Abstract
In a complementary DNA (cDNA) screening of murine Th2-skewed lymphocytes with our recently developed signal sequence trap method termed SST-REX, a novel type 1 cytokine receptor, Delta1 (δ1), was identified. Although δ1 is ubiquitously expressed in multiple tissues, the expression level is higher in Th2-skewed lymphocytes than in Th1-skewed ones. The δ1 cDNA encodes a 359–amino acid type 1 membrane protein. The extracellular domain of 206 amino acids showed 24% identity with the murine common γ receptor that is shared among the receptors for interleukin(IL)-2, IL-4, IL-7, IL-9, and IL-15. The membrane-proximal region of δ1 includes a box1 motif, which is important for association with Janus kinases (JAKs), and showed a significant homology with that of the mouse erythropoietin receptor (EPOR). A box2 motif was also found in close proximity to the box1 region. Dimerization of the cytoplasmic region of δ1 alone did not transduce proliferative signals in IL-3–dependent cell lines. However, the membrane-proximal region of δ1 could substitute for that of human EPOR in transmitting proliferative signals and activating JAK2. These results suggest that δ1 is a subunit of cytokine receptor that may be involved in multiple receptor systems and play a regulatory role in the immune system and hematopoiesis.
Cytokine systems regulate cellular growth and differentiation in hematopoiesis, immune systems, tissue development, and morphogenesis.1,2 Identification of novel members of the cytokine family and their receptors is of great importance because they play key roles in regulating a broad-range biological response. Cytokines have a highly conserved 4-helix bundle tertiary structure but have a low homology in the primary amino acid sequence.3 Therefore, identification of novel cytokines using homology-based cloning methods has been rather difficult. Cytokine signals are mediated through specific receptor complexes, including type 1 and type 2 cytokine receptors, the tumor necrosis factor receptor superfamily, the tumor growth factor receptor superfamily, and tyrosine kinase receptors.4-8 The type 1 cytokine receptors transmit signals of the group of cytokines involved in hematopoiesis and blood cell functions, and they are also known as the hemopoietin receptor family. Members of the type 1 cytokine receptor family are readily identified by a cytokine receptor module comprising 2 fibronectin type III–like domains containing the N-terminal domain with conserved cysteine residues and by the highly conserved Trp-Ser-X-Trp-Ser (WSXWS) sequence present above the transmembrane domain.9-12 Structural alterations of these receptors occur in tumorigenesis,13immunodeficiency,14 genetic susceptibility to some pathologic conditions such as allergic sensitivity,15,16and metabolic disorders such as alveolar proteinosis.17Therefore, the molecular cloning of a novel cytokine receptor may help to understand the pathogenesis of some disease and to tailor treatments accordingly. Most members of the type 1 cytokine receptor family have been cloned using ligand binding as an assay. Alternatively, oligonucleotides for the WSXWS motif were used as hybridization probes, and degenerate polymerase chain reaction (PCR) with primers for the highly conserved region of type 1 cytokine receptors was also used. Nowadays, some cytokine receptors can be identified in a search of expressed sequence tag (EST) database as a result of homology with known cytokine receptors.18 19
We developed a method termed SST-REX (signal sequence trap by retrovirus-mediated expression screening)20 that detects signal sequences in complementary DNA (cDNA) fragments based on their ability to redirect a constitutively active mutant of a cytokine receptor21 to the cell surface, thereby allowing interleukin-3 (IL-3)–independent growth of otherwise IL-3–dependent Ba/F3 cells.20 SST-REX is an improved version of the signal sequence trap methods originally described by Tashiro et al22 and modified by Klein et al23 by which cDNAs for secreted molecules and surface proteins are selectively isolated. We now report isolation and characterization of a novel type 1 cytokine receptor, Delta 1 (δ1), cloned using SST-REX.
Materials and methods
Reagents
Recombinant murine IL-3 (mIL-3) was produced in silkworm. Recombinant human erythropoietin (hEPO) was a gift from the Chugai Pharmaceutical Co (Tokyo, Japan). Recombinant human thrombopoeitin (hTPO) was obtained from R & D Systems (Minneapolis, MN). Antibodies directed against TYK2, Janus kinase (JAK)1, and JAK2 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies raised against JAK3 and the 4G10 antibody, raised against phosphotyrosine, were purchased from UBI (Lake Placid, NY). The anti-FLAG M2 antibody was purchased from Eastman Chemical Company (Kingsport, TN).
Cell lines and cell culture
Ba/F3,24 FDC-P1,25 LG, LGM,26and 32D 27 cells were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS), 2 mmol/L glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin, and 1 ng/mL mIL-3. CTLL-2 cells28 were maintained in RPMI 1640 supplemented with 10% FCS, 2 mmol/L glutamine, 50 μmol/L 2-mercaptoethanol, 100 U/mL penicillin, 100 μg/mL streptomycin, and 20 ng/ml mIL-2. NIH3T3 fibroblast cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% FCS (DMEM/10%FCS). An ecotropic retrovirus packaging cell line BOSC23 29 was maintained in DMEM containing 10% FCS (DMEM/10%FCS) and guanine phosphoribosyl transferase selection reagents (Speciality Media, Lavallette, NJ). The cells were transferred to DMEM/10%FCS without guanine phosphoribosyl transferase selection reagents 2 days before transfection.
Construction of the cDNA library for SST-REX
Induction of antigen-specific activation of Th1/Th2 cells was performed using DO11.10 transgenic mice as described,30 and poly(A)+ RNA samples were prepared from resting or activated Th1 and Th2 cells using FastTrack2.0 Kit (Invitrogen Carlsbad, CA). Complementary DNA (cDNA) was synthesized from poly(A)+ RNA of resting Th1 and Th2 cells with random hexamers using SuperScript Choice System (Gibco-BRL, Rockville, MD) according to the manufacturer's instructions. The synthesized cDNA was size-separated through SizeSep 400 Spun Column (Pharmacia, Uppsala, Sweden) and inserted intoBstXI sites of pMX-SST using BstXI adaptors (Invitrogen). The ligated DNA was electroporated into DH10B cells (Electromax, Gibco-BRL) using an E-coli Pulser (Bio-Rad, Hercules, CA) at 1.8 kV. Plasmid DNA was prepared using JETstar (Genomed, Research Triangle Park, NC) from 200-ml Escherichia coli culture. Infection of Ba/F3 cells with high-titer retroviruses derived from the library for SST and, also, isolation of cDNA fragments by PCR from factor-independent clones were performed as described.20
Construction of a cDNA library from resting Th2 cells
Complementary DNA was synthesized from poly(A)+ RNA from resting Th2 cells with oligo-dT primers, using SuperScript Choice System, as described above, and inserted into BstXI sites of pME18S,31 using BstXI adaptors. The ligated DNA was electroporated, and plasmid DNA was prepared as described above.
Cloning of δ1 cDNAs
A biotinylated probe of 407 base pairs (bp) was made by PCR from the δ1 cDNA with oligoprimers 5′GGTGATGTCACAGTCGTCTGCCATG3′ and 5′ TGCCAGGCGTCCTCATCGTCATT3′ using 50-μM Biotin-21-dUTP (Clontech Laboratories, Palo Alto, CA), 200-μM dATP, 200-μM dGTP, 200-μM dCTP, and 10-μM dTTP. An oligo-dT–primed cDNA library of resting Th2 cells was screened using RecA protein coated with biotinylated linear DNA probes as described.32-34
Northern blot analysis
A Mouse Multiple Tissue Northern Blot (Clontech) was probed with the 407-bp δ1-specific PCR fragments generated with the same set of primers as described above. The blots were hybridized overnight in 50% formamide, 5 × SSC, 1% sodium dodecyl sulfate (SDS), 6 × Denhardt's reagent, and 100 μg/ml of salmon sperm DNA at 42°C with a 32P-labeled randomly primed probe and washed at 55°C in 0.1 × SSC and 0.1% SDS, followed by exposure to radiographic film. Poly(A)+ RNA samples (3 μg) prepared from various mouse cell lines using FastTrack2.0 Kit were separated by electrophoresis through a 1% agarose formaldehyde gel and transferred to Hybond-N+ (Amersham, Arlington Heights, IL) nylon filter. The Northern blot was hybridized and washed as described above.
Construction and expression of chimeric receptors consisting of the human receptor for EPO or TPO and δ1
For expression of the wild type hEPOR (hEPO receptor), a full-length hEPOR35 was cloned into the EcoRI and NotI sites of the pMX-puro retrovirus vector.36 For construction of chimeric receptors hEPOR-δ1R and hMPL-δ1R, an EcoRI site was inserted between the extracellular and transmembrane domains of each cDNA. Then, the cytoplasmic domain of δ1 cDNA generated by PCR with high-fidelity DNA polymerase Pyrobest (Takara Otsu, Shiga, Japan) was cloned into pMX-puro at 5′-EcoRI and 3′-NotI sites, and the EcoRI fragment containing the extracellular domain of hEPOR or hMPL37 was cloned into a 5′-EcoRI site). The primers used to createEcoRI and NotI sites in the δ1 cDNA were 5′-AAAGAATTCCCGCCCCTCCTGCCCCTGGGC-3′ and 5′-GCTGGCGGCCGCACCTGCAGGCGC-3′. The primers used to createEcoRI sites in hEPOR cDNA were 5′-AAAGAATTCGGGGGCTGTATCATGGAC-3′ and 5′-AAAGAATTCGGGGTCCAGGTCGCTAGG-3′. The primers used to create EcoRI sites in hMPL cDNA were 5′-GGTAGGGAATTCCGGAATTTCCTCGAGATG-3′ and 5′-AAAGAATTCCCAGGCGGTCTCGGTGGCGGT-3′. To construct a chimeric receptor EDER, a 57–amino acid stretch covering the transmembrane domain and the box1 region of hEPOR was replaced with the corresponding region of δ1; first, an HpaI site was inserted between the extracellular and transmembrane domains, and anXhoI site was inserted downstream of the conserved tryptophan residue in the cytoplasmic domain of hEPOR. Second, the box1 region of δ1 cDNA was cloned into the EcoRI and NotI sites of the pMX-puro. Then, the cytoplasmic region of hEPOR was cloned into theXhoI and NotI sites, and the extracellular domain of hEPOR was cloned into the EcoRI and HpaI sites. The primers used to create 5′-EcoRI and HpaI sites and 3′-XhoI and NotI sites at the end of the δ1 cDNA spanning the transmembrane and the membrane-proximal region were 5′-AAAGAATTCGTTAACCCGCCCCTCCTGCCCCTGGGG-3′ and 5′-AAAGCGGCCGCCTCGAGCCAGGCCTGGAAGTTCCC-3′. The primers used to create 5′-XhoI and 3′-NotI sites in the cytoplasmic region of hEPOR were 5′-AAACTCGAGCTGTACCAGAATGATGGC-3′ and 5′-AAAGCGGCCGCTCACTTGTCGTCATCGTCCTTGTAATCAGAG- CAAGCCACATAGCT-3′. The primers used to create 5′-EcoRI and 3′-HpaI sites in the extracellular region of hEPOR were 5′-AAAGAATTCGGGGGCTGTATCATGGAC-3′ and 5′-AAAGTTAACGGGGTCCAGGTCGCTAGG-3′. All constructs were confirmed by restriction enzyme analyses and sequencing.
The resulting expression plasmids were introduced into FDC-P1 via retrovirus infection as described.38 Infected cell lines were selected in the presence of 1 μg/ml of puromycin (Sigma, St. Louis, MO) to obtain a stable transfectant.
Construction and expression of a tagged molecule of δ1
To construct the tagged δ1, δ1-FLAG was designed by fusing the FLAG-sequence (DYKDDDDK) to the C-terminus of δ1. A PCR fragment of the δ1-FLAG was cloned into pMX-puro retrovirus vector at 5′-EcoRI and 3′-NotI sites. The primers used to add the FLAG sequence to the C-terminus of δ1 was 5′-AGGGAATTCCGGAATTTCCTCGAGATC-3′ and 5′-AAAGCGGCCGCTCACTTGTCGTCATCGTCCTTGTAATCC- AGGGTCATGTAGCCGCTGTC-3′.
Immunoprecipitation and Western blotting
Cells were lysed in the lysis buffer (50 mM Tris-HCl, pH 7.5; 150 mM NaCl; 1% Triton X-100; 1 mM EDTA; 0.2 mM Na3VO4; and 2 mM PMSF). For surface labeling of the cells, NHS-LC-Biotin (Pierce, Rockford, IL) was added prior to cell lysis. Lysates were cleared of debris by centrifugation at 12 000g for 40 minutes, and the supernatants were incubated with antibodies at 4°C for 2 hours. Immune complexes were precipitated with protein A-Sepharose (Pharmacia), washed with the lysis buffer, and proteins were eluted with the sample buffer (62.5 mM Tris-HCl, pH 6.8; 2% SDS; 10% glycerol; 5% 2-mercaptoethanol; and 0.02% bromophenol blue) for SDS-PAGE (SDS-polyacrylamide gel electrophoresis). The eluted proteins were electrophoresed on an 8%-to-16% gradient gel (Gradipore, North Ryde, Australia). Membranes were probed with antibodies or streptavidin-horseradish peroxidase (Vector Laboratories, Burlingame, CA) and visualized using the enhanced chemiluminescence detection system (Amersham) as described by the manufacturer.
Chromosomal localization
Interspecific backcross progenies were generated by mating (C57BL/6J × Mus spretus) F1 females and C57BL/6J males.39 A total of 205 N2 mice were used to map the δ1 locus. DNA isolation, restriction enzyme digestion, agarose gel electrophoresis, Southern blot transfer, and hybridization were performed essentially as described.40 All blots were prepared with Hybond-N+ nylon membrane (Amersham). The probe, a 407-bp fragment of mouse cDNA, was labeled with [α32P] dCTP using a random primed labeling kit (Stratagene, La Jolla, CA); washing was done to a final stringency of 1.0 × standard saline citrate (SSC), 0.1% SDS, 65°C. C57BL/6J and M. spretus DNAs were digested with several enzymes and analyzed by Southern blot hybridization for informative restriction fragment length polymorphisms (RFLPs) using a mouse cDNA δ1 probe. An ScaI fragment of 8.7 kilobases (kb) or 11.0 kb was detected in C57BL/6J DNA or M. spretus DNA, respectively. The presence or absence of 11.0-kbScaI M. spretus–specific fragment was followed in backcross mice.
A description of the probes and RFLPs for the loci linked to δ1, including Fgf5, Dmp1, Gfi1, and Crybb2 has been reported elsewhere.41 42 Recombination distances were calculated using Map Manager, version 2.6.5. Gene order was determined by minimizing the number of recombination events required to explain the allele distribution patterns.
Results
Isolation and sequencing of the δ1 cDNA clone encoding a novel cytokine receptor
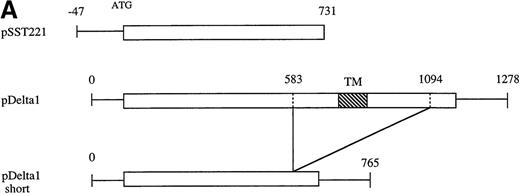
In the screening using the SST-REX system,20 Ba/F3 clones, which are transduced with cDNAs containing signal sequences, showed factor-independent growth through surface expression of a constitutively active MPL21 as a fusion protein. Among cDNA clones that we obtained from either a Th1 or a Th2 cell library using SST-REX, 1 of the clones, f215 clone, showed a significant homology with the murine common γ receptor (γc)43at the amino acid level. A full-length cDNA was obtained from an oligo-dT–primed cDNA library derived from resting Th2 cells using the RecA screening system32-34 and was confirmed to contain a sequence that was identical to f215 (Figure1A).
Complementary DNA for the murine δ1.
(A) Schematic representation of 2 independent cDNAs for murine δ1 and a short form of δ1. The box represents the open reading frame. TM represents the transmembrane region. A clone obtained by SST-REX is also shown in the panel. (B) Nucleotide and deduced amino acid sequences of the cloned δ1 cDNA. Numbers at the right margin indicate positions of nucleotides. Thick underlines indicate a putative signal sequence and a transmembrane region. Thin underlines indicate the box1 and box2 regions. The boxed region is a WSXWS box-like sequence.
Complementary DNA for the murine δ1.
(A) Schematic representation of 2 independent cDNAs for murine δ1 and a short form of δ1. The box represents the open reading frame. TM represents the transmembrane region. A clone obtained by SST-REX is also shown in the panel. (B) Nucleotide and deduced amino acid sequences of the cloned δ1 cDNA. Numbers at the right margin indicate positions of nucleotides. Thick underlines indicate a putative signal sequence and a transmembrane region. Thin underlines indicate the box1 and box2 regions. The boxed region is a WSXWS box-like sequence.
This cDNA contains an open reading frame of 1077 nucleotides, which encoded a putative protein of 359 amino acids (Figure 1B). The predicted amino acid sequence included a potential hydrophobic signal peptide, followed by a classical cytokine receptor module containing 2 of the 4 conserved cysteine residues, fibronectin type III–like domains, a WS boxlike sequence PSEWT, a 23–amino acid transmembrane domain, and a 106–amino acid cytoplasmic domain. Examination of the cytoplasmic domain of δ1 identified a membrane-proximal box1 motif (Apolar-X-X-X-Aliphatic-Pro-X-Pro) (residues 262-269),44which is known to be important for association with JAKs.
δ1 showed a moderate homology at the extracellular domain with the mouse γc (24% identical, 36% similar; Figure2A). The membrane-proximal region of δ1, including the box1 region, showed a significant homology with those of the mouse EPOR and other receptor subunits, including the common β chain (βc) (Figure 2B), and harbored the conserved tryptophan W287, which localized downstream of the box1 and is required for JAK2 association.45 The 12–amino acid sequence from A311 to L322 had a considerable similarity with the box2 region, which is also important for signal transduction in many cytokine receptors (Figure 2B).
Identity and similarity of δ1 to other known cytokine receptors.
(A) Alignment of the extracellular domains of δ1 and the mouse common γ receptor (γc). (B) Comparison of the sequences of various hematopoietic growth factor receptors. A match of the amino acid residue between δ1 and other receptors is indicated by “*” (identical) or “:” (similar). Previously defined regions of homology, termed homology box1 and 2, are shown. The box1 and box2 are short characteristic motifs found in the membrane-proximal regions of type 1 cytokine receptors.44The box1 motif is defined by the PXP motif (proline, X, proline) preceded by hydrophobic sequences. Box2 is defined as a cluster of hydrophobic amino acids harboring some negatively charged amino acids.
Identity and similarity of δ1 to other known cytokine receptors.
(A) Alignment of the extracellular domains of δ1 and the mouse common γ receptor (γc). (B) Comparison of the sequences of various hematopoietic growth factor receptors. A match of the amino acid residue between δ1 and other receptors is indicated by “*” (identical) or “:” (similar). Previously defined regions of homology, termed homology box1 and 2, are shown. The box1 and box2 are short characteristic motifs found in the membrane-proximal regions of type 1 cytokine receptors.44The box1 motif is defined by the PXP motif (proline, X, proline) preceded by hydrophobic sequences. Box2 is defined as a cluster of hydrophobic amino acids harboring some negatively charged amino acids.
A truncated form of δ1 was also cloned from an oligo-dT–primed Th2 cDNA library (Figure 1). The short form lacked membrane-proximal and transmembrane domains, and potentially encoded a soluble form of δ1.
Expression of δ1
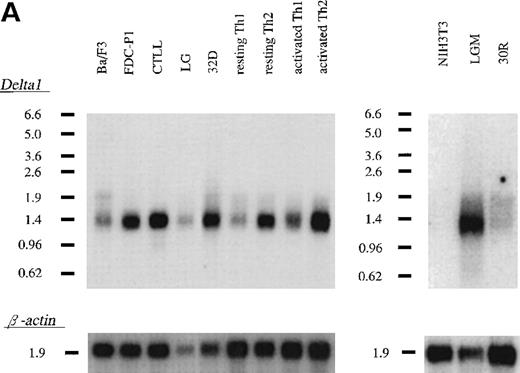
Expression of δ1 was examined by Northern blots for several mouse cell lines and Th1- and Th2-skewed cells (Figure3A). A single transcript of 1.4 kb was identified in all myeloid cell lines examined, CTLL2, and resting and activated murine Th1 and Th2 cells but not in 2 fibroblast cell lines, including NIH3T3 cells. Th2-skewed cells showed a much higher expression than Th1-skewed cells. Slight up-regulation of the expression by T-cell activation was observed in both T-cell cultures.
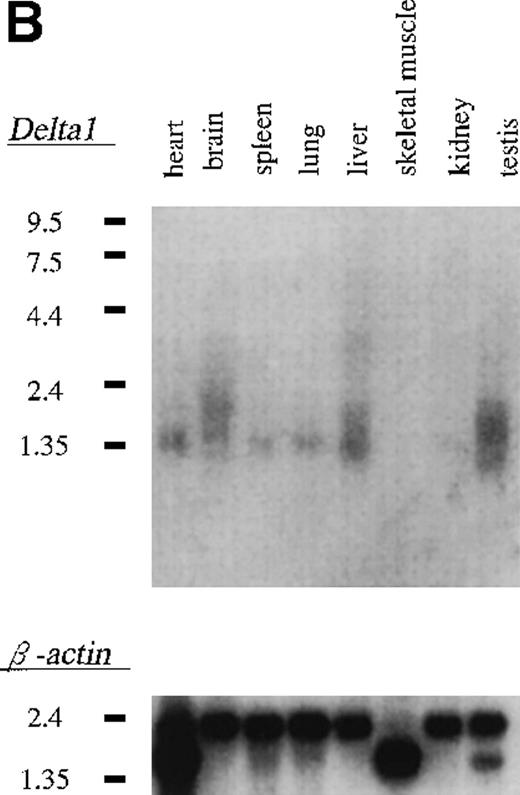
Expression of δ1 in cell lines and mouse tissues.
(A) Northern blot hybridization of murine cell lines; 3 μg of poly(A)+ RNA were separated through 1.0% agarose formaldehyde gel and then blotted using a 32P-labeled δ1 probe. Mobilities of the RNA molecular weight marker are given in kilobases. (B) Northern blot hybridization of a mouse multiple tissue blot.
Expression of δ1 in cell lines and mouse tissues.
(A) Northern blot hybridization of murine cell lines; 3 μg of poly(A)+ RNA were separated through 1.0% agarose formaldehyde gel and then blotted using a 32P-labeled δ1 probe. Mobilities of the RNA molecular weight marker are given in kilobases. (B) Northern blot hybridization of a mouse multiple tissue blot.
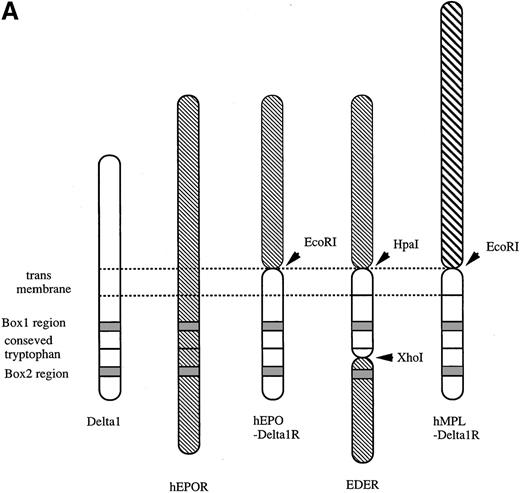
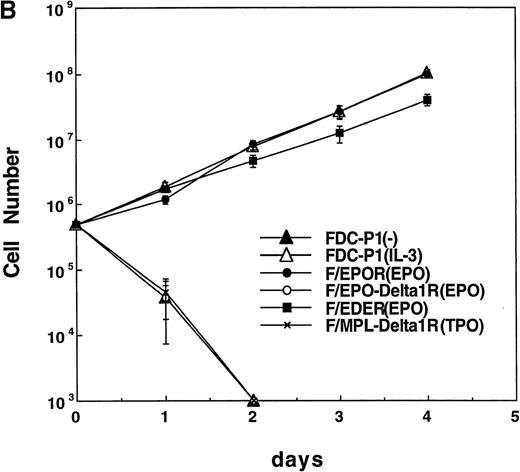
Structure of chimeric receptors and factor responsiveness of FDC-P1 transfectants.
(A) Shown schematically are the wild type δ1 and EPOR together with the chimeras hEPO-δ1R, hEDER, and hMPL-δ1R. Fused domains are designated by restriction sites generated within cognate cDNAs. (B) Time course of growth of the FDC-P1 cells retrovirally transduced with various receptor constructs. Parental FDC-P1 and FDC-P1 cells expressing hEPOR, hEPOR-δ1R, EDER, and hMPL-δ1R were cultivated in the presense of 1 ng/mL of mIL-3, 2 U/mL of hEPO, 2 U/mL of hEPO, 2 U/mL of hEPO, and 100 ng/mL of hTPO, respectively, and the cell number was counted at the indicated times.
Structure of chimeric receptors and factor responsiveness of FDC-P1 transfectants.
(A) Shown schematically are the wild type δ1 and EPOR together with the chimeras hEPO-δ1R, hEDER, and hMPL-δ1R. Fused domains are designated by restriction sites generated within cognate cDNAs. (B) Time course of growth of the FDC-P1 cells retrovirally transduced with various receptor constructs. Parental FDC-P1 and FDC-P1 cells expressing hEPOR, hEPOR-δ1R, EDER, and hMPL-δ1R were cultivated in the presense of 1 ng/mL of mIL-3, 2 U/mL of hEPO, 2 U/mL of hEPO, 2 U/mL of hEPO, and 100 ng/mL of hTPO, respectively, and the cell number was counted at the indicated times.
In Northern blot analysis of multiple tissues, a single transcript of 1.4 kb was identified in heart, brain, spleen, lung, kidney, and testis but not in skeletal muscle (Figure 3B).
Functional analysis of δ1
First, we overexpressed δ1 in Ba/F3 cells that proliferate or survive in response to IL-3 or IL-4, respectively. However, the overexpression of δ1 did not enhance Ba/F3 response to these cytokines. Moreover, these cytokines did not induce tyrosine phosphorylation of δ1-FLAG in Ba/F3 cells overexpressing δ1, suggesting that δ1 was not involved in IL-3 and IL-4 signaling (data not shown). We next examined the function of the cytoplasmic domain of δ1; chimeric receptors between δ1 and hEPOR or hMPL were generated to characterize the effect of homodimerization of the cytoplasmic domain of δ1. EDER chimera was made by substitution of the membrane-proximal region of hEPOR with that of the δ1 (Figure 4A) to evaluate function of the membrane-proximal region of δ1, including box1, as described in “Materials and methods.” With regard to mitogenic properties, FDC-P1 cells expressing the EDER proliferated efficiently in response to hEPO (Figure 4B), while FDC-P1 cells expressing a chimeric receptor hEPO-δ1R or hMPL-δ1R did not transmit proliferative signals in response to EPO or TPO, respectively.
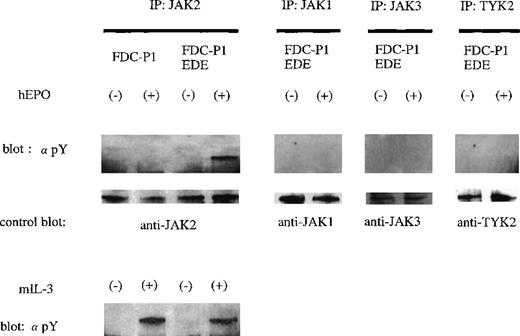
Phosphorylation of Janus kinases by the EDE chimeric receptor
The mitogenic activity of the EDER suggested that JAK was activated by hEPO-induced dimerization. Indeed, Western blot analysis detected hEPO-dependent JAK2 phosphorylation in cells expressing the EDER (Figure 5). JAK1, JAK3, and TYK2 were not phosphorylated in response to hEPO stimulation in the cells expressing the EDER. None of the 4 JAKs were phosphorylated in response to EPO or TPO stimulation in cells expressing a chimeric receptor hEPO-δ1R or hMPL-δ1R, respectively (data not shown).
Activation of JAK2 kinase through the ectopically expressed chimeric EDE receptor in FDC-P1 cells.
The ability of the chimeric receptor EDER to mediate hEPO- dependent activation of JAK kinases was assayed by immunoprecipitation with antibody to JAK2 and Western blotting with antibody to phosphotyrosine (monoclonal antibody 4G10). Reprobing was carried out using the anti-JAK2 antibody after stripping. Similar analyses were also performed using anti-JAK1, anti-JAK3, and anti-TYK2 antibodies.
Activation of JAK2 kinase through the ectopically expressed chimeric EDE receptor in FDC-P1 cells.
The ability of the chimeric receptor EDER to mediate hEPO- dependent activation of JAK kinases was assayed by immunoprecipitation with antibody to JAK2 and Western blotting with antibody to phosphotyrosine (monoclonal antibody 4G10). Reprobing was carried out using the anti-JAK2 antibody after stripping. Similar analyses were also performed using anti-JAK1, anti-JAK3, and anti-TYK2 antibodies.
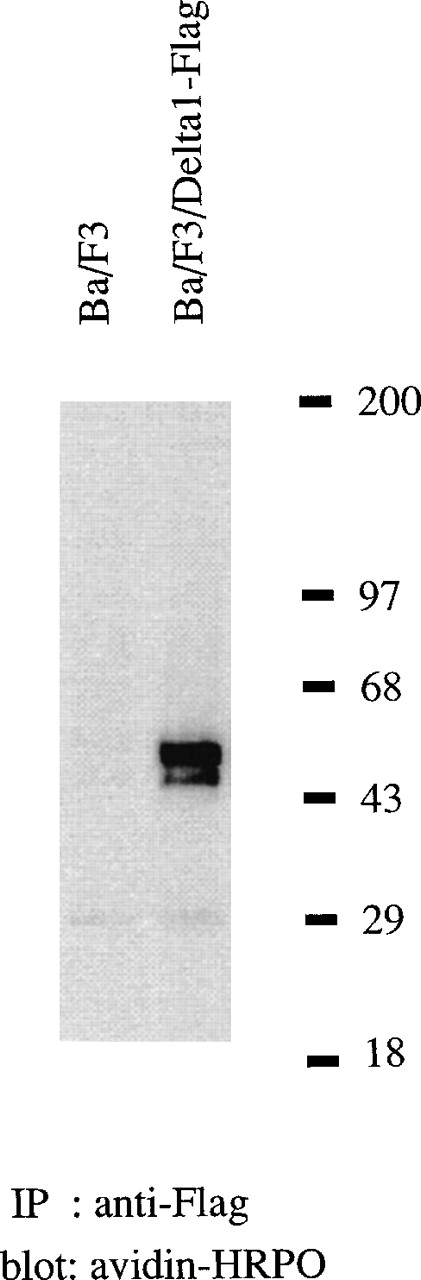
Expression of δ1 protein
A retrovirus construct, pMX-puro δ1-FLAG, was designed to express a tagged molecule of δ1 and was introduced into Ba/F3 cells via retrovirus infection. The cell lysate was immunoprecipitated by the anti-FLAG antibody (Figure 6). The expressed δ1 protein had a molecular mass of 50 kd, which is larger than the calculated molecular mass of 37 791 d. The difference is probably due to attachment of sugar moieties to some of the 2 predicted potential N-linked glycosylation sites found in the extracellular domain of δ1.
Immunoprecipitation of tagged-δ1 expressed on Ba/F3 cells, under reducing conditions.
Ba/F3 cells transduced with tagged-δ1 and parenteral cells were surface-labeled with sulfo-NHS-LC-biotin, and the cell lysates were precipitated with anti-FLAG antibody followed by the blotting with streptavidin-horseradish peroxidase. Positions of δ1-FLAG (arrowhead) and molecular weight markers are indicated.
Immunoprecipitation of tagged-δ1 expressed on Ba/F3 cells, under reducing conditions.
Ba/F3 cells transduced with tagged-δ1 and parenteral cells were surface-labeled with sulfo-NHS-LC-biotin, and the cell lysates were precipitated with anti-FLAG antibody followed by the blotting with streptavidin-horseradish peroxidase. Positions of δ1-FLAG (arrowhead) and molecular weight markers are indicated.
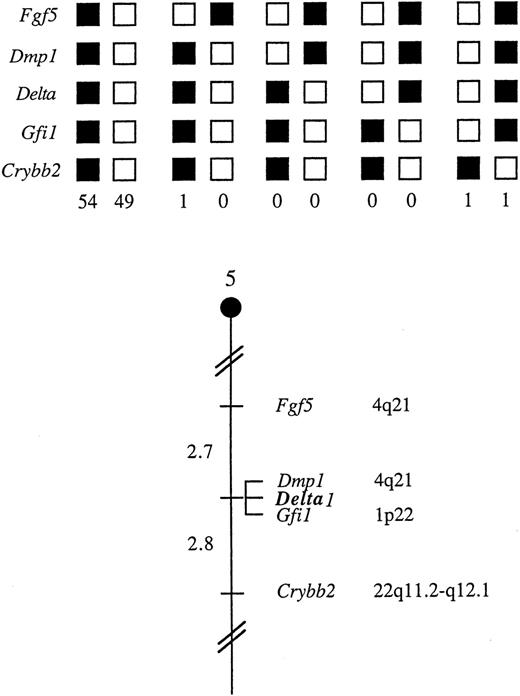
Chromosomal mapping of the δ1 gene
The mouse chromosomal location of δ1 was determined by interspecific backcross analysis using progeny derived from matings of ([C57BL/6J x M. spretus] F1 x C57BL/6J) mice. This interspecific backcross mapping panel has been typed for more than 2900 loci that are well distributed among all the autosomes as well as the X chromosome.39 The 11.0-kb ScaI M. spretusRFLP (see “Materials and methods”) was used to follow segregation of the δ1 locus in backcross mice. The mapping results indicated that δ1 is located in the central region of mouse chromosome 5 linked to Fgf5, Dmp1, Gfi1, and Crybb2. Although 106 mice were analyzed for every marker and are shown in the segregation analysis (Figure 7), up to 154 mice were typed for some pairs of markers. Each locus was analyzed in pairwise combinations for recombination frequencies using the additional data. The ratios of the total number of mice exhibiting recombinant chromosomes to the total number of mice analyzed for each pair of loci and the most likely gene order are as follows: centromere — Fgf5-4/151 — Dmp1-0/136 — δ1-0/154 — Gfi1-4/145 — Crybb2. The recombination frequencies [expressed as genetic distances in centiMorgans (cM) ± the standard error] are: centromere — Fgf5-2.7 ± 1.3 — [Dmp1, δ1, Gfi1] — 2.8 ± 1.4 — Crybb2. No recombinants were detected between Dmp1 and δ1 in 136 or between δ1 and Gfi1 in 154 mice typed in common, suggesting that 2 loci in each pair are within 2.2 and 1.9 cM of each other, respectively (upper 95% confidence limit).
δ1 maps in the central region of mouse chromosome 5.
δ1 was placed on mouse chromosome 5 by interspecific backcross analysis. The segregation patterns of δ1 and flanking genes in 106 backcross animals that were typed for all loci are shown at the top. For individual pairs of loci, more than 106 mice were typed. Each column represents the chromosome identified in the backcross progeny that was inherited from the (C57BL/6J x M. spretus) F1 parental. The shaded boxes represent the presence of a C57BL/6J allele, and white boxes represent the presense of an M. spretus allele. The number of offspring inheriting each type of chromosome is listed at the bottom of each column. A partial chromosome 5 linkage map showing the location of δ1 in relation to linked genes is shown at the bottom. Recombination distances between loci (in centimorgans) are shown to the left of the chromosome, and the positions of loci in human chromosomes, where known, are shown to the right. References for the human map positions of loci cited in this study can be obtained from GDB (Genome Data Base), a computerized database of human linkage information maintained by The William H. Welch Medical Library of The Johns Hopkins University (Baltimore, MD).
δ1 maps in the central region of mouse chromosome 5.
δ1 was placed on mouse chromosome 5 by interspecific backcross analysis. The segregation patterns of δ1 and flanking genes in 106 backcross animals that were typed for all loci are shown at the top. For individual pairs of loci, more than 106 mice were typed. Each column represents the chromosome identified in the backcross progeny that was inherited from the (C57BL/6J x M. spretus) F1 parental. The shaded boxes represent the presence of a C57BL/6J allele, and white boxes represent the presense of an M. spretus allele. The number of offspring inheriting each type of chromosome is listed at the bottom of each column. A partial chromosome 5 linkage map showing the location of δ1 in relation to linked genes is shown at the bottom. Recombination distances between loci (in centimorgans) are shown to the left of the chromosome, and the positions of loci in human chromosomes, where known, are shown to the right. References for the human map positions of loci cited in this study can be obtained from GDB (Genome Data Base), a computerized database of human linkage information maintained by The William H. Welch Medical Library of The Johns Hopkins University (Baltimore, MD).
Discussion
We isolated and characterized a novel member of the type 1 cytokine receptor family, δ1, from a cDNA library of the murine Th2-skewed cells using SST-REX; δ1 has a WS box-like sequence, PSEWT, and the box1 and box2 motifs in its membrane-proximal region. 44
When compared to other members of the type 1 cytokine receptor family, δ1 showed the closest similarity to the murine γc in the extracellular domain, with a 24% identity at the amino acid level. Its overall structure is also similar to that of γc, not only in total amino acid length (δ1 359 vs γc 369) but also in length of the extracellular domain (δ1 206 vs γc 232) and the cytoplasmic domain (δ1 106 vs γc 86). Moreover, the expression of δ1 is ubiquitous in hematopoietic cells, as is the case for γc. Although the WS box of δ1 is diversed from the typical sequence of WSXWS, the presence of box1 and box2 domains and the similarity to the γc indicated that δ1 constitutes a member of the type 1 cytokine receptor family. Because a membrane-proximal 50–amino acid segment of the cytoplasmic domain includes both box1 and box2 regions that are essential for supporting signaling and cell proliferation,45-49 it is likely that δ1 is involved in signal transduction.
We were not able to obtain a human homologue of δ1 in cross-hybridization screening using the mouse δ1 as a probe under low stringent conditions; nor did we find any human ESTs homologous to δ1 (unpublished results), suggesting that the human counterpart of δ1 has a low homology with that of the mouse or that there is not a corresponding gene in the human. The central region of mouse chromosome 5, where we identified δ1, shares regions of homology with human chromosomes 4q, 1p, and 22q (summarized in Figure 7), suggesting that the human homologue of δ1, if any, will map to 1 of these chromosomal regions.
In the functional analysis of δ1, hEPOR-δ1R or hMPL-δ1R, the 2 chimeric receptors in which the extracellular domain of δ1 was replaced by the human EPO or TPO receptors, did not transduce proliferative signals in response to hEPO or hTPO, respectively. These results suggested that homodimerization of δ1 could not transduce proliferating signals, while other cytokine receptors, including glycoprotein130, βc, and IL-2Rβ, containing both the box1 and box2 regions were able to transduce proliferating signals upon homodimerization.50-52 On the other hand, the EDER chimera, in which the box1 region of the human EPOR was replaced with that of δ1, induced phosphorylation of JAK2 and cell proliferation in response to human EPO, indicating that the membrane-proximal region of δ1 can substitute for that of the EPOR. It is known that EPO stimulation induces tyrosine phosphorylation of JAK253 and that the box1 motif of EPOR is required for association with JAK2.46 In addition, the membrane-proximal region of the EPOR specifies JAK2 activation within a heterologous IL-2 receptor complex.54 The present data indicate that the box1 motif of δ1 is capable of activating JAK2 in heterologous receptor complexes. We speculate that δ1 may be a shared receptor subunit like γc and is involved in signal transduction through JAK2 activation. Whether δ1 has its own ligand remains to be determined.
Although the expression of δ1 is ubiquitous in the immune and the hematopoietic cells, it is clearly up-regulated in Th2-skewed cells, when compared to Th1-skewed cells, suggesting that up-regulation of δ1 expression has something to do with differentiation into Th2 cells. In addition, activation of these cells by antibody
stimulation significantly enhanced the expression of δ both in Th1- and Th2-skewed cells, implicating δ1 in the regulation of the immune system. These data lead us to examine whether δ1 is involved in the IL-4 receptor system; however, overexpression of δ1 did not alter cellular response to IL-4, and tyrosine phosphorylation of δ1-FLAG was not induced in response to IL-4 (data not shown). These results suggested that δ1 was not involved in the IL-4 receptor system, but potential involvement of δ1 in IL-4 signaling cannot be ruled out at present.
Thus, it is suggested that a novel cytokine receptor δ1, which is similar to γc in its structure and ubiquitous expression, is a receptor subunit that plays some roles in signal transduction of the immune system and hematopoiesis.
Acknowledgments
We thank M. Ohara for critical reading of the manuscript and Dr S. Asano for continuous encouragement.
Supported in part by Chugai Pharmaceutical Company Ltd and grants from the Ministry of Education, Science, Sports, and Culture of Japan.
The nucleotide sequence data for this cDNA will appear in the DNA Databank of Japan/European Molecular Biology Laboratory/Genbank nucleotide sequence database with accession number AB031333.
Reprints:Toshio Kitamura, Department of Hematopoietic Factors, the Institute of Medical Science, the University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan; e-mail:kitamura@ims.u-tokyo.ac.jp.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal