Abstract
Inhibitors of the protease of human immunodeficiency virus type 1 (HIV-1) may inhibit cytoplasmic retinoic acid-binding proteins, cytochrome P450 isoforms, as well as P-glycoproteins. These features of the protease inhibitors might enhance the activity of retinoids. To explore this hypothesis, myeloid leukemia cells were cultured with all-trans retinoic acid (ATRA) either alone or in combination with the HIV-1 protease inhibitors indinavir, ritonavir, and saquinavir. Consistent with the hypothesis, the HIV-1 protease inhibitors enhanced the ability of ATRA to inhibit growth and induce differentiation of HL-60 and NB4 myeloid leukemia cells, as measured by expression of CD11b and CD66b cell surface antigens, as well as reduction of nitroblue tetrazolium. Growth of ATRA-resistant UF-1 cells was also inhibited when cultured with the combination of ATRA and indinavir. Moreover, indinavir enhanced the ability of ATRA to induce expression of the myeloid differentiation-related transcription factor C/EBPε messenger RNA in NB4 cells by 9.5-fold. Taken together, the results show that HIV-1 protease inhibitors enhance the antiproliferative and differentiating effects of ATRA on myeloid leukemia cells. An HIV-1 protease inhibitor might be a useful adjuvant with ATRA for patients with acute promyelocytic leukemia and possibly retinoid-resistant cancers.
Introduction
Recent studies have shown that a high proportion of patients with acute promyelocytic leukemia (APL) achieved complete remission after treatment with all-trans retinoic acid (ATRA).1-4 However, the duration of these remissions was generally brief, and despite the continuous treatment with ATRA, almost all patients had a relapse. Once relapse occurred, APL cells were usually resistant to ATRA.1 5-8
Previous studies proposed that one possible explanation for the resistance was reduced plasma concentrations of ATRA.5,6ATRA is rapidly cleared from plasma, with a half-life of about 40 minutes and a peak concentration of nearly 10−6 mol/L lasting for 1 to 2 hours after receiving orally about 45 mg/m2.2,9 Serial pharmacokinetic studies showed that daily ATRA treatment was associated with a marked decrease in plasma drug levels compared with those at the initiation of therapy secondary to rapid metabolism of the drug.5 This is consistent with retinoid-induced increased expression of cytochrome P450 isoforms (CYPs).10
A second explanation for resistance relates to cytoplasmic retinoic acid–binding proteins (CRABPs) binding to ATRA to facilitate its degradation by CYPs.11-13 CRABP-I and CRABP-II act as modulators of intracellular levels of ATRA.14-19 Previous clinical studies showed that increased levels of CRABP-II were found in samples of APL that were resistant to ATRA.6 Furthermore, CRABP-II levels were higher in APL samples from individuals at the time of relapse compared with their APL cells at the initiation of therapy.15 This may contribute to resistance to ATRA.
A third explanation of ATRA resistance is overexpression of P-glycoprotein (P-gp), which pumps ATRA out of the cells, resulting in decreased intracellular concentrations of ATRA.20 Previous studies showed that ATRA-resistant APL cells, but not fresh APL cells, express P-gp.20
A fourth mechanism by which APL cells can become resistant to retinoids is by acquiring a mutation of the ligand-binding region of the retinoic acid receptor α (RAR α) gene.21,22 This was found in 3 of 8 individuals with APL who were resistant to retinoic acid (RA).21 Moreover, several ATRA-resistant APL sublines have been established that harbor a point mutation in the ligand-binding domain of RAR α.23-25 Most of these lines were established in vitro in the presence of a high concentration of ATRA.
Human immunodeficiency virus type 1 (HIV-1) protease inhibitors have become important tools in the treatment of HIV infection; these include saquinavir mesylate, ritonavir, and indinavir sulfate.26,27 Adverse effects associated with this class of drugs include peripheral fat wasting, central adiposity, hyperlipidemia, and insulin resistance, which together are referred to as lipodystrophy syndrome, as well as dermatitis and dry lips.27-31 A hypothesis to explain the adverse side effects revolves around altered lipid metabolism associated with RA.31 Some of these same adverse effects are also observed in individuals treated with ATRA. These adverse effects in both circumstances potentially could be caused by the same molecular mechanisms. A previous study showed that the catalytic region of the HIV-1 protease to which HIV-1 protease inhibitors bind has approximately 60% homology to the C-terminal region of CRABP-I.31 The biosynthesis of 9-cis-retinoic acid (9-cis-RA) remains to be fully elucidated; however, CRABPs probably transfer RA to endoplasmic reticulum where ATRA is isomerized to 9-cis-RA.31 9-cis-RA is the exclusive ligand of the retinoid X receptor (RXR). Carr and colleagues hypothesized that the protease inhibitors might bind to the homologous region within CRABP-I and hinder ATRA binding to CRABP-I.31 This could result in altered metabolism of RA,31 resulting in increased levels of intracellular ATRA. Furthermore, recent studies raised the possibility that the HIV-1 protease inhibitors can inhibit CYPs20,32 and P-gp,33 both of which are associated with ATRA resistance. Taken together, we reasoned that this family of drugs could enhance the inhibition of proliferation and induction of differentiation of APL cells. To verify our hypothesis, we examined the effects of HIV-1 protease inhibitors either alone or combined with ATRA for their enhanced antiproliferative and prodifferentiation effect on myelocytic leukemia cell lines in vitro.
Materials and methods
Cell lines
This study used the HL-60,34 NB4,35and UF-136 myelocytic leukemia cell lines. HL-60 and NB4 were grown in RPMI 1640 medium (GIBCO Life Technologies, Grand Island, NY) with 10% heat-inactivated fetal calf serum (FCS, GIBCO). UF-1 was grown in RPMI 1640 medium with 20% heat inactivated FCS under standard culture conditions.
Chemicals
All-trans RA was purchased from Sigma Chemical Co (St Louis, MO) and was dissolved in 100% ethanol at a stock concentration of 10−2 mol/L, stored at −20°C, and protected from light. Saquinavir mesylate (Roche, Branchburg, NJ), ritonavir (Abbott Labs, North Chicago, IL), and indinavir sulfate (Merck, West Point, PA) were dissolved in dimethyl sulfoxide (DMSO; Burdick & Jackson, Muskegon, MI) to a stock concentration of 10−2 mol/L and stored at −80°C.
Assays for cellular proliferation
Cells (105/mL) were incubated with various concentrations of either an HIV-1 protease inhibitor (10−6-2 × 10−5 mol/L), ATRA (10−11-10−6 mol/L), or their combination for 6 days in 96-well plates (Flow Laboratories, Irvine, CA). After culture, cell number and viability were evaluated by staining with trypan blue and counting using light microscopy.
Assays for differentiation
Induction of differentiation of cell lines was measured by expression of CD11b and CD66b antigens and reduction of nitroblue tetrazolium (NBT) dye. Cells were harvested after 6 days of incubation. For detection of CD11b, phycoerythrin-conjugated mouse antihuman CD11b (DAKO, Carpinteria, CA) was used. For detection of CD66b, indirect immunostaining was performed on cells using murine antihuman CD66b (Pharmingen, San Diego, CA), followed by staining with antimurine secondary antibody conjugated with fluorescein isothiocyanate (Pharmingen). Control studies were performed with a nonbinding murine IgG antibody (DAKO). Analysis of fluorescence was performed on a FACScan flow cytometer (Becton Dickinson, Mountain View, CA).
For NBT measurements, cells (105/mL) were incubated in 24-well plates (Corning, Corning, NY) for 6 days as described above. After incubation, each cell suspension was mixed with an equal volume of RPMI 1640 containing 1 mg/mL NBT (Sigma) and 10−6 mol/L 12-0-tetradecanoyl-13-acetate (TPA; Sigma) for 30 minutes at 37°C. The cells were washed in phosphate-buffered saline (PBS), cytocentrifuged, fixed in methanol for 5 minutes, and stained with Gram safranin for 10 minutes at room temperature; 300 cells were analyzed for blue dye using light microscopy.
RNA isolation and Northern blotting
Total RNA was extracted using Trizol (GIBCO) according to the manufacturer's instructions. Total RNA (30 μg/lane) was electrophoresed on 1.1% agarose gel containing 3-(N-morpholino) propanesulfonic acid (MOPS) and formaldehyde and transferred to nylon membrane (Micron Separation, Westborough, MA). Blots were hybridized for 2 hours at 65°C in Rapid Hybrid solution (Amersham, Arlington Heights, IL) with 3 × 106 cpm/mL of full-length C/EBPε complementary DNA (cDNA) probe after labeling with α-[32P]-deoxycytidine triphosphate (dCTP) by the random primer DNA-labeling tenchnique (Stratagene, La Jolla, CA). Membranes were washed to a stringency of 0.1 × standard sodium citrate (SSC) at 65°C and exposed to Kodak XAR film (Eastman Kodak, New Haven, CT).
Results
Effect of HIV-1 protease inhibitors on proliferation of myelocytic leukemia cell lines
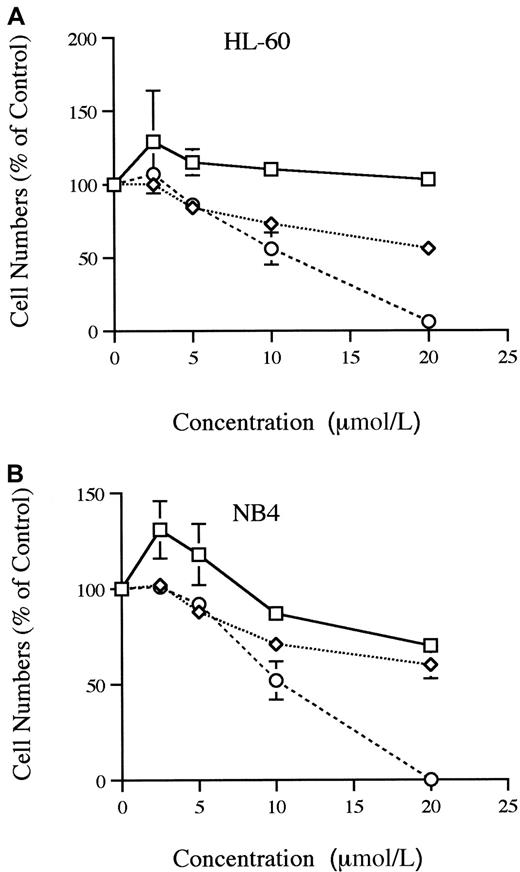
The HIV-1 protease inhibitors were examined for their effect on proliferation of HL-60 and NB4 cells in liquid culture system (Figure1A-B). Saquinavir effectively inhibited the growth by 50% (ED50) at approximately 1.1 × 10−5 mol/L for HL-60 cells and 1 × 10−5 mol/L for NB4 cells. Indinavir did not inhibit the growth of HL-60, but 2 × 10−5 mol/L indinavir inhibited the growth of NB4 by 30%. Even though an ED50was not reached, ritonavir was able to inhibit partially the growth of both HL-60 and NB4 cells in a dose-dependent manner.
Proliferation of HL-60 and NB 4 cells.
Dose-response effects of indinavir, ritonavir, and saquinavir on proliferation of HL-60 (A) and NB4 (B) cells are shown. Results are expressed as a percent of control plates containing no protease inhibitor. Data represent mean ± SD of triplicate cultures. These figures show findings of at least 3 independent experiments. ■, indinavir; ⋄, ritonavir; ○, saquinavir.
Proliferation of HL-60 and NB 4 cells.
Dose-response effects of indinavir, ritonavir, and saquinavir on proliferation of HL-60 (A) and NB4 (B) cells are shown. Results are expressed as a percent of control plates containing no protease inhibitor. Data represent mean ± SD of triplicate cultures. These figures show findings of at least 3 independent experiments. ■, indinavir; ⋄, ritonavir; ○, saquinavir.
The HL-60, NB4, and UF-1 cells were cultured with various concentrations of ATRA (10−11-10−6 mol/L) either alone or in combination with an HIV-1 protease inhibitor to evaluate the effect of the combinations of these drugs. The combination of ATRA (10−8 mol/L) and either saquinavir (10−5 mol/L) or ritonavir (10−5 mol/L) showed an enhanced inhibition of growth of HL-60 cells (Figure2A). For example, ATRA (10−8mol/L) inhibited the growth of HL-60 by 39%, and saquinavir (10−5 mol/L) inhibited proliferation by 44%. The combination of ATRA (10−8 mol/L) and saquinavir (10−5 mol/L) inhibited HL-60 growth by 86% (Figure 2A). None of the protease inhibitors potentiated the growth inhibitory activity of ATRA using NB4 as the target cells (Figure 2B). The UF-1 cell line is composed of APL cells from an individual who developed resistance to ATRA. When UF-1 cells were cultured with the combination of ATRA (10−9-10−6 mol/L) and indinavir (10−5 mol/L), a modest enhancement of growth inhibition occurred. ATRA (10−7 mol/L) inhibited growth of UF-1 by 16%, indinavir (10−5 mol/L) inhibited growth of UF-1 by 5%, simultaneous culture with both inhibited growth by 31% (Figure 2C).
Proliferation of HL-60, NB4, and UF-1 cells.
The effects of the combination of ATRA and HIV-1 protease inhibitors on proliferation of HL-60 (A), NB4 (B), and UF-1 (C) are shown. Cells were cultured with various concentrations of ATRA (10−8-10−6 mol/L for HL-60 and UF-1; 10−11-10−9 mol/L for NB4) either alone or in combination with indinavir (2 × 10−5 mol/L), ritonavir (1 × 10−5 mol/L), or saquinavir (1 × 10−5 mol/L). Data represent mean ± SD of triplicate cultures and results represent at least 3 independent experiments. ■, ATRA; ⋄, ATRA + Indinavir (2 × 10-5 mol/L; ○, ATRA + Ritonavir (1 × 10-5 mol/L); ▵, ATRA + Saquinavir (1 × 10-5 mol/L).
Proliferation of HL-60, NB4, and UF-1 cells.
The effects of the combination of ATRA and HIV-1 protease inhibitors on proliferation of HL-60 (A), NB4 (B), and UF-1 (C) are shown. Cells were cultured with various concentrations of ATRA (10−8-10−6 mol/L for HL-60 and UF-1; 10−11-10−9 mol/L for NB4) either alone or in combination with indinavir (2 × 10−5 mol/L), ritonavir (1 × 10−5 mol/L), or saquinavir (1 × 10−5 mol/L). Data represent mean ± SD of triplicate cultures and results represent at least 3 independent experiments. ■, ATRA; ⋄, ATRA + Indinavir (2 × 10-5 mol/L; ○, ATRA + Ritonavir (1 × 10-5 mol/L); ▵, ATRA + Saquinavir (1 × 10-5 mol/L).
Effect of HIV-1 protease inhibitors on differentiation of leukemic cell lines
Induction of differentiation of acute myeloid leukemia cell lines into more mature, granulocyte-like cells by HIV-1 protease inhibitors was assayed by measuring induction of expression of CD11b and CD66b antigens and ability to produce superoxide as measured by NBT reduction.
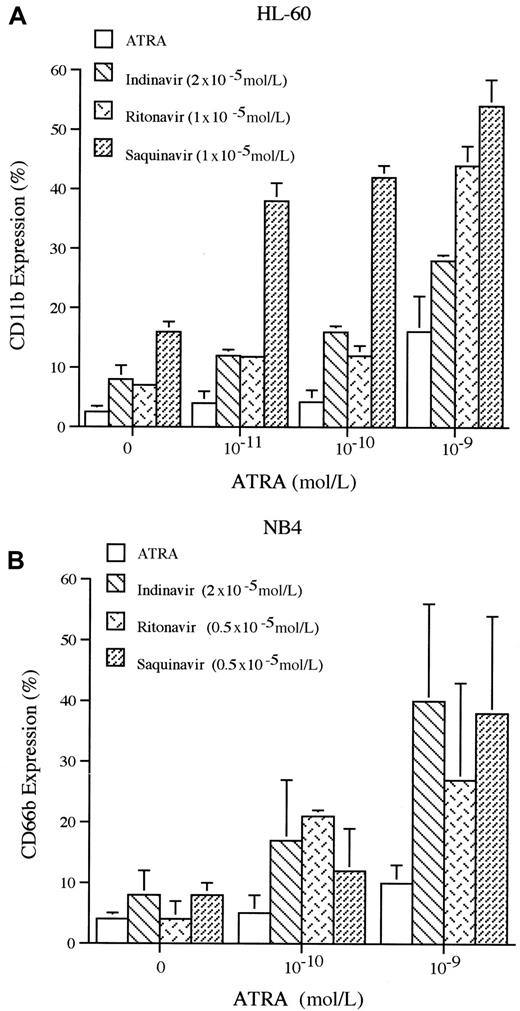
The expression of CD11b increases as myeloid cells differentiate toward either macrophages or granulocytes.37 Each of the HIV-1 protease inhibitors induced a low level of expression of CD11b on HL-60, with indinavir (2 × 10−5 mol/L), ritonavir (1 × 10−5 mol/L), and saquinavir (1 × 10−5 mol/L) inducing 8%, 7%, and 16% of HL-60 cells to express CD11b, respectively, on day 5 of culture. In contrast, each of the HIV-1 protease inhibitors prominently enhanced the ATRA-mediated induction of CD11b expression on HL-60 cells. For example, ATRA (10−9 mol/L) alone induced CD11b antigen expression on approximately 16% of HL-60 cells. When ATRA (10−9 mol/L, 5 days) was combined with either indinavir (2 × 10−5 mol/L), ritonavir (1 × 10−5 mol/L), or saquinavir (1 × 10−5 mol/L), a mean 28%, 44%, and 54% of HL-60 cells were induced to express CD11b, respectively (Figure3A).
Effects of HIV-protease inhibitors on CD11b expression of HL-60 cells and CD66b expression on NB4 cells.
(A) HL-60 cells were cultured for 5 days with either ATRA (10−11-10−9 mol/L) alone or in combination with either indinavir (2 × 10−5 mol/L), ritonavir (1 × 10−5 mol/L), or saquinavir (1 × 10−5 mol/L), and then analyzed by FACscan for expression of CD11b. (B) NB4 cells were cultured for 5 days with either ATRA (10−10-10−9 mol/L) alone or in combination with indinavir (2 × 10−5 mol/L), ritonavir (0.5 × 10−5 mol/L), or saquinavir (0.5 × 10−5 mol/L), and then analyzed by FACscan for expression of CD66b. These results represent 3 independent experiments.
Effects of HIV-protease inhibitors on CD11b expression of HL-60 cells and CD66b expression on NB4 cells.
(A) HL-60 cells were cultured for 5 days with either ATRA (10−11-10−9 mol/L) alone or in combination with either indinavir (2 × 10−5 mol/L), ritonavir (1 × 10−5 mol/L), or saquinavir (1 × 10−5 mol/L), and then analyzed by FACscan for expression of CD11b. (B) NB4 cells were cultured for 5 days with either ATRA (10−10-10−9 mol/L) alone or in combination with indinavir (2 × 10−5 mol/L), ritonavir (0.5 × 10−5 mol/L), or saquinavir (0.5 × 10−5 mol/L), and then analyzed by FACscan for expression of CD66b. These results represent 3 independent experiments.
The CD66b (formerly CD67) is a granulocyte-specific activation antigen expressed on secondary granule membranes of myeloid cells during their late stages of differentiation.38 The combination of ATRA and protease inhibitors over 5 days of culture enhanced differentiation. For example, indinavir (2 × 10−5mol/L), ritonavir (0.5 × 10−5 mol/L), and saquinavir (0.5 × 10−5 mol/L) alone induced 8%, 4%, and 8% of NB4 to express CD66b, respectively. ATRA (10−9 mol/L) alone induced approximately 10% of NB4 cells to express CD66b. The combination of ATRA (10−9 mol/L) with either indinavir (2 × 10−5 mol/L), ritonavir (0.5 × 10−5mol/L), or saquinavir (0.5 × 10−5 mol/L) induced 40%, 27%, or 38% of NB4 cells to express CD66b, respectively (Figure 3B).
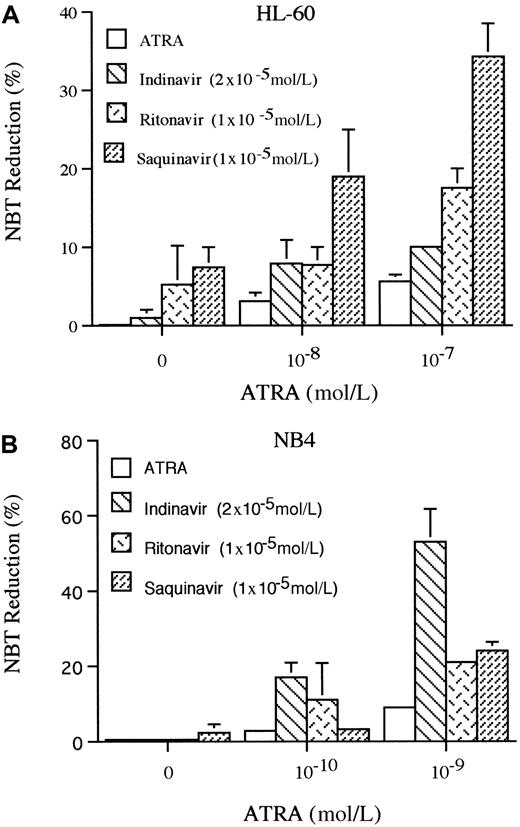
The production of superoxide is a marker of granulocyte-like differentiation, which can be measured by the ability to reduce NBT.39 Using this marker, we examined the ability of HIV-1 protease inhibitors to induce differentiation of HL-60 and NB4 cells. Indinavir (2 × 10−5 mol/L), ritonavir (1 × 10−5 mol/L), or saquinavir (1 × 10−5 mol/L) reduced NBT in 1%, 5%, or 7% of HL-60 cells, respectively, on the 5th day of culture. ATRA (10−7 mol/L) induced 6% of cells to reduce NBT; however, when it was combined with either indinavir (2 × 10−5mol/L), ritonavir (1 × 10−5 mol/L), or saquinavir (1 × 10−5 mol/L), a mean of 10%, 18%, and 34% of HL-60 cells reduced NBT, respectively (Figure4A). For NB4, only saquinavir (1 × 10−5 mol/L, 5 days) slightly increased the ability of the cells to reduce NBT, but each of these protease inhibitors enhanced the ability of ATRA to reduce NBT. ATRA (10−9mol/L) induced 9% of the cells to become NBT positive, and when combined with either indinavir (2 × 10−5 mol/L), ritonavir (0.5 × 10−5 mol/L), or saquinavir (0.5 × 10−5 mol/L), a mean of 53%, 21%, and 24% of the NB4 cells reduced NBT (Figure 4B), respectively, on the 5th day of culture.
NBT reduction.
The effects of HIV-1 protease inhibitors on NBT reduction of HL-60 (A) and NB4 (B) are shown. Cells were cultured for 5 days with various concentrations of ATRA (10−8-10−7 mol/L for HL-60; 10−10-10−9 mol/L for NB4) either alone or in combination with either indinavir (2 × 10−5mol/L), ritonavir (1 × 10−5 mol/L), or saquinavir (1 × 10−5 mol/L), and differentiation was determined by NBT reduction. Figure shows results of 3 independent experiments.
NBT reduction.
The effects of HIV-1 protease inhibitors on NBT reduction of HL-60 (A) and NB4 (B) are shown. Cells were cultured for 5 days with various concentrations of ATRA (10−8-10−7 mol/L for HL-60; 10−10-10−9 mol/L for NB4) either alone or in combination with either indinavir (2 × 10−5mol/L), ritonavir (1 × 10−5 mol/L), or saquinavir (1 × 10−5 mol/L), and differentiation was determined by NBT reduction. Figure shows results of 3 independent experiments.
Changes in expression of C/EBPε messenger RNA after exposure of NB4 to indinavir and all-trans retinoic acid
C/EBPε is a newly identified CCAAT/enhancer-binding transcriptional factor whose expression is restricted to maturing myeloid cells.40 Expression of C/EBPε occurs as either HL-60 or NB4 cells differentiate to granulocytes.40,41This transcription factor is necessary for full maturation of the granulocytic lineage.42 We examined whether indinavir enhanced the ability of ATRA to induce C/EBPε mRNA in NB4 cells (Figure 5). After 48 hours of culture with these compounds, cells were harvested, RNA extracted, Northern blotted, and sequentially hybridized with [32P]-labeled C/EBPε and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Indinavir (2 × 10−5 mol/L, lane 2) did not induce expression of C/EBPε mRNA; ATRA (10−9 mol/L, lane 3) induced a 2.5-fold increase of the transcription factor; and the combination of ATRA (10−9 mol/L) and indinavir (2 × 10−5 mol/L) enhanced levels of expression of C/EBPε mRNA by 9.5-fold (lane 4) compared to control wild-type NB4 cells (lane 1).
Enhancement of C/EBPε mRNA expression in NB4 cells by indinavir.
NB4 cells were cultured for 48 hours under various conditions: lane 1, (diluent control); lane 2, indinavir (2 × 10−5 mol/L); lane 3, ATRA (10−9 mol/L); lane 4, combination of indinavir (2 × 10−5 mol/L) and ATRA (10−9mol/L). Total RNA was extracted and analyzed by Northern blot technique (30 μg/lane) and hybridized with [32P]-labeled C/EBPε cDNA probe as described in “Materials and methods.” The same blot was rehybridized with [32P]-labeled GAPDH probe to show equality of RNA loading in each lane. Results are expressed as fold increase in expression as compared with control NB4 cells (lane 1). The band intensity was measured using a densinometer.
Enhancement of C/EBPε mRNA expression in NB4 cells by indinavir.
NB4 cells were cultured for 48 hours under various conditions: lane 1, (diluent control); lane 2, indinavir (2 × 10−5 mol/L); lane 3, ATRA (10−9 mol/L); lane 4, combination of indinavir (2 × 10−5 mol/L) and ATRA (10−9mol/L). Total RNA was extracted and analyzed by Northern blot technique (30 μg/lane) and hybridized with [32P]-labeled C/EBPε cDNA probe as described in “Materials and methods.” The same blot was rehybridized with [32P]-labeled GAPDH probe to show equality of RNA loading in each lane. Results are expressed as fold increase in expression as compared with control NB4 cells (lane 1). The band intensity was measured using a densinometer.
Discussion
The present study was based on the hypothesis that HIV-1 protease inhibitors could have 3 effects that might enhance the potency of retinoids. These inhibitors may occupy the C-terminal region of CRABPs, which is the same region where ATRA binds. In addition, the metabolizing enzymes of retinoids (CYPs) and the P-gp are inhibited by the protease inhibitors. Thus, intracellular concentrations of ATRA may increase when combined with protease inhibitors, which can enhance the potency of ATRA to inhibit proliferation and induce differentiation of myeloid leukemia cells. Consistent with our hypothesis, indinavir, ritonavir, and saquinavir enhanced the ability of ATRA to inhibit the growth and to induce the differentiation of myeloblast/promyelocyte leukemia cell lines, HL-60 and NB4. Notably, indinavir when combined with ATRA inhibited the growth of UF-1, an APL cell line. UF-1 cells were established from an individual with APL whose leukemia cells were resistant to ATRA; the cell line is also moderately resistant to ATRA but its RAR α gene is not mutated.36Recently, Lenhard and coworkers reported that indinavir enhanced the ability of ATRA to induce the murine fibloblast-like C3H10T1/2 cells to express alkaline phosphatase (ALP), which is an RA responsive gene.43 In contrast, the activity of the retinoid known as CH55 was not enhanced by indinavir. Whereas ATRA binds to CRABPs and RAR, CH55 only binds to RAR, supporting the hypothesis that the enhanced retinoid activity may result from binding and blocking the activity of CRABPs.
The CRABPs bind to ATRA, facilitating the degradative metabolism of the retinoid by CYP enzymes. ATRA is hydroxylated at the cyclohexenyl ring to form a 4-hydroxy-RA metabolite by the CYP-dependent mono-oxygenase system.44 The CYP 2C8, 2C9, as well as 3A4 play a central role in this metabolism.45,46 Induction of the activity of CYPs is associated with reduction of the plasma and intracellular concentrations of ATRA. Increased levels of both CYPs and CRABPs were observed in ATRA-treated hamsters.47 Therefore, inhibitors of CYPs may be useful therapeutic agents for ATRA-resistant APL patients whose leukemic cells often are associated with rapid metabolism of ATRA.10,45,46 Our previous study showed that clotrimazole, a CYP inhibitor, restored the responsiveness of APL cells to ATRA.20 Furthermore, a recent in vitro study using hamster liver microsomes demonstrated that HIV-1 protease inhibitors, especially ritonavir, are potent inhibitors of CYP 3A4.32Based on these in vitro studies, coadministration of ritonavir and saquinavir, the latter normally being extensively metabolized by CYP 3A4, is recognized as a clinically useful therapeutic strategy.48-50 In our experiments, the strongest CYP 3A4 inhibitor, ritonavir, was not the protease inhibitor that possessed the most potent antileukemic activity. Thus, we believe that the HIV-1 protease inhibitors probably mediate their antileukemic effect at least in part independent of their inactivation of CYP 3A4.
The P-gp is an integral plasma membrane protein encoded by the multidrug-resistant (MDR) gene, belonging to the adenosine triphosphate-binding cassette family of transporters.51,52It is an energy-dependent efflux pump for a wide variety of compounds including ATRA. Although APL cells express P-gp less frequently compared to other types of acute myeloid leukemias, the activity of P-gp is still considered to be associated with ATRA-resistance of APL cells.53-56 Therefore, inhibitors of P-gp could be a useful therapeutic agent. Indeed, verapamil, a P-gp antagonist, restored the responsiveness of the ATRA-resistant HL-60 subline and ATRA-resistant fresh APL cells in vitro.20However, the drug concentration of verapamil necessary for modulating MDR would cause severe cardiac toxicity.57 Recent in vitro studies showed that the 3 HIV-1 protease inhibitors used in this study inhibited the activity of P-gp.33 Thus, HIV-1 protease inhibitors may have a role as inhibitors of P-gp in leukemic cells.
Interestingly, HIV-1 protease inhibitors themselves also showed mild antiproliferative and differentiative effects on myeloblastic/promyelocytic leukemia cell lines. Saquinavir had a greater potency than any of the other protease inhibitors to inhibit growth of HL-60 and NB4 cells. It also was the most potent protease inhibitor of the 3 in inducing differentiation of HL-60. These results parallel a previous in vitro study that showed that saquinavir was the most potent of the 3 analogs in its ability to inhibit the viral replication of HIV.58 Indinavir was the most potent inducer of the NB4 cells. The reason that different cell types and different functions (cellular proliferation and differentiation) varied in their responsiveness to the 3 HIV-1 protease inhibitors is unclear.
C/EBPε is implicated in the differentiation of myeloid cells and its expression is directly induced by ATRA via either the RARα or PML/RARα pathway.38,40,41,58,59 Forced expression of C/EBPε in U937 myeloblasts can induce these cells to differentiate to more mature granulocytes.38 Furthermore, C/EBPε “knock-out” (deletional) mice have incomplete maturation of their granulocytes, and the cells do not have specific granule proteins.42 These findings suggest that C/ΕΒPε is pivotal to granulocytic differentiation. We have found that the combination of the HIV-1 protease inhibitor, indinavir, and ATRA markedly enhanced mRNA expression of this myeloid differentiation-related transcription factor. These results are congruent with the hypothesis that the HIV-1 protease inhibitors increase the intracellular levels of ATRA, which augment transcriptional activation of C/ΕΒPε.
Taken together, we conclude that HIV-1 protease inhibitors enhance the ability of ATRA to inhibit the growth and induce the differentiation of myeloid leukemia cells in vitro. HIV-1 protease inhibitors might act as an antagonist of CRABPs, resulting in increased intracellular concentrations of ATRA. In addition, HIV-1 protease inhibitors possess inhibitory effects on CYPs and P-gp, both of which might be involved in the acquisition of ATRA resistance in patients with APL. HIV-1 protease inhibitors could have a possible role for patients with ATRA-resistant APL and P-gp–mediated drug-resistant leukemia and other cancer cells. Furthermore, the pharmacokinetics of some anticancer drugs might be improved with concomitant administration of ritonavir, which can inhibit metabolism of selective drugs through the CYPs pathway.
Acknowledgments
We thank Patricia Lin (Flow Cytometry Core Facility, Cedars-Sinai Medical Center) and Hiroshi Kawabata (Division of Hematology/Oncology, Cedars-Sinai Medical Center) for their generous technical assistance. We also thank Kim Burgin for her excellent secretarial help.
Supported in part by National Institutes of Health grants, the Parker Hughes Trust, Horn Foundation, C. and H. Koeffler Fund, and Lymphoma Foundation of America. H.P.K. is a member of the Jonsson Comprehensive Cancer Center and holds an endowed Mark Goodson Chair of Oncology Research at Cedars-Sinai Medical Center.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Takayuki Ikezoe, Division of Hematology/Oncology, Cedars-Sinai Research Institute, UCLA School of Medicine, 8700 Beverly Blvd, Los Angeles, CA 90048.


![Fig. 5. Enhancement of C/EBPε mRNA expression in NB4 cells by indinavir. / NB4 cells were cultured for 48 hours under various conditions: lane 1, (diluent control); lane 2, indinavir (2 × 10−5 mol/L); lane 3, ATRA (10−9 mol/L); lane 4, combination of indinavir (2 × 10−5 mol/L) and ATRA (10−9mol/L). Total RNA was extracted and analyzed by Northern blot technique (30 μg/lane) and hybridized with [32P]-labeled C/EBPε cDNA probe as described in “Materials and methods.” The same blot was rehybridized with [32P]-labeled GAPDH probe to show equality of RNA loading in each lane. Results are expressed as fold increase in expression as compared with control NB4 cells (lane 1). The band intensity was measured using a densinometer.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/96/10/10.1182_blood.v96.10.3553/5/m_h82200383005.jpeg?Expires=1769093834&Signature=zC7MBIMEuuDqPV-7wlHaQYfmdPqcif0~jXKC64A85d1aJhl-gvhZULk9gpW9dNSYRj27KAibyjykOWVTVdsH9SYOZxOdnTOW8ZSnC27BW4cWTfmVzgkv2J0-6At-28QV2xHX~KBSaJ~LD5MoSQzrEiD4ID8KW~dVeo15TU63oICBLq0KUDnS95rtqy9M3ROqTkB9rqxmQjIQD8hfBKsPuV71hqvkdXCAYhDt-~3TIHFId30aE37REhYc5SriRvMpT9KQcNgkmyfHcE4qVUVpYeFSQNe2M2vghSlKPw8JrbwIIEK7AvpEXTdGEGEbVyD53WhFiBOaanBqo0f8wiPeXQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal