Abstract
Protein S deficiency is a recognized risk factor for venous thrombosis. Of all the inherited thrombophilic conditions, it remains the most difficult to diagnose because of phenotypic variability, which can lead to inconclusive results. We have overcome this problem by studying a cohort of patients from a single center where the diagnosis was confirmed at the genetic level. Twenty-eight index patients with protein S deficiency and a PROS1 gene defect were studied, together with 109 first-degree relatives. To avoid selection bias, we confined analysis of total and free protein S levels and thrombotic risk to the patients' relatives. In this group of relatives, a low free protein S level was the most reliable predictor of a PROS1gene defect (sensitivity 97.7%, specificity 100%). First-degree relatives with a PROS1 gene defect had a 5.0-fold higher risk of thrombosis (95% confidence interval, 1.5-16.8) than those with a normal PROS1 gene and no other recognized thrombophilic defect. Although pregnancy/puerperium and immobility/trauma were important precipitating factors for thrombosis, almost half of the events were spontaneous. Relatives with splice-site or major structural defects in the PROS1 gene were more likely to have had a thrombotic event and had significantly lower total and free protein S levels than those relatives having missense mutations. We conclude that persons withPROS1 gene defects and protein S deficiency are at increased risk of thrombosis and that free protein S estimation offers the most reliable way of diagnosing the deficiency.
Protein S (PS), a 69-kd vitamin K–dependent plasma glycoprotein, plays an important regulatory role in the protein C anticoagulant system.1 PS functions primarily as a nonenzymatic cofactor to activated protein C (APC) in the proteolytic inactivation of factors Va and VIIIa.2 Plasma PS circulates in 2 forms. Approximately 60% is present as a noncovalent complex with the β-chain of the complement component C4b binding protein (C4bBP), while the remaining 40% is free.3 Only free PS possesses APC cofactor activity.4
Three types of PS deficiency are currently recognized: type I is characterized by low total and free PS antigen levels, type II by normal free PS levels but reduced APC cofactor activity, and type III by a selective reduction in free PS levels.5 Evaluation of the relationship between PS and C4bBP in patients with inherited PS deficiency has shown that types I and III may be phenotypic variants of the same disease.6-8
Human DNA contains 2 PS genes: the active PROS1 gene and a closely linked pseudogene (PROS2), which shows 96.5% homology to exons 2 to 15 of the PROS1 gene.9-11 ThePROS1 gene comprises 15 exons and 14 introns spanning some 80 kb of genomic DNA.12 Despite the complexity of thePROS1 gene, the development of procedures permitting selective amplification of PROS1 gene sequences and the availability ofPROS1 mRNA in platelets have facilitated investigation of the molecular basis of PS deficiency and the identification ofPROS1 gene defects, which have recently been compiled into aPROS1 mutation database.13 Three PROS1 gene dimorphisms have been reported: an exonic A/G dimorphism in codon 626,14,15 a C/T dimorphism in nucleotide 54 in intron K (PIPS1), and a C/A dimorphism 520 bp downstream of the stop codon in the 3′UTR (PEPS2).16 Another, rare, T/C dimorphism in codon 460 causes substitution of S460 by P (single-letter amino acid code) and results in a circulating PS molecule with a lower molecular weight than normal. This Heerlen PS is thought to have a higher affinity for C4bBP than normal, causing an abnormal distribution of mutated and normal PS on C4bBP and a selective reduction in free PS.17 Although the Heerlen allele was originally demonstrated to occur at similar frequencies among thrombophilia patients and healthy blood donors,18 2 separate studies found that it occurred more often among PS-deficient patients, particularly those with type III deficiency.17 19
Venous thrombosis is a multifactorial disease resulting from the interaction of genetic and environmental risk factors.20Those patients with genetic defects are placed at increased risk by use of the oral contraceptive pill21 and by surgery.22 Patients with more than 1 genetic risk factor are also at increased risk of venous thrombosis.23-27
PS deficiency is recognized to be a risk factor for venous thrombosis and is found in 1.5% to 7% of selected groups of thrombophilic patients.28,29 Although one large prospective population-based study failed to find such an association,30 in a similarly designed case-control study of 327 consecutive patients with deep venous thrombosis, Faioni et al22 reported that PS-deficient patients had a 2.4-fold higher risk for the development of thrombosis. In addition, PS deficiency has been reported to be a risk factor for venous thrombosis at other sites, such as the mesenteric31 and cerebral veins.32 The prevalence of PS deficiency in the general population is unknown. There is an age-related increase in PS levels, independent of the influence of sex, in both normal and PS-deficient individuals.6
In this study, we report the clinical impact and laboratory results of a well-characterized cohort of PS-deficient patients and their families, in which every individual was investigated at the clinical, phenotypic, and genetic levels. By specifically studying first-degree relatives, we defined the thrombotic risk in affected and unaffected family relatives. Furthermore, the value of free and total PS estimations in making the diagnosis of PS deficiency was examined.
Patients and methods
Patients
All 137 patients in this study were investigated at the Sheffield Hemophilia and Thrombosis Centre as part of a prospective study of thrombophilia. Here we report on the clinical and laboratory findings in 28 families with PS deficiency in which a PROS1 gene defect was identified. All index patients had experienced at least 1 venous thromboembolic event and were diagnosed as having PS deficiency based on repeatedly reduced PS levels. Acquired PS deficiency was excluded through a search for relevant risk factors and by confirmation of a genetic defect in the PROS1 gene. Confirmation of thrombotic events was with Doppler scanning or venography in the case of limb thrombosis and ventilation-perfusion scanning or pulmonary angiography in the case of pulmonary emboli. This research project has received approval from the South Sheffield Ethics Committee.
Coagulation tests
Total and free PS levels were measured as described by Woodhams33 using an enzyme-linked immunosorbent assay that was standardized against the First International Standard for PS National Institute for Biological Standards and Controls (Hertfordshire, UK) (NIBSC). Normal ranges (NR), based on the mean ± 2 SD, were derived from the results obtained from 65 healthy volunteers (NR: total PS, 0.71-1.36 IU/mL; free PS, 0.64-1.31 IU/mL). A normal range for PS levels in patients taking warfarin was derived from levels in non–PS-deficient individuals who were stably anticoagulated with a therapeutic international normalized ratio (NR: total PS, 0.49-0.87 IU/mL; free PS, 0.27-0.79 IU/mL). The same normal ranges were used regardless of age and sex.
Detection of factor V Leiden and the prothrombin 20 210A allele
PROS1 gene analysis
All exons and intron-exon boundaries of the PROS1 gene were amplified from genomic DNA as described previously.8 Where possible, oligonucleotide primers were designed to have mismatches with the pseudogene sequence to selectively amplify PROS1-specific sequences. Following amplification, polymerase chain reaction (PCR) products were purified and directly sequenced as described before.8 Alternatively, PCR products were analyzed by conformation-sensitive gel electrophoresis (CSGE),35 and those forming heteroduplexes were directly sequenced to identify defects present. The Heerlen allele was detected by CSGE of amplified exon 13, and its presence was confirmed by RsaI digestion.PROS1 alleles were haplotyped using the codon 626 BstXI dimorphism, the PIPS1 and PEPS2 dimorphisms, and the extragenic D3S1251 marker, as described earlier.35
Southern blot analysis of the PROS1 gene
Genomic DNA samples were digested with XbaI, electrophoresed in 0.8% (w/v) agarose, and then blotted onto Hybond-N nylon membrane (Amersham Pharmacia Biotech, Bucks, UK). The membrane was probed with a 1719-bp fragment corresponding to exons 5 to 15 of the PROS1cDNA, which was amplified by PCR from reverse-transcribed buffy coat RNA and radiolabeled with 32P by random priming with the Ready-To-Go kit (Stratagene, Amsterdam, Netherlands).
Statistical analysis
Statistical analyses were performed using Graph Pad Prism 2.0 (GraphPad Software Inc., San Diego, CA) and Instat software. Comparison of the prevalence of thrombosis in relatives with and without PROS1 gene defects and also of the thrombotic risk associated with different types of defects was performed using Fisher's exact test, with the 95% confidence intervals (CIs) calculated using the approximation of Katz. Relative risks were calculated using only the first thromboembolic events during follow-up. Kaplan-Meier estimates were calculated, using censored data to correct for individual age of the study population, for assessment of thrombosis-free survival in relatives with and without aPROS1 gene defect. An unpaired, 2-tailed t test was used to analyze the relative distributions of total and free PS levels in relation to PROS1 gene defects.
Results
Characteristics of the cohort
Twenty-eight index patients (18 male and 10 female) registered with our center had a history of venous thromboembolism and were heterozygous for a partial or fully characterized PROS1 gene defect. At least 1 of the thromboses was confirmed in 23 of the index patients; the unconfirmed thromboses largely occurred many years earlier. One hundred nine first-degree relatives (46 male and 63 female), aged 14 to 84 years, were studied with respect to thrombotic history, total and free PS levels, and the presence of PROS1gene defects. Fifty-seven affected relatives were identified, and 6 of these had additional familial thrombophilic defects: factor V Leiden (3 cases), factor V Leiden and antithrombin gene defect (1 case), or the prothrombin 20 210A allele (2 cases). Fifty-two relatives had a normal PROS1 gene, but 7 of these had factor V Leiden.
PROS1 gene defects associated with PS deficiency
The PROS1 gene defects identified in index cases and their relatives are summarized in Table 1. Intronic mutations were identified in 12 of 28 cases. A novel G to T transversion in the invariant acceptor site AG dinucleotide upstream of exon 13 in intron L was identified in 1 case. Another patient was heterozygous for an A to G transition 7 bp downstream of exon 1 in intron A. This transition is listed in the PROS1 mutation database as a probable polymorphism because it was unclear whether it was causative in the single individual previously identified with the defect.13,36 However, because neither a family study nor RNA analysis was performed to investigate the individual reported in the database and because the defect was shown to be absent from 70 normal alleles, the evidence that this mutation is a polymorphism is weak.36 We have considered it as causative here because it was the only PROS1 gene alteration detected and it was associated with reduced free PS levels in the index and the 2 heterozygous family members investigated. A further family member without this mutation had normal PS levels.
PROS1 gene defects identified in index cases and relatives
| PROS1 gene defect . | Predicted amino acid substitution . | Index cases . | Relatives . |
|---|---|---|---|
| Intron K, A → G, exon 12 − 9 | 9 | 15 | |
| 145, TGT → TAT | 145, C → Y | 5 | 9 |
| 570, ATG → ACG | 570, M → T | 3 | 8 |
| 625, TGT → CGT | 625, C → R | 1 | 6 |
| Intron J, G → A, exon 10 + 5 | 1 | 3 | |
| 581, CTT → CGT | 581, L → R | 1 | 3 |
| Intron A, A → G, exon 1 + 7 | 1 | 2 | |
| Intron L, G → T, exon 13 − 1 | 1 | 2 | |
| 206, TGC → GGC | 206, C → G | 1 | 1 |
| MajorPROS1 gene defect* | 5 | 7 | |
| Total | 28 | 56 |
| PROS1 gene defect . | Predicted amino acid substitution . | Index cases . | Relatives . |
|---|---|---|---|
| Intron K, A → G, exon 12 − 9 | 9 | 15 | |
| 145, TGT → TAT | 145, C → Y | 5 | 9 |
| 570, ATG → ACG | 570, M → T | 3 | 8 |
| 625, TGT → CGT | 625, C → R | 1 | 6 |
| Intron J, G → A, exon 10 + 5 | 1 | 3 | |
| 581, CTT → CGT | 581, L → R | 1 | 3 |
| Intron A, A → G, exon 1 + 7 | 1 | 2 | |
| Intron L, G → T, exon 13 − 1 | 1 | 2 | |
| 206, TGC → GGC | 206, C → G | 1 | 1 |
| MajorPROS1 gene defect* | 5 | 7 | |
| Total | 28 | 56 |
One definite and 4 putative (see text).
Missense mutations were detected in 11 of 28 cases (Table 1). Two T to G transversions, 1 in exon 8 and another in exon 14, resulting in substitution of C206 by G and L581 by R, respectively, had not been reported previously. One of the patients who was heterozygous for the G to A transition causing substitution of C145 by Y was the subject of a previous report because, in addition to being heterozygous for factor V Leiden, he was heterozygous for an antithrombin gene defect causing substitution of T85 by M.26
PROS1 gene analysis was carried out in all cases for the detection of the TCC to CCC, or Heerlen, dimorphism causing substitution of S460. None of the patients reported had the Heerlen polymorphism.
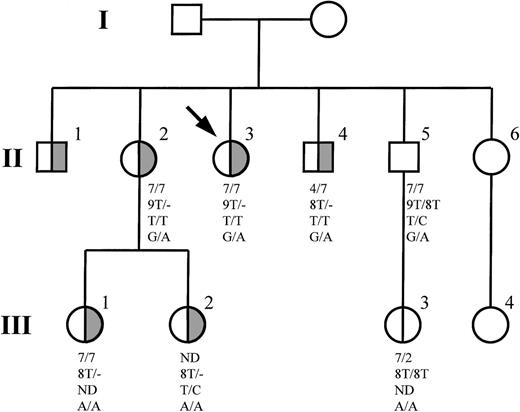
Haplotyping of PROS1 alleles, Southern blot analysis, or both provided evidence for major PROS1 gene defects in DNA from members of 5 families (D1-D5). The PS-deficient members of families D1 and D2 were homozygous for all 3 intragenic polymorphic markers studied but did not share a common allele, indicating that they were hemizygous at all the polymorphic sites and that 1 PROS1 allele was deleted. An insertion of a single T in a tract of 8T residues located 8 to 15 bp upstream of exon 8 in intron G was detected in DNA from the index case of family D3. Although the insertion was not associated with PS deficiency within the family, it was an informative marker for the presence of a partial PROS1 gene deletion in affected family members (Figure 1). Southern blotting of DNA from the index case of family D4 failed to show a reduction in intensity of any PROS1 gene fragments when compared with a control sample. However, an additional fragment of approximately 8 kb was observed, indicating that a major rearrangement occurred. Finally, Southern blot analysis revealed a reduced intensity of allPROS1 gene fragments in DNA from the index case of family D5 when compared with a control sample, consistent with the occurrence of a major PROS1 gene deletion. No extra bands were detected.
Inheritance of defective PROS1 allele in family D3.
Individuals with PS deficiency are represented by half-filled symbols. The index case is indicated by an arrow, and individuals who were not investigated are indicated by empty symbols. Haplotypes are shown in the following order: D3S1251, length of poly T tract in intron G, PIPS1, codon 626.
Inheritance of defective PROS1 allele in family D3.
Individuals with PS deficiency are represented by half-filled symbols. The index case is indicated by an arrow, and individuals who were not investigated are indicated by empty symbols. Haplotypes are shown in the following order: D3S1251, length of poly T tract in intron G, PIPS1, codon 626.
Total and free PS levels
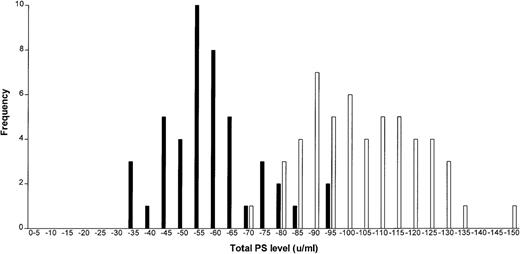
Total and free PS levels (Figures 2 and3) were assayed in all relatives, and the value of each in the phenotypic prediction of a PROS1 gene defect was determined. Twelve relatives who were taking warfarin at the time of their assay were excluded from this analysis. The mean (SD) total and free PS levels for relatives with a PROS1 defect were 0.57 (0.14) and 0.30 (0.16), respectively, which are significantly lower than those of relatives with a normal PROS1 gene, who had total and free PS levels of 1.03 (0.17) and 1.06 (0.19) (P < .0001). Total PS had a sensitivity of 82.2% and a specificity of 98.1% with respect to the presence of a gene defect. Free PS had a sensitivity of 97.7% and a specificity of 100% and therefore was a more reliable predictor of a PROS 1 gene abnormality.
Total protein S levels in relatives with/withoutPROS1 gene defects.
Frequency distribution graph shows total protein S levels in relatives with (black bars) and without (white bars) PROS1gene defects.
Total protein S levels in relatives with/withoutPROS1 gene defects.
Frequency distribution graph shows total protein S levels in relatives with (black bars) and without (white bars) PROS1gene defects.
Free protein S levels in relatives with/withoutPROS1 gene defects.
Frequency distribution graph shows free protein S levels in relatives with (black bars) and without (white bars) PROS1 gene defects.
Free protein S levels in relatives with/withoutPROS1 gene defects.
Frequency distribution graph shows free protein S levels in relatives with (black bars) and without (white bars) PROS1 gene defects.
Total and free PS levels in patients taking warfarin
We had previously established a PS “normal range” for patients who were taking oral anticoagulants. Our laboratory range for persons stably anticoagulated is 0.49 to 0.87 IU/mL for total PS and 0.27 to 0.79 IU/mL for free PS. In this study, 25 subjects (13 index cases and 12 affected relatives) were taking warfarin at the time of testing. All 25 patients had reduced total PS levels, which ranged from 0.18 to 0.35 IU/mL. Twenty-four patients had reduced free PS levels in the range of 0.02 to 0.17 IU/mL, whereas a single index case had a level of 0.37 IU/mL.
Thromboembolic events
In total, 115 thromboembolic events were identified retrospectively in the 137 individuals studied. All index case patients had suffered at least 1 episode of venous thromboembolism. Forty-nine events were identified in 27 first-degree relatives studied, with 44 (89.8%) of these occurring in those with PROS1 gene abnormalities. Venous thromboembolism was confirmed objectively in 65% of cases. At least 1 venous thrombotic event was confirmed in 44 (78.6%) of the 55 patients (28 index cases and 27 relatives) who reported previous thromboses. Thirty-three index cases and affected individuals suffered multiple thromboembolic events, but only 1 individual with a normalPROS1 gene had more than 1 thrombosis. Most (65.2%) of the thrombotic events occurred in the leg veins, 22.6% were pulmonary emboli, 1.7% were upper-limb venous thromboses, and 8.7% were episodes of thrombophlebitis.
Thrombotic events in relatives
The sex distribution, mean age, and age range of relatives with and without PROS1 gene defects and other thrombophilic defects were similar. Relatives with a PROS1 gene defect were more likely to have suffered a thrombosis (41.2%) than those with a normalPROS1 gene (6.7%) (P < .0001). Individuals with a normal PROS1 gene but with other thrombophilic defects had a higher prevalence of thrombosis (14.2%) than individuals with no thrombophilic defects (6.6%), but the difference was not significant (P = .45). Table 2 shows the data for thrombotic risk in the patients with different gene defects. The incidence of thrombosis was greater in relatives with an isolatedPROS1 gene defect (17.2 per 1000 person-years) compared with those with a normal gene (2.3 per 1000 person-years), with a relative risk for thrombosis of 5.0 (95% CI, 1.5-16.8). Relatives with aPROS1 defect and an additional thrombophilic defect also had a higher relative risk of thrombosis, at 4.0 (95% CI, 0.7-23.7), compared with those with a normal PROS1 gene, but the small number of patients in this group limits the interpretation that can be placed on these data (Table 2).
Thrombotic risk according to the presence or absence of a PROS1 gene defect and other prothrombotic defects*
| . | PROS1 gene abnormal (n = 57) . | PROS1 gene normal (n = 52) . | ||
|---|---|---|---|---|
| Without other defect (n = 51) . | With other defect (n = 6) . | Without other defect (n = 45) . | With other defect (n = 7) . | |
| Male, n (%) | 19 (37.2) | 2 (33.3) | 21 (46.6) | 4 (57.2) |
| No. (%) with thrombosis | 21 (41.2) | 2 (33.3) | 3 (6.6) | 1 (14.2) |
| Total thrombotic events | 41 | 3 | 4 | 1 |
| Years of follow-up | 2381 | 288 | 1713 | 287 |
| Age at first thrombosis, median (range) | 34.8 (17.1-69.5) | 35.1 | 24.5 (20.4-35.7) | 52.2 |
| Events per 1000 person-years, median (range) | 17.2 (12.3-23.4) | 10.4 (2.2-30.5) | 2.3 (0.6-6.0) | 3.4 (0.1-19.4) |
| Relative risk for thrombosis (95% CI) | 5.0 (1.5-16.8) | 4.0 (0.7-23.7) | 1 | 2.0 (0.2-19.1) |
| . | PROS1 gene abnormal (n = 57) . | PROS1 gene normal (n = 52) . | ||
|---|---|---|---|---|
| Without other defect (n = 51) . | With other defect (n = 6) . | Without other defect (n = 45) . | With other defect (n = 7) . | |
| Male, n (%) | 19 (37.2) | 2 (33.3) | 21 (46.6) | 4 (57.2) |
| No. (%) with thrombosis | 21 (41.2) | 2 (33.3) | 3 (6.6) | 1 (14.2) |
| Total thrombotic events | 41 | 3 | 4 | 1 |
| Years of follow-up | 2381 | 288 | 1713 | 287 |
| Age at first thrombosis, median (range) | 34.8 (17.1-69.5) | 35.1 | 24.5 (20.4-35.7) | 52.2 |
| Events per 1000 person-years, median (range) | 17.2 (12.3-23.4) | 10.4 (2.2-30.5) | 2.3 (0.6-6.0) | 3.4 (0.1-19.4) |
| Relative risk for thrombosis (95% CI) | 5.0 (1.5-16.8) | 4.0 (0.7-23.7) | 1 | 2.0 (0.2-19.1) |
Other inherited prothrombotic defects include antithrombin deficiency, factor V Leiden, or prothrombin 20 210A allele.
Precipitating factors for thrombosis
Precipitating factors for the 57 first episodes of thrombosis in index patients and relatives were analyzed. Twenty-eight (49.1%) of all episodes were spontaneous, 8 (14.0%) were related to trauma, 6 (10.5%) to pregnancy and the puerperium, 6 (10.5%) to surgery or immobility, 5 (8.8%) to combined oral contraceptive use, and in 4 cases other factors were contributing.
Thrombosis-free survival
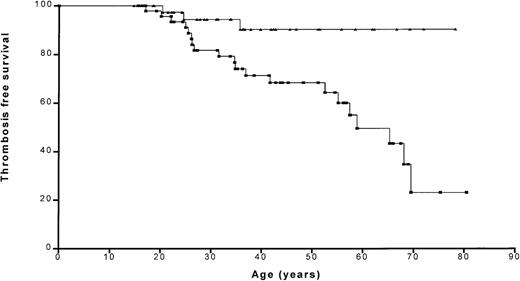
Kaplan-Meier analysis showed that the probability of remaining thrombosis-free was significantly (P = .0049) lower in relatives with a PROS1 gene defect than in those without (Figure 4). The probability of remaining thrombosis-free at age 50 years was 68.5% and 90.3% for affected and unaffected relatives, respectively.
Thrombosis-free survival in relatives with/withoutPROS1 gene defects.
Kaplan-Meier plot shows the thrombosis-free curves for relatives with (squares) and without (triangles) PROS1 gene defects.
Thrombosis-free survival in relatives with/withoutPROS1 gene defects.
Kaplan-Meier plot shows the thrombosis-free curves for relatives with (squares) and without (triangles) PROS1 gene defects.
Association of thrombotic risk with nature of the PROS1 gene defect
The thrombotic risk and phenotypic differences associated withPROS1 defects due to either splice-site/major structural defects or missense defects causing deficiency of PS were compared in the affected relatives. Those with additional thrombophilic defects were excluded from studies on thrombotic risk, and those taking warfarin were excluded from the analysis of PS levels. The thrombotic risk associated with each type of defect is summarized in Table3. The proportion of relatives with a history of thrombosis was significantly higher among those with splice-site/major structural defects (48.1%) or missense defects (37.5%) than among those with a normal PROS1 gene (6.6%) (P < .005). A higher proportion of patients with splice-site or major structural defects experienced a thrombotic event, but this was not statistically different compared with those with missense defects (P = .57). Total and free PS levels were both significantly lower in relatives with splice-site/major structural defects than in those with missense defects (Table 3): P < .05 andP < .001 for total and free PS levels, respectively.
Thrombotic risk and protein S levels according to the type of PROS1 gene defect
| . | Splice-site or major structural defect . | Missense defect . | Normal . |
|---|---|---|---|
| n | 27 | 24 | 45 |
| Male, n (%) | 9 (33.3) | 10 (41.6) | 21 (46.6) |
| No. (%) with venous thromboembolism | 13 (48.1) | 9 (37.5) | 3 (6.6) |
| Total thrombotic events | 26 | 15 | 4 |
| Years of follow-up | 1187 | 1194 | 1713 |
| Median age at first thrombosis, y (range) | 41.7 (17.1-69.5) | 34.7 (20.1-58.9) | 24.5 (20.4-35.7) |
| Thromboses per 1000 person-years, median (range) | 21.9 (14.3-32.0) | 12.6 (7.0-20.7) | 2.3 (0.6-6.0) |
| Relative risk for thrombosis (95% CI) | 6.3 (1.8-22.0) | 4.3 (1.2-15.8) | 1.0 |
| Total PS, mean (SD) in IU/mL | 0.496 (0.13) | 0.578 (0.08) | 1.03 (0.18) |
| Free PS, mean (SD) in IU/mL | 0.196 (0.11) | 0.349 (0.13) | 1.07 (0.20) |
| . | Splice-site or major structural defect . | Missense defect . | Normal . |
|---|---|---|---|
| n | 27 | 24 | 45 |
| Male, n (%) | 9 (33.3) | 10 (41.6) | 21 (46.6) |
| No. (%) with venous thromboembolism | 13 (48.1) | 9 (37.5) | 3 (6.6) |
| Total thrombotic events | 26 | 15 | 4 |
| Years of follow-up | 1187 | 1194 | 1713 |
| Median age at first thrombosis, y (range) | 41.7 (17.1-69.5) | 34.7 (20.1-58.9) | 24.5 (20.4-35.7) |
| Thromboses per 1000 person-years, median (range) | 21.9 (14.3-32.0) | 12.6 (7.0-20.7) | 2.3 (0.6-6.0) |
| Relative risk for thrombosis (95% CI) | 6.3 (1.8-22.0) | 4.3 (1.2-15.8) | 1.0 |
| Total PS, mean (SD) in IU/mL | 0.496 (0.13) | 0.578 (0.08) | 1.03 (0.18) |
| Free PS, mean (SD) in IU/mL | 0.196 (0.11) | 0.349 (0.13) | 1.07 (0.20) |
Patients with other thrombophilic defects were excluded. Relative risk is compared with that of relatives with a normal PROS1gene.
Discussion
Of all the recognized inherited thrombophilic defects, PS deficiency is the most difficult to diagnose accurately. Phenotypic diagnosis is difficult because plasma levels are influenced by age, sex, liver disease, oral contraceptive use, pregnancy, lupus anticoagulants, and coumarin therapy.37 The situation is further complicated by the fact that, unlike protein C or antithrombin, PS is complexed with C4bBP in plasma and only the free moiety is physiologically active. The primary strength of our study was that all patients reported were from a single center and all persons with the deficiency had theirPROS1 gene defect characterized. This allowed us to assess more accurately both the thrombotic risk and the relative value of the free and total PS levels in making the diagnosis of PS deficiency.
We have confirmed that PS deficiency is a risk factor for venous thrombosis. Relatives with a PROS1 gene defect were found to have a 5.0 times higher relative risk for thrombosis compared with those without a defect. Although PS deficiency is generally accepted as a risk factor for thrombosis,22,28 this has not been confirmed in all studies.30 We found that relatives without a PROS1 gene defect had a 90.3% chance of being thrombosis-free at the age of 50, compared with 68.5% in those withPROS1 gene defects (Figure 4). Patients with other genetic defects in addition to PS deficiency did not appear to have a worse thrombotic history, but the number in this group was small (n = 6) and only 2 of these patients had a thrombosis.
The annual incidence of thrombosis in relatives with a PROS1gene defect, based on a cumulative observation period of 2381 years, was 17.2 per 1000 person-years of follow-up. We found the incidence of thrombosis in relatives without a PROS1 gene defect or any other prothrombotic abnormality to be 2.3 per 1000 person-years (observation period 1713 years), a figure close to that seen in unselected populations. This suggests either that the PS gene defect is the primary abnormality in these families or that if additional defects are present, they are closely linked to thePROS1 gene.
In some studies, the effect of prothrombotic risk factors on thrombotic risk has been biased by the inclusion of index cases originally referred because of venous thrombotic events. We have reduced this bias by studying first-degree relatives with and without PROS1 gene defects, although some bias is evident in this approach because the families themselves are selected by presentation with thrombosis.
The availability of family members with and without PROS1 gene defects has allowed us to assess the sensitivity and specificity of total and free PS in making the phenotypic diagnosis of the deficiency. By using the detection of a genetic defect as the gold standard, we found free PS to be superior to the total level in making the diagnosis. Although the specificity of both total and free levels was good at 98.1% and 100%, respectively, the sensitivity of the free estimation was superior at 97.7%, compared with 82.2% for the total PS level (Figures 2 and 3).
Because PS is a vitamin K–dependent clotting factor, diagnosis of its deficiency is difficult in anticoagulated patients. The 2 methods most widely used to make the diagnosis of PS deficiency in these patients are to compare levels with those of another vitamin K–dependent factor such as factor X or to use normal ranges for patients taking warfarin. In our laboratory, the normal range for free PS in stably anticoagulated patients is 0.27 to 0.79 U/mL, compared with 0.64 to 1.31 IU/mL for adult patients not taking anticoagulants. Our normal range for total PS on oral anticoagulants is 0.49 to 0.87 IU/mL. Twenty-four of 25 patients with PROS1 defects taking warfarin in our study had both total and free levels below our normal range, indicating the potential value of establishing a separate normal range for anticoagulated patients.
In the patients studied, most (49.1%) of the thrombotic events were apparently spontaneous, with only 8.8% related to the use of the combined contraceptive pill. In view of the high proportion of spontaneous events, regardless of the effectiveness of prophylactic anticoagulant therapy at times of increased thrombotic risk, we are unlikely to eliminate most of the thromboses in these families. The earliest thrombotic event in this study was at the age of 17.1 years, indicating that thrombosis in childhood is rare in this population. On the basis of our study, there is no necessity to test children of affected individuals before their teenage years.
Potential limitations of this study are its retrospective nature and the fact that not all thrombotic events were confirmed. Although venous thrombotic events are now routinely investigated, this was not universally practiced in the past (especially when they were pregnancy related), and therefore a number of older, not fully investigated events were included in this study. We believe that including only confirmed events would have biased the study by excluding thrombotic events that occurred in older patients when they were younger. Ideally, a prospective study including only confirmed events is required to confirm our observations.
A wide spectrum of PROS1 gene defects associated with PS deficiency was seen in the cohort. One novel splice-site defect was detected. This was the G to T transversion in the invariant AG dinucleotide of the acceptor splice site upstream of exon 13 in intron L, which is likely to result in abnormal splicing of PROS1 RNA and an unstable mRNA that is rapidly degraded. Unfortunately, samples were not available from the patient to investigate this further at the RNA level. For the same reason, we could not examine whether the A to G transition at nucleotide position +7 in intron A affected the processing of PROS1 RNA. The sequence preceding the transition is +1GTGAGT+6, which closely matches the donor splice-site consensus sequence of +1GTAAGT+6. It is possible that although the A+7G transition occurs outside the consensus sequence, it may result in aberrant RNA processing, a hypothesis that is supported by our failure to detect any other PROS1 gene defect in the patient with this transition.
Two novel missense mutations, predicted to result in substitution of L581 by R and C206 by G, were identified. Both mutations were associated with reduced total and free PS levels, supporting the likelihood that they were causative. L581 is located in the sex hormone-binding globulin–like domain of PS, important in the interactions of PS with C4bBP and with factor V during APC-mediated inactivation of factor VIIIa.38 It is conserved across species39 and, as a nonpolar hydrophobic residue, is likely to be internal. Its substitution by a bulky charged residue, in this case R, might therefore be predicted to disrupt protein folding and the release of PS. C206 forms a disulfide bridge with C215 in the fourth epidermal growth factor (EGF) domain of PS.40Substitution by G would therefore be predicted to interfere with folding of the EGF module and to cause a reduction in its affinity for Ca2+, increasing the susceptibility of PS to proteases.41
Only 3 of the 58 families with type I PS deficiency listed in thePROS1 mutation database have large gene deletions, 2 of which are identical. Our study found a high prevalence of major PROS1gene structural defects among PS-deficient members of the cohort. Five of 28 index cases (17.9%) had either a large gene deletion or a rearrangement. The precise nature of these defects and the mechanisms underlying them remain to be established. Characterization of majorPROS1 gene defects is complicated by the presence of the highly homologous PROS2 pseudogene at the same locus, which makes interpretation of Southern blots difficult. The difference in prevalence of major PROS1 gene defects between this study and the mutation database is likely to reflect these difficulties.
We have studied the effect of the type of genetic defect on thrombotic tendency. Patients with splice-site and major structural defects were found to be at the same risk for thrombosis as patients with missense mutations. This analysis encompassed a range of defects, so it is possible that differences in patients with specific defects were not detected. Splice-site and major structural defects can be predicted to result in quantitative PS deficiency. However, we cannot be certain of the effects of missense mutations identified in this study. Molecular modeling and in vitro expression of mutated PS molecules will be required to further elucidate the effects of the missense mutations identified here on PS structure and function and may in the future permit associations between genetic defect and clinical phenotype.
In conclusion, we have demonstrated in a genetically defined PS-deficient population at a single center that relatives with aPROS1 gene defect are at a higher risk for thrombosis compared with those without a defect, in whom the thrombotic risk is similar to the “background” of the general population. In our study, free PS estimation was the preferred test for PS deficiency, identifying 97.7% of persons with a genetic defect. The use of total PS estimation in isolation failed to identify 17.7% of relatives with PROS1gene defects as having PS deficiency.
Supported by British Heart Foundation grant no. PG/95039.
Reprints:F. E. Preston, Division of Molecular and Genetic Medicine, University of Sheffield, Royal Hallamshire Hospital, Glossop Rd, Sheffield, S10 2JF, United Kingdom; e-mail:f.e.preston@Sheffield.ac.UK
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal