Abstract
The c-myc proto-oncogene encodes a short-lived transcription factor that plays an important role in cell cycle regulation, differentiation and apoptosis. c-myc is often rearranged in tumors resulting in deregulated expression. In addition, mutations in the coding region of c-myc are frequently found in human lymphomas, a hot spot being the Thr58 phosphorylation site, a mutation shown to enhance the transforming capacity of c-Myc. It is, however, still unclear in what way this mutation affects c-Myc activity. Our results show that proteasome-mediated turnover of c-Myc is substantially impaired in Burkitt's lymphoma cells with mutated Thr58 or other mutations that abolish Thr58 phosphorylation, whereas endogenous or ectopically expressed wild type c-Myc proteins turn over at normal rates in these cells. Myc Thr58 mutants expressed ectopically in other cell types also exhibit reduced proteasome-mediated degradation, which correlates with a substantial decrease in their ubiquitination. These results suggest that ubiquitin/proteasome-mediated degradation of c-Myc is triggered by Thr58 phosphorylation revealing a new important level of control of c-Myc activity. Mutation of Thr58 in lymphoma thus escapes this regulation resulting in accumulation of c-Myc protein, likely as part of the tumor progression.
The myc family of proto-oncogenes encodes transcription factors that play a central role in the regulation of various aspects of cell growth (for review see Henriksson and Lüscher1). Chromosomal aberrations involvingmyc family loci have been implicated in the generation of a variety of human tumors and are often strongly correlated to a poor prognosis. Among these are the translocations involving c-mycand the immunoglobulin loci that occur in a high percentage of human Burkitt's lymphoma, AIDS-associated lymphomas, and certain acute lymphoblastic leukemias, resulting in deregulated expression of c-myc (for review see Klein and Klein2 and Marcu et al3). In addition, as shown in Figure 1, point mutations are frequently found in the coding region of c-myc both in primary tumors and in cell lines derived from these lymphomas.4-12 Hot spots for these mutations are found within or near the evolutionary conserved Myc box 1, the residue most frequently mutated being Thr58, a mutation also common to 3 avian viral myc genes and which has been shown to enhance the transforming activity of both v- and c-Myc proteins.13-17Thr58 is an in vivo phosphorylation site suggested to be targeted by glycogen synthase kinase 3 (GSK3).16-18 The phosphorylation is abolished by mutation of Pro57 and is dependent on prior phosphorylation of Ser62 by MAP and/or cyclin-dependent kinases,16-20 2 other frequently mutated residues in the lymphomas (Figure 1). Although it seems clear that phosphorylation of Thr58 mediates a negative regulatory signal to c-Myc and that mutation of this residue may be involved in lymphoma tumor progression it is still unclear in what way the mutations affect the activity of c-Myc. Recent reports have suggested that Myc-family proteins are degraded by the ubiquitin/proteasome pathway in vivo.21-23Because proteasome-mediated turnover of several transcription factors and cell cycle regulatory proteins are modulated by phosphorylation/dephosphorylation,24 25 we investigated the possibility that Thr58 phosphorylation may be involved in ubiquitin/proteasome-mediated degradation of c-Myc.
c-myc missense mutations in lymphomas.
Upper panel, summary of reported missense mutations in the c-myc coding region in Burkitt's lymphomas, AIDS-related lymphomas, B-cell acute lymphoblastic leukemias and the 3 avian v-myc-containing acute transforming retroviruses MC29, OK10, and MH2, representing 32 primary tumors and 28 cell lines.4-13 Amino acid positions of the most frequently mutated residues are indicated. Below is shown the c-Myc structure with indicated conserved regions. MB1; Myc box 1, MB2; Myc box 2, TAD; transactivation domain, Acidic; central acidic region, NLS; nuclear localization signal, BR; basic region, HLH; helix-loop-helix motif, Zip, leucine zipper motif, P; clusters of in vivo phosphorylation sites.1 The box at upper right shows the amino acid sequence of the conserved Myc box 1, including the Thr58 and Ser62 phosphorylation sites. GSK3; glycogen synthase kinase 3, MAPK; mitogen activated protein kinase, CDK; cyclin-dependent kinase. Asterisk (*) denotes the positions of frequent missense mutations. Lower panel, amino acid positions of c- or v-myc missense mutations in cell lines or constructs used in the study.
c-myc missense mutations in lymphomas.
Upper panel, summary of reported missense mutations in the c-myc coding region in Burkitt's lymphomas, AIDS-related lymphomas, B-cell acute lymphoblastic leukemias and the 3 avian v-myc-containing acute transforming retroviruses MC29, OK10, and MH2, representing 32 primary tumors and 28 cell lines.4-13 Amino acid positions of the most frequently mutated residues are indicated. Below is shown the c-Myc structure with indicated conserved regions. MB1; Myc box 1, MB2; Myc box 2, TAD; transactivation domain, Acidic; central acidic region, NLS; nuclear localization signal, BR; basic region, HLH; helix-loop-helix motif, Zip, leucine zipper motif, P; clusters of in vivo phosphorylation sites.1 The box at upper right shows the amino acid sequence of the conserved Myc box 1, including the Thr58 and Ser62 phosphorylation sites. GSK3; glycogen synthase kinase 3, MAPK; mitogen activated protein kinase, CDK; cyclin-dependent kinase. Asterisk (*) denotes the positions of frequent missense mutations. Lower panel, amino acid positions of c- or v-myc missense mutations in cell lines or constructs used in the study.
Materials and methods
Cell culture and transfection
The Burkitt's and BJAB lymphoma cell lines and the U-937-myc-6 monoblasts were cultured in RPMI-1640 medium supplemented with 10% fetal calf serum and antibiotics. The U2OS osteosarcoma cells were grown in DMEM containing 10% calf serum. Daudi, Jijoye, Raji, Mutu 1148 and Rael are EBV-positive African Burkitt's lymphoma, CA46 and ST486 are EBV negative American Burkitt's lymphoma cell lines.7,11,12 BJAB is a non-Burkitt's human B-cell lymphoma cell line12 and U-937-myc-6 is an OK10 v-myc-containing subclone of the human monoblastic cell line U-937.26 For transfection of Raji, 2 × 107cells were mixed with 20 μg of DNA in a 0.4 cm electroporation cuvette (Biorad), and electroporated at 960 μF, 300 V in a Biorad gene pulser as described.26 For transfection of U2OS, subconfluent cells grown on 10-cm cell culture dishes were incubated with 2 μg of DNA in 6 μL of FuGENE6 (Boehringer Mannheim) per dish. Cells were harvested 16 hours after transfection. The following DNA constructs were used: CMV-Myc and CMV-MycT58A were generated by subcloning EcoRI-fragments containing full-length human c-myccDNAs from the plasmids pspMyc0/1 and pspMycC,16respectively, kindly provided by Drs M. Henriksson and B. Lüscher, into the CMV-driven expression vector pCIneo. The CMV-MycT58A c-myc gene contains a mutation leading to substitution of Thr58 to Ala. pcDNA3-FLAG-Myc-wt, kindly provided by Dr B. Lüscher, contains an N-terminal FLAG-epitope fused in frame with a full-length human c-myc cDNA in the CMV-driven expression vector pcDNA3. The CMV-His6-Ub and CMV-HA-Ub constructs contain His6-and HA-tagged ubiquitin gene octamers,27 respectively, kindly provided by Dr D. Bohmann.
Pulse chase, immunoprecipitation, and immunoblot analysis
Five × 106 cells were pulsed for 40 minutes in 1 mL of methionine-free RPMI-1640 medium containing 5.55 MBq of 35S-methionine followed by chase in medium containing 1 mmol/L cold methionine and 10 μg/mL cycloheximide. Cells were lysed in AB buffer and immunoprecipitated with specific antibodies, whereafter the samples were analyzed on 10% SDS-PAGE gels as described.28 The signals were quantitated using a Fuji bas image analyzer. For studies of turnover of unlabeled proteins, 5 × 106 cells were treated with 100 μg/mL cycloheximide before harvest, whereafter cell lysates were immunoprecipitated as above and subjected to immunoblot analysis as described.28 IG-13 rabbit pan-Myc antiserum26and biotinylated C-33 monoclonal pan-Myc antibodies (Santa Cruz Biotechnology [SCB]) were used for immunoprecipitation and immunoblot analysis, respectively, of Myc. M2 FLAG antibodies (Sigma) was used to detect FLAG-epitope tagged c-Myc. For analysis of β-catenin, p27Kip1 and β-actin, H102, C19, and H196 antisera (SCB), respectively, were used.
Proteasome inhibitors and purification of ubiquitin conjugates
The proteasome inhibitors Z-leu-leu-NVA-H aldehyde (MG132), Z-leu-leu-leu-H aldehyde (MG115) (Peptides International) and N-acetyl-leucinyl-leucinyl-norleucinal-H (LLnL) (Sigma), the lysosome inhibitor chloroquine, the cysteine protease inhibitor L-trans-epoxysuccinic acid (E64), the acid protease inhibitor pepstatin A (Boehringer) or dimethyl sulfoxide (DMSO) (Sigma) vehicle were added to the cells 2 hours before harvest at a concentration of 50 μmol/L or for pepstatin, 10 μg/mL. For detection of ubiquitin conjugates, U2OS cells from three 10-cm dishes transfected with CMV-His6-Ub were lysed in 2 mL of 6 mol/L guanidinium-HCl, 0.1 mol/L Na2HPO4/NaH2PO4 pH 8.0, 5 mmol/L imidazole and sonicated whereafter His6-Ub-conjugated proteins were purified on a Ni2+-NTA-agarose (Qiagen) column as described.27 After washing, eluted proteins were analyzed by immunoblotting using C33 Myc antiserum (SCB) and monoclonal ubiquitin antibodies (Zymed).
Results
Decreased turnover of c-Myc mutated at Thr58 or Pro57 in Burkitt's lymphoma cells
The top panel of Figure 1 shows point mutations in the coding region of c-myc found to date in primary Burkitt's and AIDS-associated lymphomas and certain acute lymphoblastic leukemias and in cell lines derived from these tumors.4-12 The most frequently mutated residue is the in vivo phosphorylation site Thr58 followed by Pro57, which is required for Thr58 phosphorylation, both located in the evolutionary conserved Myc box 1. c-Myc has a short half-life of about 30 minutes.29 To investigate whether the hot spot mutations in c-myc might affect protein turnover, the Burkitt's lymphoma (BL) cell lines Daudi (wild type [wt] c-myc), Raji, and CA46 (c-myc mutations, including Thr58 and Pro57, see Figure 1), were treated with cycloheximide to block protein synthesis during which the decay of the c-Myc protein was measured by immunoblot analysis. Figure 2 shows that the steady state level of c-Myc in Daudi declined rapidly as expected. In contrast, the mutated c-Myc proteins in Raji and CA46 decayed at a much slower rate. To ensure that the prolongation of c-Myc half-life in these cells was not a result of a general impairment of proteasomal degradation, we measured the stability of β-catenin and p27Kip1, 2 other proteasomal substrates.30-32 The ubiquitin/proteasome-mediated degradation of β-catenin is regulated through phosphorylation by GSK3,31,32 the same kinase suggested to phosphorylate c-Myc at Thr58, suggesting that the degradation of these 2 proteins potentially may occur through the same pathway. The half-life of β-catenin seemed to be less than 1 hour in agreement with previous reports31,32 (Figure2, middle panel). No major differences in its rate of degradation were observed between the 3 cell lines, thus excluding a general defect in ubiquitin/proteasome-mediated turnover in the BL cells. The cyclin-dependent kinase inhibitor p27Kip1 turns over at a slower rate33 and was essentially stable in all 3 cell lines during this time frame (Figure 2, lower panel).
Stabilization of endogenous mutated c-Myc proteins in Burkitt's lymphoma cells.
Immunoblot analysis of c-Myc (upper panel), β-catenin (middle panel) and p27Kip1 (lower panel) after cycloheximide (CHX) treatment of Daudi, Raji, and CA46 Burkitt's lymphoma cells for the indicated time points was performed using specific antibodies as described in “Materials and Methods.”
Stabilization of endogenous mutated c-Myc proteins in Burkitt's lymphoma cells.
Immunoblot analysis of c-Myc (upper panel), β-catenin (middle panel) and p27Kip1 (lower panel) after cycloheximide (CHX) treatment of Daudi, Raji, and CA46 Burkitt's lymphoma cells for the indicated time points was performed using specific antibodies as described in “Materials and Methods.”
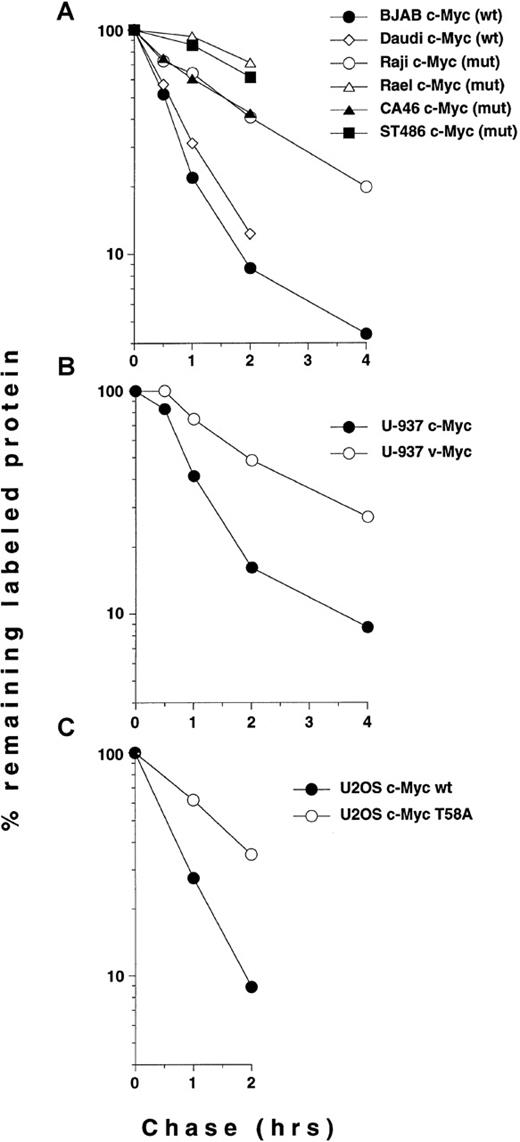
To measure c-Myc turnover more precisely, we performed35S-methionine pulse chase experiments using the lymphoma cell lines listed in Figure 1. Immunoprecipitation of c-Myc, followed by SDS-PAGE analysis showed an expected short half-life of c-Myc in Daudi and BJAB cells (both wt c-myc, Figure3), estimated to be 35 and 25 minutes, respectively, by image analysis (see also Figure 5A). In contrast, the mutated c-Myc in Raji cells exhibited a significantly slower turnover rate (half-life of approximately 105 minutes). Further, c-Myc was at least as stabilized in the endemic Epstein-Barr virus (EBV)-positive BL cell lines Mutu and Rael, both with a single mutation of Thr58, and in the sporadic EBV-negative BL cell lines CA46 and ST486 with common mutations in Pro57 (Figures 1, 3, and 5A). In Jijoye BL cells with wt c-myc, the turnover rate of c-Myc was normal. We concluded from these experiments that c-Myc was stabilized in the 5 BL cell lines that have in common the 2 most frequent c-mycmissense mutations in lymphomas, affecting amino acid positions 58 or 57, both resulting in abolishment of Thr58 phosphorylation.
Pulse chase analysis of c-Myc turnover in Burkitt's and BJAB lymphoma cells.
Cells were pulse labeled with 35S-methionine followed by a chase in medium containing cold methionine for indicated time points. Cell lysates were immunoprecipitated with pan-Myc antiserum and analyzed by SDS-PAGE as described in “Materials and Methods.”
Pulse chase analysis of c-Myc turnover in Burkitt's and BJAB lymphoma cells.
Cells were pulse labeled with 35S-methionine followed by a chase in medium containing cold methionine for indicated time points. Cell lysates were immunoprecipitated with pan-Myc antiserum and analyzed by SDS-PAGE as described in “Materials and Methods.”
Ectopically expressed wt c-Myc is degraded at a normal rate in Burkitt's lymphoma cells with mutated endogenous c-Myc
To provide further evidence that the stabilization of c-Myc in BL cells is due to the mutations in c-myc rather than a result of other oncogenic events or cell-type specific factors in these cells, we transfected Raji cells with an expression vector containing a FLAG-epitope tagged wt c-myc gene. The turnover of the transfected wt c-Myc protein as well as of the endogenous mutated c-Myc protein was determined by CHX treatment, followed by sequential immunoprecipitation of cell lysates by α-FLAG and α-Myc sera and immunoblot analysis of the immunoprecipitates using Myc antiserum. Figure 4A shows that the FLAG-wt c-Myc protein turned over at the expected normal rate, whereas the half-life of the endogenous mutated protein was prolonged as shown previously. We therefore conclude that the stabilization of c-Myc observed in the BL cells is indeed a result of the mutations in c-myc. Whether in addition also other genetic alterations are required for the stabilization of mutated c-Myc in these cells remains an open question (see also “Discussion”).
Turnover of ectopically expressed Myc proteins in Burkitt's and non-Burkitt's cells.
(A), Stability of FLAG-epitope tagged wild type (wt) c-Myc in Raji Burkitt's lymphoma cells. 5 μg of pcDNA3-FLAG-Myc-wt (FLAG-wt c-Myc) or pcDNA3 (Mock) were electroporated into 2 × 107Raji cells together with 15 μg of carrier DNA. 16 hours after electroporation aliquots of cells were treated with CHX for the indicated times or left untreated. FLAG-tagged wt c-Myc was immunoprecipitated from the cell extracts with M2 FLAG antibodies, followed by immunoprecipitation of the endogenous mutated c-Myc protein from the same extracts with IG-13 α-Myc serum. The immunoprecipitated material was then subjected to immunoblot analysis using C33 Myc antibodies. Note that the exposure time of the film presented at the left α-FLAG panel is longer for than for the right α-Myc panel. (B, C), pulse chase analysis of v- and c-Myc turnover in U-937 monoblasts stably expressing the OK10 v-Myc protein (B) and of wt c-Myc and the T58A mutant after transient transfections of U2OS osteosarcoma cells (C). The pulse chase was performed at the indicated time points as described in the legend to Figure 3. In (C), semiconfluent U2OS osteosarcoma cells were transfected with 2 μg of CMV-driven c-myc per 10-cm dish. The pulse chase was performed 16 hours after transfection.
Turnover of ectopically expressed Myc proteins in Burkitt's and non-Burkitt's cells.
(A), Stability of FLAG-epitope tagged wild type (wt) c-Myc in Raji Burkitt's lymphoma cells. 5 μg of pcDNA3-FLAG-Myc-wt (FLAG-wt c-Myc) or pcDNA3 (Mock) were electroporated into 2 × 107Raji cells together with 15 μg of carrier DNA. 16 hours after electroporation aliquots of cells were treated with CHX for the indicated times or left untreated. FLAG-tagged wt c-Myc was immunoprecipitated from the cell extracts with M2 FLAG antibodies, followed by immunoprecipitation of the endogenous mutated c-Myc protein from the same extracts with IG-13 α-Myc serum. The immunoprecipitated material was then subjected to immunoblot analysis using C33 Myc antibodies. Note that the exposure time of the film presented at the left α-FLAG panel is longer for than for the right α-Myc panel. (B, C), pulse chase analysis of v- and c-Myc turnover in U-937 monoblasts stably expressing the OK10 v-Myc protein (B) and of wt c-Myc and the T58A mutant after transient transfections of U2OS osteosarcoma cells (C). The pulse chase was performed at the indicated time points as described in the legend to Figure 3. In (C), semiconfluent U2OS osteosarcoma cells were transfected with 2 μg of CMV-driven c-myc per 10-cm dish. The pulse chase was performed 16 hours after transfection.
Stabilization of ectopically expressed v- and c-Myc with mutated Thr58
To investigate whether Myc proteins mutated at amino acid position 58 were stabilized also in other cells than BL, we used human U-937-myc-6 monocytic cells containing the avian OK10 virus v-myc gene,28 which is mutated at Thr58 and at 1 C-terminal residue not found in other v-myc genes or in tumors (Figure 1, lower panel). Pulse chase experiments showed that the OK10 v-Myc protein, which migrates slightly faster than the human protein, exhibited a prolonged half-life similar to that of the mutated c-Myc proteins in BL (Figures 4B and 5B), whereas the endogenous c-Myc protein was degraded at a normal rate (half-lives 120 and 40 minutes, respectively). Unlike v-Myc, normal chicken c-Myc was not stabilized when transiently transfected into human HeLa cervical carcinoma or U2OS osteosarcoma cells (data not shown).
To provide further evidence for a general role of Thr58 in c-Myc turnover, we transiently transfected U2OS cells with full-length c-myc cDNA-constructs with a single mutation substituting Thr58 for Ala (T58A) or with the wt sequence. Pulse chase experiments showed that the T58A mutant was stabilized (estimated half-life 80 minutes), whereas the wt c-Myc protein turned over at a normal rate (half-life 30 minutes) (Figures 4C and 5C). Quantitations of some of the Myc pulse chase experiments in Figures 3 and 4 are summarized in Figure 5. We conclude from these results that Myc proteins with common mutations at Thr58 or Pro57 in BL cells have significantly slower turnover rates than the wt proteins and that Thr58 mutants have increased half-lives also in 2 other cell types.
Quantitation of c- and v-Myc turnover in Burkitt's and non-Burkitt's cells.
(A), Burkitt's lymphoma cells lines, (B) U-937-myc-6 cells, (C) transiently transfected U2OS cells. The results shown in Figures 3 and4B, C were quantitated by image analysis. The cell lines and the status of their respective endogenous c-Myc proteins or constructs used for transfection are indicated in the figure. The quantitation is presented in a logarithmic scale as percentage of remaining labeled protein during the times of chase indicated.
Quantitation of c- and v-Myc turnover in Burkitt's and non-Burkitt's cells.
(A), Burkitt's lymphoma cells lines, (B) U-937-myc-6 cells, (C) transiently transfected U2OS cells. The results shown in Figures 3 and4B, C were quantitated by image analysis. The cell lines and the status of their respective endogenous c-Myc proteins or constructs used for transfection are indicated in the figure. The quantitation is presented in a logarithmic scale as percentage of remaining labeled protein during the times of chase indicated.
Decreased proteasome-mediated turnover and inefficient ubiquitination of mutated Myc proteins
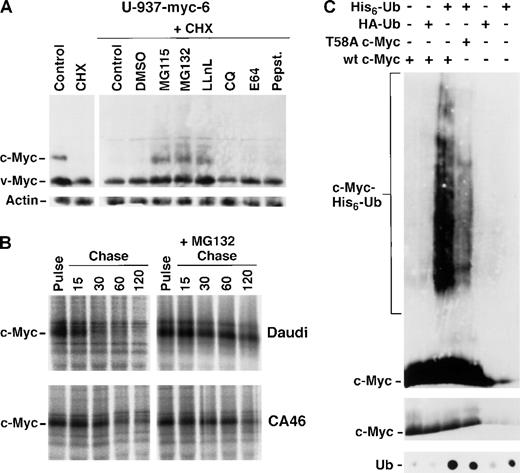
c- and N-Myc, like many other transcription factors and cell cycle regulatory molecules, are degraded in vitro by the ubiquitin-proteasome pathway,34 recently shown also in vivo.21-23 We therefore investigated whether inhibitors of the proteasome affected the turnover of Thr58 and Pro57 Myc mutants. Immunoblot analysis of v- and c-Myc in U-937-myc-6 cells showed that c-Myc decayed rapidly after blocking protein synthesis with cycloheximide, whereas a substantial amount of v-Myc was still present (Figure6A, upper panel) in agreement with the pulse chase experiments in Figure 4B. Addition of the proteasome inhibitors MG115, MG132, or LLnL together with cycloheximide stabilized c-Myc, whereas the lysosome inhibitor chloroquine (CQ), the cysteine protease inhibitor E64, the acid protease inhibitor pepstatin, or the vehicle (DMSO) had little effect, thus confirming the involvement of the proteasome in Myc turnover. A faint smear of high molecular weight bands was observed in cells treated with proteasome inhibitors, which might represent polyubiquitinated forms of c-Myc. Although v-Myc was more stable than c-Myc, the addition of proteasome inhibitors lead to some further accumulation of v-Myc. Again chloroquine, E64 and pepstatin had little effect. Immunoblot analysis of β-actin (Figure6A, lower panel) confirmed that equal amounts of protein were applied to the gel.
Retarded proteasome-mediated turnover and inefficient ubiquitination of Myc mutants.
(A) Immunoblot analysis of v- and c-Myc (upper panel) and β-actin (lower panel) in U-937-myc-6 cells after 2 hours treatment with the proteasome inhibitors MG115, MG132, and LLnL, the protease inhibitors chloroquine (CQ), E64, and pepstatin or DMSO vehicle in the presence of cycloheximide (CHX) as indicated. (B) Pulse chase analysis of c-Myc turnover in the presence of the proteasome inhibitor MG115. Daudi (wt c-Myc) and CA46 (mutated c-Myc) Burkitt's lymphoma cells were treated with the proteasome inhibitor for 1.5 hours, pulsed with35S methionine followed by chase in the presence of MG115 for the indicated time points or were pulse labeled in the absence of the inhibitor. Cell lysates were then immunoprecipitated with pan-Myc antibodies and analyzed as in Figure 3. (C) Reduced ubiquitination of the T58A c-Myc mutant. U2OS cells were transfected with wt c-Myc, the T58A c-Myc mutant, His6-Ub or HA-Ub vectors alone or in combination as indicated in the figure. The transfected cells were treated with MG115 during the last 2 hours before harvest. His6-Ub-conjugated proteins were purified as described in “Materials and Methods” and subjected to Western blot analysis using Myc antibodies (upper panel) or dot blot analysis using ubiquitin antibodies (lower panel). The smears of c-Myc-His6-Ub-conjugates are indicated. The middle panel shows c-Myc immunoblot analysis of 1/10 of the input material (note that the exposure time for this blot is shorter than for the upper panel).
Retarded proteasome-mediated turnover and inefficient ubiquitination of Myc mutants.
(A) Immunoblot analysis of v- and c-Myc (upper panel) and β-actin (lower panel) in U-937-myc-6 cells after 2 hours treatment with the proteasome inhibitors MG115, MG132, and LLnL, the protease inhibitors chloroquine (CQ), E64, and pepstatin or DMSO vehicle in the presence of cycloheximide (CHX) as indicated. (B) Pulse chase analysis of c-Myc turnover in the presence of the proteasome inhibitor MG115. Daudi (wt c-Myc) and CA46 (mutated c-Myc) Burkitt's lymphoma cells were treated with the proteasome inhibitor for 1.5 hours, pulsed with35S methionine followed by chase in the presence of MG115 for the indicated time points or were pulse labeled in the absence of the inhibitor. Cell lysates were then immunoprecipitated with pan-Myc antibodies and analyzed as in Figure 3. (C) Reduced ubiquitination of the T58A c-Myc mutant. U2OS cells were transfected with wt c-Myc, the T58A c-Myc mutant, His6-Ub or HA-Ub vectors alone or in combination as indicated in the figure. The transfected cells were treated with MG115 during the last 2 hours before harvest. His6-Ub-conjugated proteins were purified as described in “Materials and Methods” and subjected to Western blot analysis using Myc antibodies (upper panel) or dot blot analysis using ubiquitin antibodies (lower panel). The smears of c-Myc-His6-Ub-conjugates are indicated. The middle panel shows c-Myc immunoblot analysis of 1/10 of the input material (note that the exposure time for this blot is shorter than for the upper panel).
We next investigated the effects of proteasome inhibitors on c-Myc stability in BL cells. Pulse chase experiments revealed that wt c-Myc in Daudi was unstable with a half-life of 30 minutes, whereas mutated c-Myc in CA46 turned over slowly with a half-life of 95 minutes (Figure6B). The addition of MG132 stabilized wt c-Myc in Daudi significantly, although only a small effect was seen in CA46 cells. In both cases the increased half-life was in the order of 125 minutes. Together these results suggested that Myc proteins with mutations in amino acid positions 58 or 57 have an impaired proteasome-mediated turnover but are still degraded by this pathway albeit at a slower rate.
Because degradation by the 26S proteasome is dependent on covalent ubiquitination of target proteins by ubiquitin (Ub)-conjugating enzymes (for review see references 24 and 25), we investigated whether the retarded proteasome-mediated turnover of mutated Myc proteins was correlated with decreased ubiquitination. Ubiquitinated c-Myc proteins are very difficult to detect due to their rapid turnover. To visualize these intermediates, U2OS cells were transiently transfected with wt or T58A mutant c-Myc together with an expression vector containing histidine-tagged ubiquitin (His6-Ub) octamers27and treated with the proteasome inhibitor MG115 before harvest. Proteins conjugated with His6-Ub were purified over a Ni-agarose column and the presence of His6-Ub-conjugated c-Myc was analyzed by immunoblotting. Figure 6C shows that a high molecular weight smear indicative of polyubiquitinated proteins was detected by c-Myc antibodies in cells transfected with wt c-Myc together with His6-Ub, but not in cells transfected with c-Myc plus hemagglutinin-tagged ubiquitin (HA-Ub), with c-Myc alone or with His6-Ub or HA-Ub alone. We therefore conclude that c-Myc became ubiquitinated in vivo in agreement with recent reports.21-23 The c-Myc band migrating at its normal molecular weight represents unmodified c-Myc protein binding unspecifically to the column, since it was present also in lanes lacking His6-Ub. In contrast to the prominent smear of polyubiquitinated wt c-Myc protein, significantly less His6-Ub-c-Myc was detected in cells transfected with the T58A c-Myc mutant together with His6-Ub (Figure 6C). Immunoblot analysis of ubiquitin showed that approximately equal amounts of total ubiquitinated proteins from His6-Ub transfected cells were eluted from the column (Figure 6C, lower panel). The middle panel of Figure 6C shows that approximately equal amounts of transfected c-Myc were applied to the column. We conclude from these results that mutation of amino acid position 58 results in inefficient ubiquitination of c-Myc.
Discussion
In summary, our results show that endogenous c-Myc proteins with mutated Thr58, with or without additional mutations, or with Pro57 mutation as the common denominator display substantially decreased proteasome-mediated turnover in Burkitt's lymphoma cells. Because phosphorylation of Thr58 is dependent on an intact Pro57,17this mutation probably has the same consequence as mutation of Thr58. Our experiments further show that transfected wt c-Myc turns over at a normal rate in BL cells containing mutated endogenous c-Myc and that the half-lives of other proteasomal substrates are not altered. This rules out a general impairment of the ubiquitin/proteasome-mediated degradation or other genetic alterations as the main explanation for the c-Myc stabilization in these cells. Ectopically expressed v- and c-Myc with Thr58 mutations are stabilized also in 2 other cell types, in monoblastic tumor cells and osteosarcoma cells, respectively, which is in agreement with recent observations in U2OS cells,23published during the preparation of this manuscript. We further show that the decreased turnover rate of the Thr58 mutant protein correlates with inefficient ubiquitination, thus providing a rationale for the stabilization.
Our results suggest that phosphorylation of Thr58 triggers ubiquitin/proteasome-mediated turnover. Escape from this regulation may be sufficient to explain the increased transforming activity of Thr58 mutants 14-17 and their insensitivity to negative regulation by the retinoblastoma-family protein p107.20,35p107-associated cyclin/CDK complexes are suggested to phosphorylate Ser62, another mutation in some lymphomas. This phosphorylation seems to be a prerequisite for subsequent phosphorylation of Thr58.16,18 Because incoming growth stimulatory or inhibitory signals may affect phosphorylation of both these phosphorylation sites, one can envisage a quite complex regulation of Myc function in this region, which potentially could be influenced by other oncogenic events. In agreement with this notion, a recent report suggests that oncogenes like ras may affect c-Myc stability.36 Mutation of Thr58 does, however, not completely abolish ubiquitination and proteasome inhibitors somewhat further stabilize the protein, indicating that there may be other signals for ubiquitination of c-Myc. Recent studies of c-Myc degradation in yeast37 and in HeLa cells23indicate that there may be several independent signals for proteasome-mediated turnover in the N-terminus. Such additional signals in combination with differences in genetic and cellular backgrounds might account for the apparent slight variations in the half-lives of both wt and mutant c-Myc proteins in different cells (Figures 2 to 5). Further studies are required to define these possible alternative degradation motifs and how they are related to Thr58 and other occurring mutations in c-Myc. Although other regions of c-Myc may also participate in the ubiquitination and degradation of c-Myc our results clearly suggest that Thr58 phosphorylation/dephosphorylation is a main regulator of this process.
Recent studies suggest that phosphorylation is a frequently used mechanism for controlling the degradation of cell cycle and growth regulatory proteins through the ubiquitin/proteasome pathway.24,25 It is intriguing that several viral forms of oncoproteins, such as v-Jun and v-Fos like shown here for v-Myc, escape degradation by the proteasome due to mutated destruction motifs regulated by phosphorylation.27 38 Our results suggest that development of Burkitt's and other lymphomas involves activation of c-Myc by 2 sequential steps. First, the deregulation of expression of the c-myc gene through translocation to the immunoglobulin loci and, second, the mutation of a regulatory phosphorylation site leading to escape from ubiquitin/proteasome-mediated turnover and accumulation of c-Myc protein.
Acknowledgments
We thank Dr George Klein for providing Burkitt's lymphoma cell lines and Drs Dirk Bohmann, Marie Henriksson, and Bernhard Lüscher for plasmid constructs. We are also thankful to Drs Göran Akusjärvi, Kenneth Nilsson, Bernhard Lüscher, FredrikÖberg, Catharina Svensson, and Hans Ronne for valuable discussions and criticism.
Supported by grants from the Swedish Cancer Society, the Children Cancer Foundation of Sweden and the Lovisa & Thielman Foundation.
Reprints:Lars-Gunnar Larsson, Uppsala Genetic Center, Department of Plant Biology, Swedish University of Agricultural Sciences, Box 7080, 750 07 Sweden; e-mail:lars-gunnar.larsson@vbiol.slu.se.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal