Cytokines exert pleiotropic effects on target cells in a manner dependent on the cell type or stage of differentiation. To determine how instinctive cell properties affect biological effects of cytokine, we introduced an erythroid/megakaryocyte lineage-specific transcription factor, GATA-1, into a murine myeloid cell line M1, which is known to undergo macrophage differentiation in response to interleukin 6 (IL-6). Overexpression of GATA-1 changed the phenotype of M1 cells from myeloid to megakaryocytic lineage. Furthermore, GATA-1 blocked both IL-6-induced macrophage differentiation and apoptosis of M1 cells. Although STAT3 is essential for IL-6-induced macrophage differentiation of M1 cells, GATA-1 had little or no effect on tyrosine phosphorylation, DNA binding, and transcriptional activities of STAT3 in Western blot analysis, electropholic mobility shift assay (EMSA), and luciferase assays. During IL-6-induced macrophage differentiation of M1 cells, IL-6 down-regulated cyclin D1 expression and induced p19INK4D expression, leading to reduction in cdk4 activities. In contrast, sustained expression of cyclin D1 and a significantly lesser amount of p19INK4D induction were observed in IL-6-treated M1 cells overexpressing GATA-1. Furthermore, although bcl-2 expression was severely reduced by IL-6 in M1 cells, it was sustained in GATA-1-introduced M1 cells during the culture with IL-6. Both IL-6-induced macrophage differentiation and apoptosis were significantly abrogated by coexpression of cyclin D1 and bcl-2, whereas overexpressions of cyclin D1 or bcl-2 inhibited only differentiation or apoptosis, respectively. These results suggested that GATA-1 may not only reprogram the lineage phenotype of M1 cells but also disrupt the biologic effects of IL-6 through the sustained expression of cyclin D1 and bcl-2.
Growth, differentiation, and survival of hematopoietic cells are regulated by a number of cytokines and growth factors that activate multiple signal transduction pathways through binding to their cognate receptors. Among various hematopoietic growth factors, interleukin 6 (IL-6) exerts pleiotropic effects on hematopoietic cells (for reviews, see 1 and 2). IL-6 has been shown, for example, to augment growth of multipotential hematopoietic progenitor cells and plasmacytoma/myeloma cells.3-5 IL-6 has also been implicated in differentiation of various cell types, including normal B cells, megakaryocytes, and myeloid cell lines M1, Y6, and 1A9-M.6-10
The IL-6 receptor is a heterodimeric complex, consisting of an IL-6 specific ligand-binding subunit, α chain, and a signal-transducing subunit, gp130. Structural analysis has revealed that both subunits belong to the cytokine receptor superfamily, and gp130 is shared by the receptors for ciliary neurotrophic factor, leukemia inhibitory factor, oncostatin M, and cardiotropin 1. The binding of IL-6 to α chain leads to the formation of receptor complexes, followed by tyrosine phosphorylation and activation of Janus protein tyrosine kinases (JAKs) and various cellular proteins, including gp130 itself. The activated JAKs, in turn, phosphorylate and activate latent signal transducer and activator of transcription (STAT) family proteins, especially STAT1 and STAT3 (for reviews, see 1 and 11). Yamanaka et al12 showed that the YXXQ motif of gp130 was critical not only for STAT3 activation but also for growth arrest and macrophage differentiation of M1 cells. In accord with this finding, dominant negative (dn)-STAT3 was reported to inhibit IL-6-induced macrophage differentiation in M1 and 1A9-M cells, suggesting that STAT3 is a key regulator of IL-6-induced macrophage differentiation.10,13,14 Furthermore, by using murine IL-3-dependent Ba/F3 cells expressing chimeric receptors composed of the extracellular domain of granulocyte colony-stimulating factor receptor and the transmembrane and cytoplasmic domain of gp130, Fukada et al15,16 described that STAT3 was linked with anti-apoptotic signals through the induction of bcl-2 and with G1/S transition in cell cycle control. In contrast, IL-6 was recently shown to down-regulate bcl-2 messenger RNA (mRNA) expression during IL-6-induced macrophage differentiation of 1A9-M cells, thereby leading to apoptosis of the cells.10 Thus, the biologic activities of IL-6 appear to be modulated by the instinctive cell properties. However, molecular mechanisms underlying the differential biological activities of IL-6 remain largely unknown.
It has become increasing apparent that transcription factors play a key role in hematopoiesis. Transcription factors of the GATA-family are composed of six members and are essential for the development and subsequent growth and differentiation of diverse cell types (for a review, see 17). The first cloned member of this family, GATA-1, was identified as an erythroid nuclear protein that binds to consensus GATA motifs, (A/T)GATA(A/G), in globin gene promoters, enhancers, and locus control regions (LCRs).18,19 It has been shown that GATA-1 is expressed at high levels in erythroid cells, megakaryocytes, and mast cells and at lower levels in hematopoietic progenitor cells and Sertoli cells of testis.20-22 In contrast, GATA-2 is ubiquitously expressed, and GATA-3 is exclusively expressed in T lymphocytes.23 The functional roles of GATA-1 in the hematopoietic system have been elucidated by gene-targeting experiments. In chimeric mice generated by mutant embryonic stem (ES) cells lacking GATA-1, the mutant cells did not contribute to erythropoiesis.24 In addition, GATA-1-null ES cells were unable to differentiate into mature erythroid cells in vitro.25,26 Thus, GATA-1 has been implicated in regulating terminal differentiation of erythroid progenitor cells. Recently, Shivdasani et al27 reported that lineage-selective GATA-1 knock out mice exhibited striking thrombocytopenia as well as severe anemia because of the decreased proliferation and impaired cytoplasmic maturation of megakaryocytes. Furthermore, Takahashi et al28 demonstrated that heterozygous mutant mice chimeric for GATA-1 gene expression displayed marked splenomegaly, anemia, and thrombocytopenia because of progenitor proliferation in the spleen and consequently differentiation arrest of immature erythroid and megakaryocytic cells. Those results suggest that GATA-1 plays essential roles not only in erythropoiesis but also in megakaryopoiesis or thrombopoiesis.
Previous studies have also shown that GATA-1 is unique in its ability to influence the lineage phenotype of hematopoietic cells. Enforced expression of GATA-1 reprograms transformed chicken myeloblasts into erythroblasts, thromboblasts, or eosinophiles.29 Moreover, overexpression of GATA-1 in the early myeloid 416B cells provoked megakaryocytic differentiation accompanied by a marked decrease in myeloid surface phenotype.30 Recently, Yamaguchi et al31 reported that forced GATA-1 expression in the murine myeloid M1 cells, which differentiate into macrophage and undergo apoptosis in response to IL-6, led to megakaryocytic or erythroid differentiation. Those findings led us to speculate that GATA-1 might also affect cytokine-mediated signaling pathways that are involved in gene transcription necessary for growth, differentiation, and survival of hematopoietic stem or progenitor cells. In this study, therefore, we introduced GATA-1 into murine myeloid M1 cells and examined the effect of GATA-1 on IL-6-induced macrophage differentiation and apoptosis of the cells. Over-expression of GATA-1 was found to block IL-6-induced macrophage differentiation and apoptosis of M1 cells without affecting the STAT3 pathway. The effects of GATA-1 were mediated, at least partially, through the sustained expression of cyclin D1 and bcl-2. Thus, we here provide evidence that GATA-1 is capable of modulating biological activities of IL-6 through regulating specific members of cell cycle and apoptosis regulating molecules.
Materials and methods
Reagents and antibodies
Highly purified recombinant human (rh) IL-6 and rabbit anti-c-Mpl antiserum were provided by Kirin Brewery Company Ltd (Tokyo, Japan). Anti-CD61 monoclonal antibody (mAb) was purchased from Dako Japan Company Ltd (Tokyo, Japan). Anti-CD32 and anti-F4/80 mAbs were purchased from PharMingen (San Diego, CA) and Serotec Ltd (Oxford, UK), respectively. Anti-phosphotyrosine mAb was supplied from Dr. B. Druker (Oregon Health Science University, Portland, OR). Anti-GATA-1 (N1), anti-STAT3 (C-20), anti-bcl-2 (ΔC 21), and anti-cdk4 (C-22-G) polyclonal antibodies were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Anti-cyclin D1 mAb was purchased from Medical Biological Laboratories (MBL) Company Ltd (Nagoya, Japan).
Plasmid construct and complementary DNAs
Murine cyclin D1, cyclin D2, and cyclin D3 complementary DNAs (cDNAs) were provided from Dr. H. Matsusime (Nippon Roche Research Institute, Kanagawa, Japan). Murine bcl-2 cDNA was a generous gift from Dr. Y. Tsujimoto (Osaka University, Osaka, Japan). To construct expression vectors of GATA-1, cyclin D1, and bcl-2, full length of human GATA-1, murine cyclin D1, and bcl-2 cDNAs were subcloned into an expression vector, pcDNA3 (Invitrogen, De Schelp, The Netherlands). An expression vector of hemagglutinin (HA)-tagged dn-STAT3 (pCAGGS-neo-HA-STAT3D) was described previously.13 Other cDNAs were kindly provided from the investigators as follows: murine p21WAF1 from Dr. B. Vogelstein (Johns Hopkins Oncology Center, Baltimore, MD); human p27Kip1 from Dr. H. Kiyokawa (University of Illinois, Cancer Center, Chicago, IL); murine p18INK4C and p19INK4D from Dr. C. Sherr (Howard Hughes Medical Institute, TN); and murine bcl-xL, bax, and bak from Dr. T. Tsujimura (Hyogo College of Medicine, Nishinomiya, Japan).
Cell lines and cultures
M1, a murine myeloid leukemia cell line originally established by Ichikawa32 was cultured in RPMI (Nakarai Tesq, Kyoto, Japan) supplemented with 10% fetal calf serum (FCS) (Flow, North Ryde, Australia). HepG2 and NIH3T3 cells were cultured in DMEM supplemented with 10% FCS.
Morphological analysis
The morphological characteristics of cultured cells were determined by staining the cytospin preparations (Shandon, Pittsburgh, PA) with May-Grünwald-Giemsa.
Flow cytometry
The surface phenotypes of cells were examined with the indirect immunofluorescent method by using rabbit anti-c-Mpl antiserum, anti-CD61, anti-CD32, or anti-F4/80 mAb as previously reported.33 The cell cycle analysis of cultured cells was performed by staining with propidium iodide as previously described.34
Northern blot analysis
The isolation of total cellular RNA and the method for Northern blot were described previously.35
Immunoprecipitation and immunoblotting
The isolation of total cellular lysates, immunoprecipitation, gel electrophoresis, and immunoblotting were performed according to the methods described previously.36 Immunoreactive proteins were visualized with the enhanced chemiluminescence detection system (DuPont NEN, Boston, MA).
Metabolic labeling and measurement of protein turnover
To examine the half-lives of cyclin D1 and bcl-2 proteins, 1 × 107 cells for each sample were radiolabeled with 200 μCi of 35S]methionine in 1 mL of methionine-free DMEM for 30 minutes or 6 hours. The cells were then washed and resuspended in DMEM containing 2 mmol/L of unlabeled methionine. After the culture with or without IL-6, total cellular lysates were prepared at the time indicated. Cyclin D1 and bcl-2 were immunoprecipitated from the lysates and subjected to SDS-PAGE, respectively. The gels were dried and subjected to the autoradiography. The radioactivities of the bands corresponding to cyclin D1 or bcl-2 protein were measured by a densitometric analysis.
Preparation of M1 clones expressing GATA-1, cyclin D1, bcl-2, or a combination
M1 cells were transfected with 30 μg of pcDNA3-GATA-1, pcDNA3-cyclin D1, pcDNA3-bcl-2, or an empty pcDNA3 by electroporation (250 V, 960 μFD) (Bio-Lad Laboratory, Richmond, CA). The transfected cells were screened by the culture with 1.2 mg/mL of G418 (Sigma). Of several G418-resistant clones, expression levels of each transgene were examined by Northern blot and Western blot analyses. To prepare a stable transformant designated M1-W, in which both cyclin D1 and bcl-2 are overexpressed, cyclin D1-transfected M1 (M1-D1) was further cotransfected with 50 μg of pcDNA3-bcl-2 and 5 μg of pSV2bsr, an expression vector of blasticidin S deaminase (Kaken Pharmaceutical Co, Tokyo, Japan) and screened by the culture with 30 μg/mL of blasticidin S hydrochloride (Funakoshi, Tokyo, Japan).
Luciferase assays
Luciferase assays were performed with a reporter gene for STAT3, named 4 × APRE-Luc.13 NIH3T3 or HepG2 cells were transfected with various amounts of an effector gene of pcDNA3-GATA-1 or pCAGGS-neo-HA-STAT3D along with 1 μg of a 4 × APRE-Luc reporter gene and 10 ng of pRL-CMV-Rluc, an expression vector of renilla luciferase, by calcium phosphate coprecipitation method. Total amounts of DNA for each transfection were equalized by the addition of an empty pcDNA3 or pCAGGS. After 12 hours of culture, the cells were washed, serum starved for 24 hours, and then stimulated with 20 ng/mL of rhIL-6 for 5 hours. Luciferase assays were performed by using the Dual-Luciferase Reporter System (Promega, Madison, WI) in which relative firefly luciferase activities were calculated by normalizing transfection efficiency according to the renilla luciferase activities.
cdk4-associated GST-Rb kinase assay
In vitro cdk4-associated GST-Rb (Rb: retinoblastoma protein) kinase assay was performed as previously described.37 Briefly, cdk4 was immunoprecipitated from equal amounts of cell lysates prepared from cultured cells. Immune complex kinase assay was performed in kinase buffer containing 5 μg of GST-Rb fusion protein (Santa Cruz) and 20 μCi of γ-32P]ATP for 30 minutes at 30°C. After the addition of protein loading buffer, samples were boiled and subjected to SDS-PAGE. The gels were stained with Coomassie blue to confirm the amounts of immunoprecipitates, then destained, dried, and subjected to autoradiography.
Electropholic mobility shift assay (EMSA)
The isolation of nuclear extracts was performed as previously described.37 A double-stranded oligonucleotide containing STAT3-binding sequence (APRE) was synthesized and used as a probe or a competitor (5′-AGCTTCCTTCTGGGAATTCCT-3′, APRE sequence is underlined).38 Also, one more double-stranded oligonucleotide, which contains mutated APRE sequence, was used as a competitor (5′-AGCTTCCTGCTGGGACTTCCT-3′, mutated recognition site is underlined). Nuclear extract (15 μg of each sample) was incubated in 20 μL of binding buffer containing 2 μg of poly (dI-dC) (Pharmacia) and labeled probe (30 000 cpm) for 20 minutes at 4°C. The reaction mixture was loaded onto 4% polyacrylamide gel, electrophoresed, dried, and subjected to autoradiography.
Results
Preparation and characterization of GATA-1introduced M1 cells
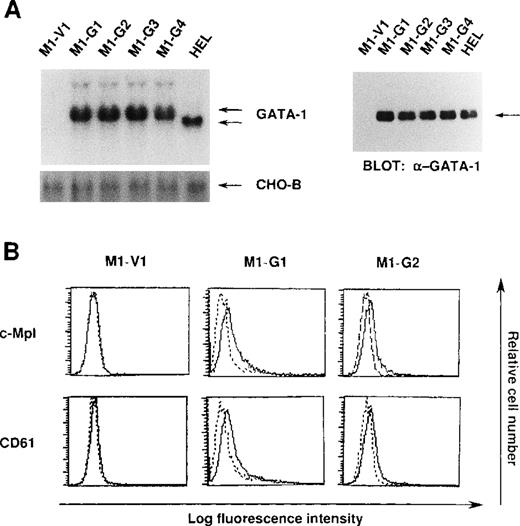
We initially introduced an expression vector of GATA-1 into M1 cells. After the selection with G418, expression of GATA-1 transgene was examined by Northern blot analysis. As shown in Figure1A, GATA-1 mRNA was hardly detected in a control clone (M1-V1) transfected with an empty vector. In contrast, expression of GATA-1 mRNA was observed in GATA-1-transfectants (M1-G1, M1-G2, M1-G3, and M1-G4) at high levels almost similar to that in a human erythroleukemia cell line (HEL), whereas GATA-1 transgene showed more slowly migrating bands. In addition, Western blot analysis showed that comparable amounts of GATA-1 proteins were expressed in M1-G1, M1-G2, M1-G3, M1-G4, and HEL cells, but not in M1-V1 cells (Figure 1A). Because the ectopic overexpression of GATA-1 was reported to induce erythroid or megakaryocytic differentiation, or both,29-31 we examined the effects of GATA-1 on surface expression of two megakaryocytic lineage markers, TPO receptor (c-Mpl) and CD61 (GPIIb/IIIa). As shown in Figure 1B, although c-Mpl and CD61 were scarcely expressed on M1-V1, a weak but easily detectable level of c-Mpl and CD61 expression was observed on M1-G1 and M1-G2. These results suggested that ectopic overexpression of GATA-1 reprogrammed M1 cells, to some degree, toward megakaryocytic lineage.
Expression of GATA-1-transfected M1 cells.
(A) Expression of GATA-1 in M1-V1, GATA-1-transfected M1 (M1-G1, -G2, -G3, and -G4), and a human erythrocytic cell line in Northern blot (left panel) and Western blot (right panel) analyses. Total cellular RNA was isolated from each clone, and 15 μg of each sample was electrophoresed on formaldehyde agarose gels. The filter was hybridized with 32P-labeled full length of human GATA-1 complementary DNA or CHO-B (left panel). Total cellular lysates (15 μg per each lane) were subjected to SDS-PAGE and probed with rabbit anti-GATA-1 polyclonal antibody. Immunoreactive proteins were visualized with the enhanced chemiluminescence detection system (right panel). (B) Expression of c-Mpl and CD61 in M1-V1, M1-G1, and M1-G2. Expression of c-Mpl and CD61 was examined by staining with rabbit anti-c-Mpl antiserum or CD61 monoclonal antibody (—) with a reference to rabbit preimmune serum or control antibody of the same isotype (---).
Expression of GATA-1-transfected M1 cells.
(A) Expression of GATA-1 in M1-V1, GATA-1-transfected M1 (M1-G1, -G2, -G3, and -G4), and a human erythrocytic cell line in Northern blot (left panel) and Western blot (right panel) analyses. Total cellular RNA was isolated from each clone, and 15 μg of each sample was electrophoresed on formaldehyde agarose gels. The filter was hybridized with 32P-labeled full length of human GATA-1 complementary DNA or CHO-B (left panel). Total cellular lysates (15 μg per each lane) were subjected to SDS-PAGE and probed with rabbit anti-GATA-1 polyclonal antibody. Immunoreactive proteins were visualized with the enhanced chemiluminescence detection system (right panel). (B) Expression of c-Mpl and CD61 in M1-V1, M1-G1, and M1-G2. Expression of c-Mpl and CD61 was examined by staining with rabbit anti-c-Mpl antiserum or CD61 monoclonal antibody (—) with a reference to rabbit preimmune serum or control antibody of the same isotype (---).
Effect of GATA-1 on IL-6-induced growth arrest and subsequent macrophage differentiation of M1 cells
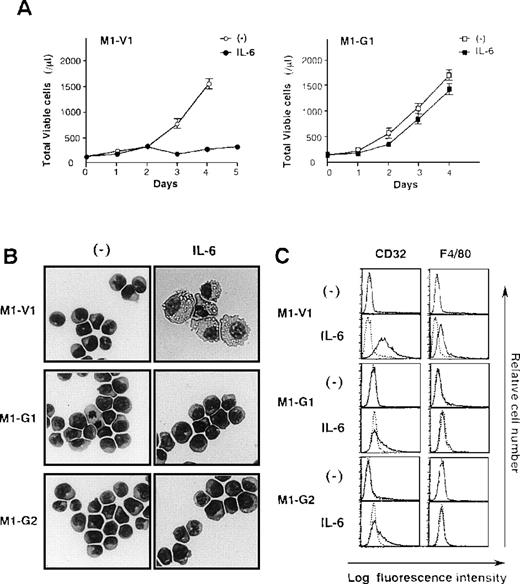
Next, we examined the growth and differentiation of M1-V1 and M1-G1 in the presence or absence of rhIL-6 (Figure2). Without the addition of rhIL-6, no significant difference was seen between growth curves of M1-G1 and M1-V1 (Figure 2A). However, when cultured with rhIL-6, M1-V1 ceased to grow after 3 days in the same way as parental M1, whereas M1-G1 showed continuous growth during the 4-day culture. Similar results were observed in other GATA-1-transfected clones, M1-G2, M1-G3, and M1-G4 (data not shown).
Biologic responses of M1-V1 and M1-G1 to IL-6.
(A) Changes in total viable cell number during the culture with or without recombinant human (rh) interleukin 6 (IL-6). M1-V1 and M1-G1 cells were seeded at a cell density 100/μL and cultured with or without 20 ng/mL rhIL-6. Total viable cell number was counted by trypan blue dye exclusion method. M1-V1, open circle, IL-6 (-), closed circle, IL-6 (+); M1-G1, open square, IL-6 (-), closed square, IL-6 (+). The results are shown as the mean ± SD of triplicate cultures. (B) Light micrograph of M1-V1, M1-G1, and M1-G2 before and after 72-hour culture with rhIL-6. Cytocentrifugation preparation from each culture was stained with May-Gr ¸nwald-Giemsa (magnification × 100). (C) Flow cytometric analyses on expression of F4/80 and CD32 before and after 72-hour culture with rhIL-6. Expression of F4/80 and CD32 in the cultured cells was examined by staining with anti-F4/80 or anti-CD32 monoclonal antibody (—) with a reference to control antibody of the same isotype (---).
Biologic responses of M1-V1 and M1-G1 to IL-6.
(A) Changes in total viable cell number during the culture with or without recombinant human (rh) interleukin 6 (IL-6). M1-V1 and M1-G1 cells were seeded at a cell density 100/μL and cultured with or without 20 ng/mL rhIL-6. Total viable cell number was counted by trypan blue dye exclusion method. M1-V1, open circle, IL-6 (-), closed circle, IL-6 (+); M1-G1, open square, IL-6 (-), closed square, IL-6 (+). The results are shown as the mean ± SD of triplicate cultures. (B) Light micrograph of M1-V1, M1-G1, and M1-G2 before and after 72-hour culture with rhIL-6. Cytocentrifugation preparation from each culture was stained with May-Gr ¸nwald-Giemsa (magnification × 100). (C) Flow cytometric analyses on expression of F4/80 and CD32 before and after 72-hour culture with rhIL-6. Expression of F4/80 and CD32 in the cultured cells was examined by staining with anti-F4/80 or anti-CD32 monoclonal antibody (—) with a reference to control antibody of the same isotype (---).
Although a modest level of c-Mpl and CD61 expression was detectable only on GATA-1 transfectants (Figure 1B), morphological analysis showed that each M1-V1, M1-G1, and M1-G2 clone was exclusively composed of undifferentiated blast cells before the culture with rhIL-6 (Figure 2B, left panel). After a 3-day culture with rhIL-6, M1-V1 revealed morphological changes indicative of macrophage differentiation that is characterized by enlarged cell size and vacuoles in the cytoplasm, whereas M1-G1 and M1-G2 still remained to be immature blast cells (Figure 2B, right panel). Furthermore, a noticeable proportion of M1-V1, but not of M1-G1 or M1-G2, was found to undergo apoptotic cell death characterized by shrunk cell size, nuclear fragmentation, or both after a 3-day culture with rhIL-6 (data not shown).
We also examined the surface expression of macrophage antigens, CD32 and F4/80, before and after treatment with rhIL-6 for 48 hours. In agreement with morphological characteristics, CD32 or F4/80 were not expressed on M1-V1, M1-G1, or M1-G2 before the culture with rhIL-6, but they were significantly up-regulated by rhIL-6 on M1-V1; these up-regulated expressions of CD32 and F4/80 were significantly suppressed in M1-G1 and M1-G2 (Figure 2C). These results indicated that overexpression of GATA-1 blocked IL-6-induced macrophage differentiation and apoptosis of M1 cells.
Effect of GATA-1 on IL-6 signaling
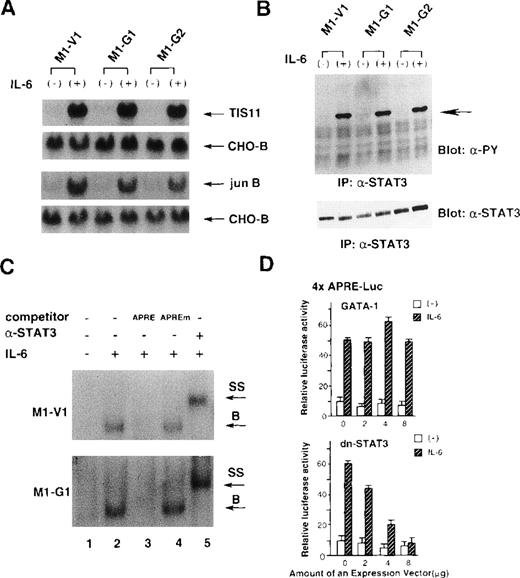
Because IL-6 failed to exert biologic effects on M1-G1 and M1-G2, we examined as to which signaling pathway(s) of IL-6 was perturbed by GATA-1. To first determine whether IL-6 signaling could be transduced into the nucleus, we investigated the induction of IL-6-responsive immediate early genes, TIS11 and jun B,39 in M1-V1, M1-G1, and M1-G2. As shown in Figure 3A, the treatment with rhIL-6 resulted in rapid induction of Tis11 and Jun B mRNA in M1-V1, M1-G1, and M1-G2 at a comparable level, suggesting that IL-6 signaling to the nucleus was not disrupted in GATA-1 transfectants. We next examined tyrosine phosphorylation of STAT3 before and after treatment with rhIL-6, because STAT3 was reported to be essential for IL-6-induced macrophage differentiation of M1.12-14 As shown in Figure 3B, rhIL-6 was capable of inducing tyrosine phosphorylation of STAT3 in M1-G1, M1-G2, and M1-V1 in a similar fashion.
Effects of GATA-1 on IL-6 signaling.
(A) Induction of TIS11 and jun B genes by recombinant human (rh) interleukin 6 (IL-6) in M1-V1, M1-G1, and M1-G2. Total cellular RNA was extracted from each clone before and after the stimulation with rhIL-6 (20 ng/mL) for 30 minutes and subjected to Northern blot analysis. The filters were hybridized with 32P-labeled probes for TIS11, jun B, and CHO-B. (B) Changes in tyrosine phosphorylation of STAT3 in response to rhIL-6 in M1-V1, M1-G1, and M1-G2. The cells of each clone were serum starved for 12 hours and then stimulated with rhIL-6 (20 ng/mL) for 15 minutes. Total cell lysates were isolated before and after the stimulation with rhIL-6, immunoprecipitated with anti-STAT3 polyclonal antibody, and subjected to SDS-PAGE. The blots were probed with anti-phosphotyrosine monoclonal antibody. Immunoreactive proteins were visualized with the enhanced chemiluminescence detection system. Then, the filters were stripped and reprobed with anti-STAT3 antibody. (C) Effects of GATA-1 on DNA-binding activities of STAT3. M1-V1 and M1-G1 were serum deprived for 12 hours then unstimulated or stimulated with rhIL-6 (20 ng/mL) for 15 minutes, and nuclear extracts were isolated. The nuclear extract was incubated in binding buffer containing 2 μg of poly(dI-dC) and labeled probe (30 000 cpm) for 20 minutes at 4°C. The reaction mixture was electrophoresed, dried, and subjected to autoradiography. In competition assays, nuclear extracts were preincubated with a 200-fold molar excess of unlabeled competitor oligonucleotide before the binding reaction. In supershift assays, the nuclear proteins were preincubated with 1 μg of rabbit anti-STAT3 polyclonal monoclonal at 4°C for 30 minutes, and then the binding reaction was performed. (D) Effects of GATA-1 on transcriptional activities of STAT3. NIH3T3 cells were transfected with 1 μg of 4 × APRE-Luc, 10 ng of pRL-CMV-Rluc, and various amounts of pcDNA3-GATA-1 or pCAGGS-neo-dn-STAT3 by calcium phosphate coprecipitation method. After 12 hours, the cells were serum starved for 24 hours and then stimulated with rhIL-6 (20 ng/mL) for 5 hours. The cells were lysed in lysis buffer, followed by the measurement of the firefly and the renilla luciferase activities. The relative firefly luciferase activities were calculated by normalizing transfection efficiency according to the renilla luciferase activities. The results are shown as the mean ± SD of triplicate experiments.
Effects of GATA-1 on IL-6 signaling.
(A) Induction of TIS11 and jun B genes by recombinant human (rh) interleukin 6 (IL-6) in M1-V1, M1-G1, and M1-G2. Total cellular RNA was extracted from each clone before and after the stimulation with rhIL-6 (20 ng/mL) for 30 minutes and subjected to Northern blot analysis. The filters were hybridized with 32P-labeled probes for TIS11, jun B, and CHO-B. (B) Changes in tyrosine phosphorylation of STAT3 in response to rhIL-6 in M1-V1, M1-G1, and M1-G2. The cells of each clone were serum starved for 12 hours and then stimulated with rhIL-6 (20 ng/mL) for 15 minutes. Total cell lysates were isolated before and after the stimulation with rhIL-6, immunoprecipitated with anti-STAT3 polyclonal antibody, and subjected to SDS-PAGE. The blots were probed with anti-phosphotyrosine monoclonal antibody. Immunoreactive proteins were visualized with the enhanced chemiluminescence detection system. Then, the filters were stripped and reprobed with anti-STAT3 antibody. (C) Effects of GATA-1 on DNA-binding activities of STAT3. M1-V1 and M1-G1 were serum deprived for 12 hours then unstimulated or stimulated with rhIL-6 (20 ng/mL) for 15 minutes, and nuclear extracts were isolated. The nuclear extract was incubated in binding buffer containing 2 μg of poly(dI-dC) and labeled probe (30 000 cpm) for 20 minutes at 4°C. The reaction mixture was electrophoresed, dried, and subjected to autoradiography. In competition assays, nuclear extracts were preincubated with a 200-fold molar excess of unlabeled competitor oligonucleotide before the binding reaction. In supershift assays, the nuclear proteins were preincubated with 1 μg of rabbit anti-STAT3 polyclonal monoclonal at 4°C for 30 minutes, and then the binding reaction was performed. (D) Effects of GATA-1 on transcriptional activities of STAT3. NIH3T3 cells were transfected with 1 μg of 4 × APRE-Luc, 10 ng of pRL-CMV-Rluc, and various amounts of pcDNA3-GATA-1 or pCAGGS-neo-dn-STAT3 by calcium phosphate coprecipitation method. After 12 hours, the cells were serum starved for 24 hours and then stimulated with rhIL-6 (20 ng/mL) for 5 hours. The cells were lysed in lysis buffer, followed by the measurement of the firefly and the renilla luciferase activities. The relative firefly luciferase activities were calculated by normalizing transfection efficiency according to the renilla luciferase activities. The results are shown as the mean ± SD of triplicate experiments.
We also examined the binding activities of STAT3 to APRE oligonucleotides by EMSA (Figure 3C). The rhIL-6-treated nuclear extracts from M1-V1 became to possess binding activity to APRE oligonucleotide (upper panel, lane 2), and this binding was effectively competed by the APRE competitor (lane 3) but not by equimolar amounts of the mutated APRE oligonucleotide (lane 4). Furthermore, the DNA-binding complex was completely supershifted by preincubation with anti-STAT3 antibody (lane 5). These findings indicated that the IL-6-induced APRE-binding complex was formed in a sequence-specific manner and contained STAT3 proteins. The similar patterns of DNA-binding and supershift were observed in the nuclear extract from M1-G1 (Figure 3C, lower panel), indicating that the DNA-binding activities of STAT3 induced by rhIL-6 were not inhibited in M1-G1.
To further investigate the effects of GATA-1 on transcriptional activities of STAT3, we performed luciferase assays with a reporter gene for STAT3, 4 × APRE-Luc.13 As shown in Figure4D, the treatment with rhIL-6 stimulated 4 × APRE-Luc about fivefold in NIH3T3 cells. When dn-STAT3 was cotransfected, rhIL-6-stimulated 4 × APRE-Luc activities were inhibited by dn-STAT3 in a dose-dependent manner (Figure 3D, lower panel). In contrast, cotransfected GATA-1 had little or no effect on rhIL-6-stimulated reporter gene activities (Figure 3D, upper panel). The similar results were also observed in HepG2 cells (data not shown). These results suggested that GATA-1 did not affect IL-6-induced STAT3 activities.
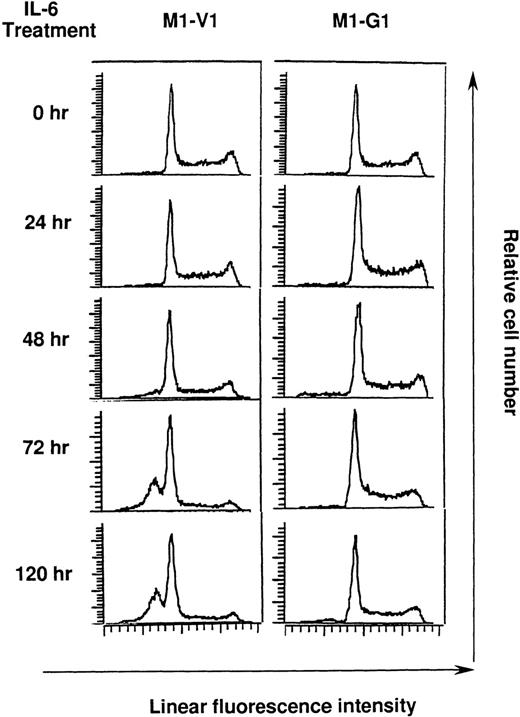
Cell cycle analysis on M1-V1 and M1-G1 cells during the culture with recombinant human (rh) interleukin 6 (IL-6).
M1-V1 and M1-G1 were cultured with 20 ng/mL of rhIL-6, and the cultured cells were subjected to propidium iodide staining at the time indicated. DNA content was analyzed on FACSort. Cell cycle analysis was performed with the program Modfit LT2.0.
Cell cycle analysis on M1-V1 and M1-G1 cells during the culture with recombinant human (rh) interleukin 6 (IL-6).
M1-V1 and M1-G1 were cultured with 20 ng/mL of rhIL-6, and the cultured cells were subjected to propidium iodide staining at the time indicated. DNA content was analyzed on FACSort. Cell cycle analysis was performed with the program Modfit LT2.0.
Cell cycle analysis on M1-V1 and M1-G1 cells during the culture with rhIL-6
Next, we performed cell cycle analysis during the culture with rhIL-6 (Figure 4). Before the culture with rhIL-6, M1-V1 and M1-G1 did not show an apparent difference in cell cycle distributions: M1-V1, G0/G1 50%, S 39%, and G2/M 11%; M1-G1, G0/G1 48%, S 41%, and G2/M 11%. After 72-hour culture of M1-V1 with rhIL-6, the proportions of cells in S and G2/M phases were reduced to 11% and 5%, respectively. In addition, the rhIL-6-treatment induced apoptosis of M1-V1, which was detected as a subdiploid fraction (% subdiploid fraction: 40% at 72 hours; 43% at 120 hours). In contrast, rhIL-6 showed only a minimal effect on cell cycle distribution of M1-G1 at 72 hours (G0/G1 53%, S 38%, and G2/M 9%) and did not apparently increase the subdiploid fraction of M1-G1 from 72 to 120 hours. These results suggested that GATA-1 could prevent M1 cells from IL-6-induced cell cycle arrest and apoptosis.
Expression of cell cycle regulatory and apoptosis-related molecules in M1-V1 and M1-G1 cells during the culture with IL-6
To clarify the mechanism underlying the differential biological responses of M1-V1 and M1-G1 to rhIL-6, we examined the expression of cell cycle regulatory molecules, including cyclins, cyclin-dependent kinases, and cyclin-dependent kinase inhibitors, and apoptosis-related genes during 72-hour culture with rhIL-6. As shown in Figure 5A, Northern blot analysis showed that expression of cyclin D1 mRNA decreased gradually to an undetectable level after 24 hours in M1-V1, whereas it was up-regulated at 4 hours and retained at a detectable level throughout the culture in M1-G1. The expression of p19INK4D mRNA was up-regulated by rhIL-6 in M1-V1 and, to a lesser extent, in M1-G1 from 4 hours to 24 hours. By contrast, the expression levels of cyclin D2, cyclin D3, p18INK4C, and p27Kip1 mRNA did not show apparent difference between M1-V1 and M1-G1. Expression of p21WAF1 was not detectable in both clones during the test period. With reference to apoptosis-related genes, expression of bcl-2 mRNA was down-regulated markedly in M1-V1 after 12 hours, whereas it was detected constantly for up to 72 hours in M1-G1. However, an apparent difference was not observed in expression levels of bax, bak, or bcl-xL mRNA in M1-V1 and M1-G1. In addition to mRNA levels, Western blot analysis demonstrated that cyclin D1 and bcl-2 proteins were expressed in M1-G1 during 72-hour culture with rhIL-6, whereas their expression was severely reduced in M1-V1 at 72 hours (Figure 5B upper panel and Figure 5C). Because D-type cyclins and the INK4 family of cyclin-dependent kinase inhibitors have been reported to complex with cdk4 and to regulate its activities, we examined the changes in cdk4 activities with an immune complex kinase assay by using GST-Rb as a substrate (Figure 5B, lower panel). Although expression of cyclin-dependent D2 and cyclin D3 was retained in M1-V1 during 72-hour culture with rhIL-6, cdk4 activities were gradually reduced in M1-V1, possibly owing to the decreased expression of cyclin D1 and to the induction of p19INK4D. By contrast, cdk4 activities were sustained at an almost constant level for up to 72 hours in M1-G1. These results raised the possibility that GATA-1 might protect M1 cells from IL-6-induced cell cycle arrest and apoptosis through the sustained expression of cyclin D1 and bcl-2.
Changes in expression of cell cycle regulatory molecules and apoptosis-related genes during IL-6 treatment in M1-V1 and M1-G1.
(A) Northern blot analysis on expression of cell cycle regulatory molecules and apoptosis-related genes in M1-V1 and M1-G1 during the culture with recombinant human (rh) interleukin 6 (IL-6). M1-V1 and M1-G1 were cultured with 20 ng/mL of rhIL-6, and total cellular RNA was isolated at the time indicated. Fifteen μg of each sample was electrophoresed on formaldehyde agarose gels, blotted onto the filters, and hybridized with 32P-labeled probes as indicated. (B) Western blot analysis on cyclin D1 expression (upper panel) and a cdk4-associated immune complex kinase assay (lower panel) in M1-V1 and M1-G1 during the culture with rhIL-6. Total cellular lysates (15 μg per each lane) obtained from the cultured cells were subjected to SDS-PAGE and probed with rabbit anti-cyclin D1 monoclonal antibody. Immunoreactive proteins were visualized with the enhanced chemiluminescence detection system (upper panel). cdk4 was immunoprecipitated from equal amounts of the cell lysates prepared from the cultured cells. Immune complex kinase assay was performed in kinase buffer containing 5 μg of GST-Rb fusion protein and 20 μCi of γ-[32P]ATP for 30 minutes at 30°C. After the addition of a protein-loading buffer, samples were subjected to SDS-PAGE. The gels were stained with Coomassie blue, destained, dried, and subjected to autoradiography (lower panel). (C) Western blot analysis on bcl-2 expression in M1-V1 and M1-G1 during the culture with rhIL-6. Fifteen μg of total cellular lysates obtained from the cultured cells were subjected to SDS-PAGE and probed with rabbit anti-bcl-2 polyclonal antibody.
Changes in expression of cell cycle regulatory molecules and apoptosis-related genes during IL-6 treatment in M1-V1 and M1-G1.
(A) Northern blot analysis on expression of cell cycle regulatory molecules and apoptosis-related genes in M1-V1 and M1-G1 during the culture with recombinant human (rh) interleukin 6 (IL-6). M1-V1 and M1-G1 were cultured with 20 ng/mL of rhIL-6, and total cellular RNA was isolated at the time indicated. Fifteen μg of each sample was electrophoresed on formaldehyde agarose gels, blotted onto the filters, and hybridized with 32P-labeled probes as indicated. (B) Western blot analysis on cyclin D1 expression (upper panel) and a cdk4-associated immune complex kinase assay (lower panel) in M1-V1 and M1-G1 during the culture with rhIL-6. Total cellular lysates (15 μg per each lane) obtained from the cultured cells were subjected to SDS-PAGE and probed with rabbit anti-cyclin D1 monoclonal antibody. Immunoreactive proteins were visualized with the enhanced chemiluminescence detection system (upper panel). cdk4 was immunoprecipitated from equal amounts of the cell lysates prepared from the cultured cells. Immune complex kinase assay was performed in kinase buffer containing 5 μg of GST-Rb fusion protein and 20 μCi of γ-[32P]ATP for 30 minutes at 30°C. After the addition of a protein-loading buffer, samples were subjected to SDS-PAGE. The gels were stained with Coomassie blue, destained, dried, and subjected to autoradiography (lower panel). (C) Western blot analysis on bcl-2 expression in M1-V1 and M1-G1 during the culture with rhIL-6. Fifteen μg of total cellular lysates obtained from the cultured cells were subjected to SDS-PAGE and probed with rabbit anti-bcl-2 polyclonal antibody.
Sustained expression of cyclin D1 and bcl-2 proteins during IL-6 treatment in M1-G1 predominantly regulated at the transcriptional level
We next investigated the mechanisms by which expression of cyclin D1 and bcl-2 was regulated in M1-V1 and M1-G1. At first, we examined changes in expression levels of cyclin D1 and bcl-2 mRNA in the presence of RNA synthesis inhibitor, actinomycin D. M1-V1 and M1-G1 cells were pretreated with 10 μg/mL of actinomycin D for 2 hours and then subjected to the cultures with or without rhIL-6 in the presence of actinomycin D. As shown in Figure 6A, the kinetics of cyclin D1 and bcl-2 mRNA disappearance was not affected by rhIL-6 in M1-V1 and M1-G1, and a significant difference was not detected between M1-V1 and M1-G1 (Figure 6A). These results suggested that the stability of cyclin D1 and bcl-2 mRNA was almost the same in M1-V1 and M1-G1 and that rhIL-6 treatment shows little or no effect on their stability. Therefore, we speculated that sustained expression of cyclin D1 and bcl-2 mRNA in M1-G1 during rhIL-6 treatment may result from the continued transcription of these genes but not from the stabilization of these mRNAs. Next, we assessed the half-life of cyclin D1 and bcl-2 proteins in M1-V1 and M1-G1 during the culture with or without rhIL-6. M1-V1 and M1-G1 cells were pulsed with35S-methionin, and changes in expression levels of35S-labeled cyclin D1 and bcl-2 proteins were examined. Degradation of cyclin D1 was very rapid and was not affected by rhIL-6 in M1-V1 and M1-G1, and an apparent difference in the kinetics was not observed between M1-V1 and M1-G1 (Figure 6B). Also, bcl-2 protein was degraded in M1-V1 and M1-G1 in a similar time course regardless of the treatment with rhIL-6 (Figure 6C). These results suggested that the stability of cyclin D1 and bcl-2 proteins was almost identical in M1-V1 and M1-G1, respectively, and that rhIL-6 treatment hardly affects their stability in these clones. Thus, it was assumed that the continued expression of cyclin D1 and bcl-2 proteins was due to the sustained expression of their mRNA but not their stabilization.
Mechanisms of the sustained expression of cyclin D1 and bcl-2 in M1-G1 during the treatment with IL-6.
(A) Changes in expression levels of cyclin D1 and bcl-2 messenger RNA during the culture in the presence of actinomycin D. M1-V1 and M1-G1 cells were pretreated with 10 μg/mL of actinomycin D for 2 hours and then subjected to the cultures with or without recombinant human (rh) interleukin 6 (IL-6) in the presence of actinomycin D. Expression of cyclin D1 and bcl-2 was examined by Northern blot analysis at the time indicated. (B) Protein turnover of cyclin D1 and bcl-2 in M1-V1 and M1-G1 during the culture with or without rhIL-6. To examine the half-lives of cyclin D1 and bcl-2 proteins, the cells (1 × 107 cells for each sample) were radiolabeled with 35S]methionine for 30 minutes (for cyclin D1) or 6 hours (for bcl-2). Then, the cells were washed, resuspended in DMEM containing 2 mmol/L of unlabeled methionine and cultured with or without rhIL-6 for the time indicated. Cyclin D1 and bcl-2 were immunoprecipitated from total cellular lysates and subjected to SDS-PAGE. The gels were dried and subjected to the autoradiography. The amounts of the radioactivity of cyclin D1 and bcl-2 were measured by a densitometric analysis.
Mechanisms of the sustained expression of cyclin D1 and bcl-2 in M1-G1 during the treatment with IL-6.
(A) Changes in expression levels of cyclin D1 and bcl-2 messenger RNA during the culture in the presence of actinomycin D. M1-V1 and M1-G1 cells were pretreated with 10 μg/mL of actinomycin D for 2 hours and then subjected to the cultures with or without recombinant human (rh) interleukin 6 (IL-6) in the presence of actinomycin D. Expression of cyclin D1 and bcl-2 was examined by Northern blot analysis at the time indicated. (B) Protein turnover of cyclin D1 and bcl-2 in M1-V1 and M1-G1 during the culture with or without rhIL-6. To examine the half-lives of cyclin D1 and bcl-2 proteins, the cells (1 × 107 cells for each sample) were radiolabeled with 35S]methionine for 30 minutes (for cyclin D1) or 6 hours (for bcl-2). Then, the cells were washed, resuspended in DMEM containing 2 mmol/L of unlabeled methionine and cultured with or without rhIL-6 for the time indicated. Cyclin D1 and bcl-2 were immunoprecipitated from total cellular lysates and subjected to SDS-PAGE. The gels were dried and subjected to the autoradiography. The amounts of the radioactivity of cyclin D1 and bcl-2 were measured by a densitometric analysis.
Effects of cyclin D1 or bcl-2 overexpression or both on IL-6-induced macrophage differentiation and apoptosis
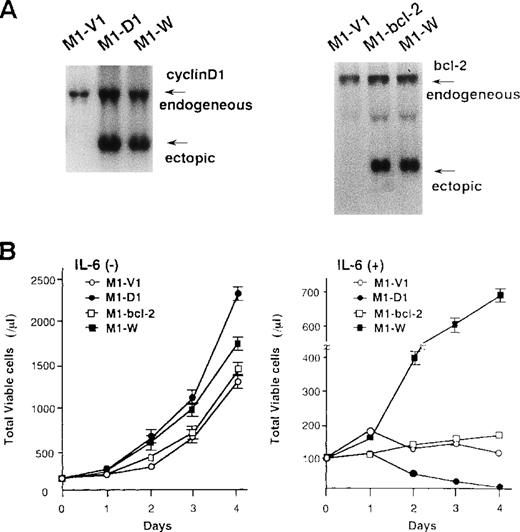
To examine the roles of cyclin D1 and bcl-2 in rhIL-6-induced differentiation and apoptosis, we introduced expression vectors of cyclin D1, bcl-2, or both into M1; M1 clones expressing cyclin D1, bcl-2, and both vectors were designated M1-D1, M1-bcl-2, and M1-W, respectively. As shown by Northern blot analysis (Figure7A), the expression of cyclin D1, bcl-2, or both transgenes was more abundant than that of endogenous genes in each transfectant. Furthermore, Western blot analysis showed more abundant expression of the transgene products in each transfectant (data not shown). After culture with or without rhIL-6, growth potential of each clone was assessed by counting viable cells (Figure 7B). In the absence of rhIL-6, all clones showed consistent growth, although M1-D1 and M1-W grew slightly faster than M1-V1 and M1-bcl-2 (Figure 7B, left panel). When rhIL-6 was added to the culture medium, however, viable cell number of M1-V1 or M1-bcl-2 did not increase and that of M1-D1 decreased significantly after 1-2 days (Figure 7B, right panel). By contrast, M1-W showed continuous cell growth for 4 days in the presence of rhIL-6, although its growth was rather slow as compared with that of M1-G1 (Figure 2A right panel vs Figure 7B right panel).
Preparation of M1 cells overexpressing cyclin D1 and/or bcl-2.
(A) Northern blot analysis on expression of cyclin D1, bcl-2, or both in each transfectant. Total cellular RNA was extracted from each clone, and Northern blot analysis was performed with 32P-labeled probe for cyclin D1 or bcl-2. (B) Changes in total viable cell number during the culture with (lower panel) or without (upper panel) recombinant human (rh) interleukin 6 (IL-6). M1-V1, M1-D1, M1-bcl-2, and M1-W cells were seeded at a cell density 100/μL and subjected to the culture in the absence or presence of rhIL-6 (20 ng/mL). Total viable cell number was counted by trypan blue dye exclusion method: M1-V1, open circle; M1-D1, closed circle; M1-bcl-2, open square; M1-W, closed square. The results are shown as the mean ± SD of triplicate cultures.
Preparation of M1 cells overexpressing cyclin D1 and/or bcl-2.
(A) Northern blot analysis on expression of cyclin D1, bcl-2, or both in each transfectant. Total cellular RNA was extracted from each clone, and Northern blot analysis was performed with 32P-labeled probe for cyclin D1 or bcl-2. (B) Changes in total viable cell number during the culture with (lower panel) or without (upper panel) recombinant human (rh) interleukin 6 (IL-6). M1-V1, M1-D1, M1-bcl-2, and M1-W cells were seeded at a cell density 100/μL and subjected to the culture in the absence or presence of rhIL-6 (20 ng/mL). Total viable cell number was counted by trypan blue dye exclusion method: M1-V1, open circle; M1-D1, closed circle; M1-bcl-2, open square; M1-W, closed square. The results are shown as the mean ± SD of triplicate cultures.
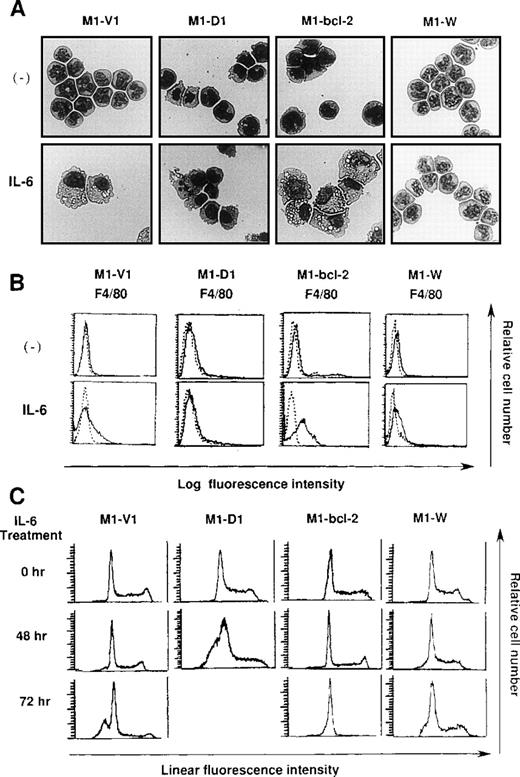
Morphological analysis demonstrated that most of the cells in these four clones had blastoid features before rhIL-6 treatment (Figure8A, upper panels). After 48-hours culture with rhIL-6, the morphological changes indicating macrophage differentiation were observed in M1-V1 and M1-bcl-2 but not in M1-D1 and M1-W (Figure 8A, lower panels). Moreover, a part of M1-D1, but not of M1-V1, M1-bcl-2, or M1-W, underwent apoptotic cell death after 48-hour culture with rhIL-6. Flow cytometric analysis also demonstrated that the rhIL-6 treatment for 48 hours led to up-regulation of F4/80 expression in M1-bcl-2 in the same way as in M1-V1, whereas its expression was scarcely induced in M1-D1 or M1-W (Figure 8B), indicating that IL-6-induced macrophage differentiation of M1 cells was inhibited by overexpression of cyclin D1. Moreover, cell cycle analysis of these clones revealed that, before the culture with rhIL-6, a substantial fraction of these clones were in cycling, whereas M1-D1 and M1-W showed slightly higher proportions of S and G2/M phase than M1-V1 and M1-bcl-2: % (S + G2/M) phase was as follows: M1-D1, 63% ; M1-W, 61%; M1-V1, 50% ; and M1-bcl-2, 51% (Figure 8C). After rhIL-6 treatment, a significant proportion (42%) of M1-D1 cells underwent apoptosis at 48 hours (Figure 8C), and ∼40% of M1-V1 yielded to apoptosis at 72 hours (Figure 4). In contrast, M1-bcl-2 showed cell cycle arrest and did not show apoptosis even after 72 hours of rhIL-6-treatment (Figure 8C). In the case of M1-W, a considerable proportion (39%) of cells were still in S and G2/M phase after 72hours of culture with rhIL-6; an apoptotic fraction (6%) at 72 hours was considerably lower than that in M1-V1 (Figure 8C and Figure 4). Therefore, overexpression of cyclin D1 together with bcl-2 was supposed to render M1 cells resistant to IL-6-induced cell cycle arrest and apoptosis.
Effects of overexpression of cyclin D1 and/or bcl-2 on IL-6-induced differentiation (A, B) and apoptosis (C) in each transfectant.
(A) Light micrograph of M1-V1, M1-D1, M1-bcl-2, and M1-W before and after 2-day culture with recombinant human (rh) interleukin 6 (IL-6). M1-V1, M1-D1, M1-bcl-2, and M1-W were cultured with 20 ng/mL of rhIL-6 for 2 days and cytocentrifugation preparation from each culture was stained with May-Gr ¸nwald-Giemsa (magnification × 100).(B) Flow cytometric analyses on expression of F4/80 before and after 2-day culture with rhIL-6. Expression of F4/80 was examined by staining with anti-F4/80 monoclonal antibody (—) with a reference to control antibody of the same isotype (---). (C) Cell cycle analysis on M1-V1, M1-D1, M1-bcl-2, and M1-W during the culture with rhIL-6. The cultured cells were subjected to propidium iodide staining, and DNA content was analyzed on FACSort. Cell cycle analysis was performed with Modfit LT2.0.
Effects of overexpression of cyclin D1 and/or bcl-2 on IL-6-induced differentiation (A, B) and apoptosis (C) in each transfectant.
(A) Light micrograph of M1-V1, M1-D1, M1-bcl-2, and M1-W before and after 2-day culture with recombinant human (rh) interleukin 6 (IL-6). M1-V1, M1-D1, M1-bcl-2, and M1-W were cultured with 20 ng/mL of rhIL-6 for 2 days and cytocentrifugation preparation from each culture was stained with May-Gr ¸nwald-Giemsa (magnification × 100).(B) Flow cytometric analyses on expression of F4/80 before and after 2-day culture with rhIL-6. Expression of F4/80 was examined by staining with anti-F4/80 monoclonal antibody (—) with a reference to control antibody of the same isotype (---). (C) Cell cycle analysis on M1-V1, M1-D1, M1-bcl-2, and M1-W during the culture with rhIL-6. The cultured cells were subjected to propidium iodide staining, and DNA content was analyzed on FACSort. Cell cycle analysis was performed with Modfit LT2.0.
Discussion
During the past decade, a number of growth factors have been identified, and their roles in hematopoiesis have been elucidated. In those studies, a single cytokine was shown to reveal pleiotropic effects dependently on the target cell types. However, it is not well understood as to which instinctive cell properties are associated with biological effects of each cytokine. By using a murine M1 myeloid cell line that is a suitable model for elucidating cytokine-induced cell differentiation accompanied by growth arrest, we investigated the effects of GATA-1 on IL-6-induced macrophage differentiation and apoptosis.
In accord with the previous findings,31 the overexpression of GATA-1 led to a change in the phenotype of M1 cells from myeloid to megakaryocytic lineage. Furthermore, we found that GATA-1 expression inhibited IL-6-induced macrophage differentiation and apoptosis. Because the STAT3 pathway was supposed to play key roles in regulating the IL-6-induced growth arrest and macrophage differentiation of M1 cells, we initially examined the effects of GATA-1 on tyrosine phosphorylation, DNA-binding, and transcriptional activation of STAT3. Western blot analysis, EMSA, and luciferase assays revealed that IL-6-induced STAT3 activities were not affected by overexpression of GATA-1, indicating that the STAT3 pathway was not involved in the suppressive effects of GATA-1 on the IL-6-induced macrophage differentiation and apoptosis of M1 cells.
Cell growth and differentiation are tightly regulated by a series of cell cycle regulatory molecules such as cyclins, cyclin-dependent kinases, and cyclin-dependent kinase inhibitors (for a review, see 40). Although GATA-1 did not significantly affect the growth of M1 cells in the absence of IL-6, IL-6-induced cell cycle arrest was apparently blocked by GATA-1. In agreement with the previous finding41 that IL-6 yielded cell cycle arrest through the induction of p19INK4D, expression of p19INK4D was found to be up-regulated by IL-6 in parental and vector-transfected M1 cells. Furthermore, we found that cyclin D1 expression was severely down-regulated by IL-6 in M1 cells, thereby leading to the decreased cdk4 activities. In contrast, GATA-1 overexpression led to a significantly lesser amount of p19INK4D induction and to sustained expression of cyclin D1 in M1-G1 cells after treatment with IL-6. Dubart et al42previously reported that NIH3T3 cells transfected with GATA-1 showed continuous growth even in a serum-deprived condition, whereas parental NIH3T3 cells exhibited growth arrest at G0/G1phase of the cell cycle. Moreover, the expression of cyclin D1, cyclin A, and cyclin B1 mRNA was retained under the serum-starved culture or the confluent condition in the GATA-1-transfected NIH3T3 cells but was severely down-regulated in parental NIH3T3 cells. In addition, Whyatt et al43 reported that overexpression of GATA-1 inhibits DMSO-induced erythroid differentiation in MEL cells. In their analyses, although cyclin D-dependent kinase activities were decreased in both control and GATA-1-transfected MEL cells, cyclin E-dependent kinase activities were retained in GATA-1-transfected MEL cells as compared with control cells. Furthermore, they reported that GATA-1 bound to tumor suppressor Rb protein in vitro.43Together, these lines of evidence including ours suggest that GATA-1 may positively contribute to cell cycle progression through the control of the expression and function of cell cycle regulating molecules.
A number of previous studies revealed that cell cycle arrest was closely related with and, in some cases, was sufficient for inducing terminal differentiation. It has been reported that expression of p21WAF1, p27Kip1, or p18INK4C is induced or up-regulated in muscle cells, keratinocytes, and B lymphoblastoid cells during the terminal differentiation. Furthermore, ectopic overexpression of p21WAF1, p18INK4C, and p19INK4D was shown to induce terminal differentiation of human CMK and UT7 megakaryoblastic leukemia cells, murine 32D-cl3 myeloid cells, and B-lymphoblastoid cells, respectively.37,44-46 Moreover, deregulation of cell cycle machinery by an inappropriate overexpression of positive cell cycle regulators, such as cyclin, cdk4, or E2F, was reported to result in differentiation arrest or apoptosis.47-49 In the case of M1 cells, IL-6-induced macrophage differentiation was found to accompany cell cycle arrest, possibly through the reduced expression of cyclin D1 and the induced expression of p19INK4D that led to decreased cdk4 activities. In contrast, GATA-1 overexpression resulted in the sustained expression of cyclin D1 and only modest induction of p19INK4D as well as abrogated IL-6-induced macrophage differentiation of M1 cells. Because the enforced expression of cyclin D1 could also significantly inhibit IL-6-induced macrophage differentiation of M1 (ie, M1-D1 and M1-W) cells, it was suggested that GATA-1 might inhibit IL-6-induced differentiation through the sustained expression of cyclin D1. However, our result seems to conflict with the previous report by Kato and Sherr,47 showing that overexpression of cyclin D2 or cyclin D3 but not of cyclin D1 inhibits G-CSF-induced granulocytic differentiation of 32D cells. In 32D cells, cyclin D2 and cyclin D3 were expressed during IL-3-induced proliferation, and only cyclin D3 was expressed during G-CSF-induced granulocytic differentiation, whereas cyclin D1 expression was not observed in both conditions. In contrast, M1 cells expressed all of the D-type cyclins in proliferating conditions, and only cyclin D1 was down-regulated during IL-6-induced differentiation, whereas expression of cyclin D2 and cyclin D3 was retained. It was, therefore, speculated that hematopoietic cells utilize D-type cyclins in various combinations to control cell growth and differentiation and also that D-type cyclins might exert distinct roles according to the cell types, the stage of differentiation, or both.
It has been reported that GATA-1-null ES cells fail to differentiate into mature erythrocytes and that GATA-1-/-proerythroblasts underwent apoptosis in a p53-independent manner.26 In addition, the reduced expression or function of GATA-1 or both was suggested to account, at least partially, for inducing apoptotic cell death in erythrocytic cell lines MEL and K562.50 51However, little information was available about how GATA-1 displayed anti-apoptotic function. In this study, IL-6-induced apoptosis was observed in control M1-V1 cells transfected with vector alone but not in GATA-1-transfected M1-G1 cells. During the culture with IL-6, the expression of bcl-2 mRNA was sustained in M1-G1, whereas its expression was severely reduced in M1-V1. In contrast, no apparent difference was seen between M1-V1 and M1-G1 cells in expression of other apoptosis-related genes, including bcl-xL, bax, and bak, suggesting that bcl-2 was involved in the anti-apoptotic effect of GATA-1. In fact, IL-6-induced apoptosis was completely canceled by the overexpression of bcl-2 as noted in M1-bcl-2. Therefore, it was suggested that GATA-1 may exert an anti-apoptotic effect through preventing down-regulation of bcl-2 in IL-6-treated M1 cells.
In summary, we demonstrated here that overexpression of GATA-1 in M1 cells resulted in phenotypic change from myeloid to megakaryocytic lineage and also in abrogation of IL-6-induced macrophage differentiation and apoptosis. These effects of GATA-1 appeared to be mediated, at least in part, by the sustained expression of cyclin D1 and bcl-2. These findings suggest that GATA-1 may affect cell fate and cytokine responses by regulating apoptosis and cell cycle machinery. Further studies will be necessary to identify the target molecule(s) of GATA-1 by using our experimental system, and precise information from such studies may add greatly to our understanding of hematopoietic stem cell biology.
Supported in part by grants from the Japanese Ministry of Education, Science, Sports and Culture; the Japanese Ministry of Health and Welfare; Senri Life Science Foundation; Uehara Memorial Foundation; Naito Foundation; and the Japan Medical Association.
Reprints:Itaru Matsumura, Department of Hematology and Oncology, Osaka University Medical School, 2-2, Yamada-oka, Suita, Osaka 565, Japan; e-mail: matumura@bldon.med.osaka-u.ac.jp.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Fig. 5. Changes in expression of cell cycle regulatory molecules and apoptosis-related genes during IL-6 treatment in M1-V1 and M1-G1. / (A) Northern blot analysis on expression of cell cycle regulatory molecules and apoptosis-related genes in M1-V1 and M1-G1 during the culture with recombinant human (rh) interleukin 6 (IL-6). M1-V1 and M1-G1 were cultured with 20 ng/mL of rhIL-6, and total cellular RNA was isolated at the time indicated. Fifteen μg of each sample was electrophoresed on formaldehyde agarose gels, blotted onto the filters, and hybridized with 32P-labeled probes as indicated. (B) Western blot analysis on cyclin D1 expression (upper panel) and a cdk4-associated immune complex kinase assay (lower panel) in M1-V1 and M1-G1 during the culture with rhIL-6. Total cellular lysates (15 μg per each lane) obtained from the cultured cells were subjected to SDS-PAGE and probed with rabbit anti-cyclin D1 monoclonal antibody. Immunoreactive proteins were visualized with the enhanced chemiluminescence detection system (upper panel). cdk4 was immunoprecipitated from equal amounts of the cell lysates prepared from the cultured cells. Immune complex kinase assay was performed in kinase buffer containing 5 μg of GST-Rb fusion protein and 20 μCi of γ-[32P]ATP for 30 minutes at 30°C. After the addition of a protein-loading buffer, samples were subjected to SDS-PAGE. The gels were stained with Coomassie blue, destained, dried, and subjected to autoradiography (lower panel). (C) Western blot analysis on bcl-2 expression in M1-V1 and M1-G1 during the culture with rhIL-6. Fifteen μg of total cellular lysates obtained from the cultured cells were subjected to SDS-PAGE and probed with rabbit anti-bcl-2 polyclonal antibody.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/4/10.1182_blood.v95.4.1264.004k09_1264_1273/6/m_bloo00409005w.jpeg?Expires=1769153557&Signature=hfedNuN2kG4oe6xmBZW7UkIhFUi0IH1qyjbuxAc1SCYwvxEra1IcaMCC74zKWACYzQYrwAe26tbPi01tc3EOwruZknomgB6jjSyuAZBSDB5PbrLjchDYEpvaelE4mCKrKZ5ljL6uHKKvBtJjtlvnVFmdqYzeXLpi79FM7UJmnGk4nlJkN8NdYvGR~wxTLC8tcpDPk~YSCjOo1Zz6Ib7zjxnAC5RmdV600PYl7lpohpUXEv-bZFA3-d5rAY5gewJDQUqzWykseYd9G2JZ2ZRWaV8un8konJg7mI37XKzf2NwmT~XPAxsga9s23y0CcTG-0Ahpscmv5a6YPfH~-u0Z7g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 6. Mechanisms of the sustained expression of cyclin D1 and bcl-2 in M1-G1 during the treatment with IL-6. / (A) Changes in expression levels of cyclin D1 and bcl-2 messenger RNA during the culture in the presence of actinomycin D. M1-V1 and M1-G1 cells were pretreated with 10 μg/mL of actinomycin D for 2 hours and then subjected to the cultures with or without recombinant human (rh) interleukin 6 (IL-6) in the presence of actinomycin D. Expression of cyclin D1 and bcl-2 was examined by Northern blot analysis at the time indicated. (B) Protein turnover of cyclin D1 and bcl-2 in M1-V1 and M1-G1 during the culture with or without rhIL-6. To examine the half-lives of cyclin D1 and bcl-2 proteins, the cells (1 × 107 cells for each sample) were radiolabeled with 35S]methionine for 30 minutes (for cyclin D1) or 6 hours (for bcl-2). Then, the cells were washed, resuspended in DMEM containing 2 mmol/L of unlabeled methionine and cultured with or without rhIL-6 for the time indicated. Cyclin D1 and bcl-2 were immunoprecipitated from total cellular lysates and subjected to SDS-PAGE. The gels were dried and subjected to the autoradiography. The amounts of the radioactivity of cyclin D1 and bcl-2 were measured by a densitometric analysis.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/4/10.1182_blood.v95.4.1264.004k09_1264_1273/6/m_bloo00409006w.jpeg?Expires=1769153557&Signature=D4NgQEdS~HgyBYmEjUhLtl0qbNtnzXiUwdCQDbkfwdhOack1wLzqUYNVTTwVa4GNa4g-iicm89ZH89EFOi~uhR-A70VfTWpF2aVeC0fAb~MJwcDb8jldvivSeN8CzgD2qYyiy1~Q0eW02ZVXyn49ikGmlywKfk5D~y1JEUvuA-dwtNQh9FX66QSo1fdyH92edmsV7VoJyZnmyMUsW~2ytDnmVOLhkrgbCxkLgW~S4oUUYTk2-NEYjxxab5UDSnnJ3oVIoWwyqt35PU63Di4KSbiW96iw~y-aQj2JJTUdUN22CH4c2oT82a-eqJJgWx3ZMYs7JRVbv~xyxyE2rare1g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal