp55 forms a high-molecular-weight complex with the EpoR and perhaps SF-Stk, both at the cell surface and within an intracellular compartment. This binding interaction leads to the constitutive activation of the EpoR. SF-Stk may contribute to this mechanism in one or more ways. For instance, SF-Stk may increase the affinity of EpoR-gp55 interaction, increase the expression of cell surface EpoR, act as a coreceptor that activates additional downstream signaling events, or activate unique downstream signaling events that are required for erythroblast transformation. (B) In the second stage of the disease, proviral integrations cause up-regulation of Sfpi1/PU.1 expression and inactivation of wild-type p53 as well as other events (not shown).101 These mutations confer a selective advantage. Although this remains speculative, Sfpi1/PU.1 may activate SF-Stk as well as other target genes, resulting in progression toward erythroleukemia.
Molecular events associated with progression of Friend virus-induced erythroleukemia
| Gene . | Origin of cell line or tumor . | Genetic change . | Frequency . | References . |
|---|---|---|---|---|
| PU.1(Sfpi1) | FVP/FVA | Proviral insertion, ↑Pu.1 | 70%-95% | 15, 16 |
| Fli1 | F-MuLV | Proviral insertion, ↑Fli-1 | 75%-90% | 97, 98 |
| Fli2(Nfe2) | FVP, F-MuLV | Proviral insertion, ↓Nfe2 | 10% | 101 |
| Epo | F-MuLV | Genomic rearrangement, ↑Epo | 80% | 100 |
| EpoR | FVP | Proviral insertion, ↑EpoR | Unknown | 46, 47 |
| p53(Trp53) | FVP/FVA, F-MuLV | 1. Proviral insertion, ↓p53 | ∼ 100% | 12-14 |
| 2. Genomic rearrangement, ↓p53, truncation of p53 | ||||
| 3. Point mutation, loss of normal p53 function |
| Gene . | Origin of cell line or tumor . | Genetic change . | Frequency . | References . |
|---|---|---|---|---|
| PU.1(Sfpi1) | FVP/FVA | Proviral insertion, ↑Pu.1 | 70%-95% | 15, 16 |
| Fli1 | F-MuLV | Proviral insertion, ↑Fli-1 | 75%-90% | 97, 98 |
| Fli2(Nfe2) | FVP, F-MuLV | Proviral insertion, ↓Nfe2 | 10% | 101 |
| Epo | F-MuLV | Genomic rearrangement, ↑Epo | 80% | 100 |
| EpoR | FVP | Proviral insertion, ↑EpoR | Unknown | 46, 47 |
| p53(Trp53) | FVP/FVA, F-MuLV | 1. Proviral insertion, ↓p53 | ∼ 100% | 12-14 |
| 2. Genomic rearrangement, ↓p53, truncation of p53 | ||||
| 3. Point mutation, loss of normal p53 function |
It is interesting to contrast the progression of FVP-induced disease with that of F-MuLV, which is gp55-independent. F-MuLV–induced erythroleukemia has a longer latency (6 weeks) than FVP-induced disease (1-2 weeks), is oligoclonal rather than polyclonal and is associated with the activation of Fli1 rather than Sfpi1/PU.1 expression.97-99 Most MEL cell lines derived from Fli1-expressing primary erythroid tumors are Epo-dependent. However, with additional passages in vivo Epo-independent MEL lines are obtained, which show a high frequency of genomic rearrangements inEpo and autonomous Epo expression.100Thus, whether it occurs early in the course of disease (FVP) or late (F-MuLV), deregulation of EpoR signaling appears to be a common step in the progression of Friend virus–induced erythroleukemia.
Summary
In summary, Friend virus–induced erythroleukemia has provided a model system for the molecular understanding of the multistage nature of leukemogenesis. Critical elements of the disease involve the interaction of host gene products (EpoR, Sfpi1/PU.1, and SF-Stk) and retroviral gene products (gp55P or gp55A). The recent demonstration of a specific role of the cell surface tyrosine kinase SF-Stk in the process of leukemogenesis underscores the importance of multiple signal transduction pathways in normal hematopoiesis and leukemogenesis.
Stk is a member of a large family of cell surface tyrosine kinase receptors, while the EpoR is a member of a large family of cytokine receptors. Accordingly, it will be interesting to determine whether other transmembrane tyrosine kinases and cytokine receptors interact at the cell surface of hematopoietic cells. These tyrosine kinases may have a more general role as modifiers (coreceptors) of cytokine receptor signaling. In this regard, it is interesting thatKit encodes a receptor tyrosine kinase that is essential for normal erythropoiesis. Mgf and Epo are potent comitogens for early erythroid progenitor cells, and this effect may be due to direct interaction between Kit and the EpoR.71
Finally, identification of SF-Stk as the product of the Fv2gene provides general insights into the mechanism of cancer resistance in mice and perhaps in humans. Friend disease develops when the proliferation of infected erythroblasts outpaces the ability of the host immune system to suppress the infection.76 Mice that are deficient in SF-Stk, such as the C57BL/6 strain, have lost little or none of their normal erythropoietic response; however, gp55-mediated erythroid proliferation is slowed. As a result, they gain resistance to Friend virus–induced erythroleukemia.
Acknowledgments
We thank James Ihle, Dwayne Barber, and Maurice Bondurant for helpful discussions and advice on the manuscript.
Supported by National Institutes of Health (NIH) grant RO1 CA84214 (P.A.N.), NIH Cancer Center Support Grant P30 CA21765, and the American Lebanese Syrian Associated Charities.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Alan D. D'Andrea, Dana-Farber Cancer Institute, Department of Pediatric Oncology, Harvard Medical School, 44 Binney St, Boston, MA 02115; e-mail:alan_dandrea@dfci.harvard.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal