The t(4;14) translocation occurs in 25% of multiple myeloma (MM) and results in both the ectopic expression of fibroblast growth factor receptor 3 (FGFR3) from der4 and immunoglobulin heavy chain-MMSET hybrid messenger RNA transcripts from der14. The subsequent selection of activating mutations of the translocated FGFR3 by MM cells indicates an important role for this signaling pathway in tumor development and progression. To investigate the mechanism by which FGFR3 overexpression promotes MM development, interleukin-6 (IL-6)-dependent murine B9 cells were transduced with retroviruses expressing functional wild-type or constitutively activated mutant FGFR3. Overexpression of mutant FGFR3 resulted in IL-6 independence, decreased apoptosis, and an enhanced proliferative response to IL-6. In the presence of ligand, wild-type FGFR3-expressing cells also exhibited enhanced proliferation and survival in comparison to controls. B9 clones expressing either wild-type FGFR3 at high levels or mutant FGFR3 displayed increased phosphorylation of STAT3 and higher levels of bcl-xL expression than did parental B9 cells after cytokine withdrawal. The mechanism of the enhanced cell responsiveness to IL-6 is unknown at this time, but does not appear to be mediated by the mitogen-activated protein kinases SAPK, p38, or ERK. These findings provide a rational explanation for the mechanism by which FGFR3 contributes to both the viability and propagation of the myeloma clone and provide a basis for the development of therapies targeting this pathway.
Multiple myeloma (MM) is a terminally differentiated and uniformly fatal B-cell malignancy that accounts for 10% of all hematopoietic neoplasms.1 The pleotropic cytokine interleukin-6 (IL-6) plays a central role in disease pathobiology as the major myeloma growth factor. Accumulated data suggest that IL-6 contributes to both the viability and proliferation of the malignant clone2-4 in which the mitogenic signal is probably transmitted through the mitogen-activated protein kinase (MAPK) pathway, particularly via ERK25 and the anti-apoptotic signal through the Jak-STAT pathway.6,7 Mechanisms by which IL-6 regulates myeloma cell survival include the up-regulation of an anti-apoptotic gene, bcl-xL8, and the down-regulation of SAPK in response to cell death stimuli.9Nevertheless, the mechanism by which IL-6 signaling becomes dysregulated in myeloma remains unknown.
In this regard we and others have previously reported that translocations into the immunoglobulin heavy (IgH) chain switch region are ubiquitous in MM cells.10-18 These translocations involve several heterogeneous translocation partners, includingc-myc,10MUM1/IRF4,11cyclin D1,12c-maf,13 andFGFR3/MMSET.14,15,18 These recurrent translocations have been identified with a similar high frequency in primary patient samples by reverse transcriptase (RT)-polymerase chain reaction,14 dual color interphase fluorescence in situ hybridization,17 and multicolor spectral karyotypes.16 They are present in patients with monoclonal gammopathy of undetermined significance (MGUS),14 and we hypothesize that these translocations are an early event in the development of MGUS or MM.
FGFR3 is a receptor tyrosine kinase normally expressed in cartilage, the central nervous system, and the brain. Mice lacking the wild-type gene develop an overgrowth of the long bones,19,20suggesting that this gene normally negatively regulates bone growth. In humans, activating mutations of FGFR3 cause various forms of dwarfism when the mutations arise within the germline.21-23Interestingly, some of the same activating mutations of FGFR3known to cause dwarfism have been found to be involved in the IgH translocations of MM. We have previously identified activating mutations of FGFR3 in 3 of 6 cell lines and in 1 of 3 primary patient samples.14 One identified mutation results in ligand-independent activation of the receptor and is known to cause thanatophoric dysplasia type 2 (TD II) when it arises in the germline.24
Currently, the consequences of translocated wild-type FGFR3 or constitutively active forms of FGFR3 on MM cells are unknown. We postulated that inappropriate expression of FGFR3 by illegitimate isotype switch recombination would enable myeloma cells to phosphorylate STATs as previously reported in chondrocytes.25,26 We also reasoned that one of the downstream targets of STATs was bcl-xL, as we have previously shown that IL-6, via gp130, causes bcl-xLup-regulation,8 and because leukemia inhibitory factor (LIF) signaling through gp130 up-regulates bcl-xL via STAT1 in cardiac myocytes.27 We report here the results of the investigation of this hypothesis.
Materials and methods
Retroviral vector construction
Two pcDNA3 plasmids containing the full-length human complementary DNA (cDNA) of wild-type or TD II mutant form of FGFR3 were obtained from Daniel. J. Donoghue (San Diego, CA). FGFR3 was released by restriction cutting with HindIII and SpeI, also removing the 3′ untranslated region. The recessed 3′ termini ends of the cDNA fragments were filled, and the fragments ligated into an HpaI/SnaBI-digested MINV retrovirus28 containing the neomycin-resistance gene as the selectable marker. The resulting vectors were termed MFINV-WT (wild-type FGFR3) and MFINV-TD (TD K650E mutant form of FGFR3). The parental MINV vector (neor only) was used as a negative vector control.
Retrovirus production
Twenty micrograms of the constructed retroviral vectors and the parental MINV (neor only) vector were electroporated into 1.5 × 106 PA317 amphotropic packaging cells.29 After 2 days of culture, the supernatant was collected, passed through a 0.45-μm filter, and overlaid on subconfluent monolayers of GP+E ecotropic packaging cells30 in the presence of 8 μg/mL of polybrene (Sigma, St. Louis, MO). On day 3 and 4 of culture, this process was repeated. Transduced GP+E cells were selected in fresh medium containing 1.2 mg/mL of G418 (Gibco, Grand Island, NY) over a period of 14 days. Vector supernatant was collected from GP+E producer cells 24 hours after the medium was changed to Iscove's Modified Dulbecco's Medium (IMDM) plus 5% fetal calf serum (FCS) (Gibco).
Generation of B9 lines expressing wild-type FGFR3 and FGFR3-TD mutant
Supernatant from the G418 selected GP+E producer lines was added to B9 cells along with 2% IL-6 conditioned media from the SP2/mIL-6 cell line31 and 8 μg/mL of polybrene (Sigma). After 2 days, fresh retroviral supernatant was added. After an additional 72 hours of incubation, B9 cells were selected in 2% IL-6 conditioned medium and 1.2 mg/mL G418 for 2 weeks, generating the cell lines B9-MINV (empty vector), B9-WT (wild-type FGFR3), and B9-TD (TD II mutant form of FGFR3). After selecting for stably transduced cells, a limiting cell dilution was performed to generate single-cell clones of B9-WT and B9-TD. Subsequent Western blot analysis used individual high- or low-expressing clones of B9-WT and B9-TD.
Tissue culture
Cells were cultured at 37°C in a humidified atmosphere containing 5% CO2. B9 cells were grown in IMDM supplemented with 5% FCS, 2% IL-6 conditioned medium, and penicillin-streptomycin (Gibco). KMS11, a human myeloma cell line containing an FGFR3 translocation, was kindly provided by Masayoshi Namba (Okayama, Japan). KMS11 was grown in RPMI Medium 1640 supplemented with 10% FCS and penicillin-streptomycin. U266, a human myeloma cell line, was grown in IMDM with 10% FCS. GP+E and PA317 cells were grown in Dulbecco modified Eagle medium supplemented with 10% FCS and penicillin-streptomycin.
Cell viability and proliferation
To examine cell viability and proliferation, B9, B9-MINV, B9-WT, and B9-TD were washed 4 times in IMDM without FCS and then plated in triplicate in a 96-well plate at a density of 1 × 104 cells/well in varying concentrations of IL-6 and cultured for 48-96 hours. Total cell number and percent viability were determined every 24 hours by trypan blue staining and enumeration with a hemocytometer. For proliferation assays, 0.5 μCi of 3H-thymidine (Amersham, Arlington Heights, IL) was added to each well in the last 18 hours of the assay. Cells were harvested onto glass filter paper (Gelman Science, Ann Arbor, MI), and3H-thymidine counts determined by liquid scintillation.
Ligand stimulated proliferation assay
B9-WT cells were plated at a concentration of 2 × 105 cells/mL in a 96-well plate in the presence of increasing concentrations of acidic fibroblast growth factor (aFGF) (R&D Labs, Minneapolis, MN) and heparin (Sigma). Cells were subsequently incubated for 2 or 3 days and then analyzed with the use of the chromogenic dye 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Boehringer Mannheim, Mannheim, Germany). Absorbance at 570 nm-650 nm was read on an enzyme-linked immunosorbent assay plate reader (Molecular Devices, Sunnyvale, CA). After optimal concentrations of aFGF (40 ng/mL) and heparin (30 μg/mL) for the support of cell proliferation were determined, B9, B9-MINV, B9-WT, and B9-TD were washed four times in IMDM lacking FCS and plated in triplicate at a concentration of 2 × 105 cells/mL in 96-well plates. Each cell line was analyzed under 4 different conditions: 40 ng/mL aFGF and 30 μg/mL of heparin, 40 ng/mL aFGF and 30 μg/mL of heparin plus 2% IL-6, 2% IL-6 without ligand, and 0% IL-6 without ligand. Cells were incubated for 48-72 hours and then analyzed by MTT.
Cell cycle and apoptosis analysis
Cell lines were washed and then plated in IMDM plus 5% FCS at a density of 0.5 × 106 in 0% IL-6 or 0.1 × 106 in the presence of IL-6. Cells were cultured for 48 or 72 hours. After harvesting the cells, 500 μL of a hypotonic fluorochrome solution (0.1% sodium citrate, 0.1% Triton X-100, 50 μg/mL of propidium iodide) was added to each cell pellet. The samples were incubated overnight in the dark at 4°C. Flow cytometry was performed on a FACScan (Becton Dickinson, Meylan, France) the following day, using Cell Quest software to acquire data and ModFit LT software for analysis. Tunel assays (Promega, Madison, WI) were performed according to the manufacturer's protocol. In brief, 4 × 106 cells were fixed with 1% paraformaldehyde for 20 minutes on ice. Cells were incubated at 37°C for 1 hour with terminal deoxynucleotidyl transferase (TdT) enzyme and nucleotide mix containing fluorescein-12-dUTP to end label the 3′OH end of the fragmented DNA. The reaction was ceased by addition of 20 mM ethylenediaminetetraacetic acid, and cells were washed twice in 0.1% Triton X-100 and stained with propidium iodide for 30 minutes. Analysis was performed by flow cytometry, and apoptotic cells were fluorescein-12-dUTP positive.
Western blots
Western blot analysis was carried out according to previously published procedures.32 Membranes were probed with antibodies to the C-terminus end of FGFR3, to murine bcl-xS/L, to murine bcl-2, to murine bax, anti-phosphotyrosine (all Santa Cruz Biotechnology, Santa Cruz, CA), anti-phospho-STAT1 (Upstate Biotechnology, Lake Placid, NY), anti-STAT3 and anti-STAT1 (Transduction Laboratories, Lexington, KY), or with polyclonal rabbit anti-phospho-STAT1/STAT5, anti-phospho-STAT5, anti-phospho-STAT3 (all 3 kindly provided by David A. Frank, Dana-Farber Cancer Institute, Boston, MA), and anti-STAT5 (kindly provided by Dr J. N. Ihle, Memphis, IN). Goat antirabbit IgG horseradish peroxidase (PharMingen, Mississauga, ON) or sheep antimouse IgG horseradish peroxidase (Amersham, Arlington Heights, IL) were used as secondary antibodies, and blots were developed by enhanced chemiluminescence (Amersham) according to manufacturer's instructions.
Electrophoretic mobility shift assay (EMSA)
EMSA was carried out as previously described.33 After generation of nuclear lysates from cells stimulated with and without IL-6, the gel shift assays were performed with the use of double-stranded 32P-labeled oligonucleotide derived from the IRF-1 promoter (CTGATTTCCCCGAAATGAC).34 Five micrograms of nuclear lysates was incubated with 0.25 ng of32P-labeled oligonucleotide in 20 μL of binding buffer,13 mmol/L HEPES (pH 7.9; 65 mmol/L NaCl, 1 mmol/L DTT, 15 mmol/L ethylenediaminetetraacetic acid, 8% glycerol, 50 μg of poly (dI-dC)/mL) for 15 minutes at 4°C. For competition experiments, 50-fold molar excess of either unlabeled IRF-1 or an oligonucleotide derived from T14 promoter (TTCTTGAGGTAATGAAAGCC)35 was added to the binding reaction. For supershifting experiments, a peptide-specific STAT3 (Zymed, San Francisco, CA) was added at the completion of the binding reaction and incubated for an additional 30 minutes at 4°C.
Immunoprecipitation and in vitro kinase assay
Immunoprecipitation and in vitro kinase assay were carried out as previously described.32 Twenty-five microliters/reaction of 20% (v/v) Protein-A Sepharose (Pharmacia, Uppsala, Sweden) was incubated with 12 μL of anti-FGFR3, anti-ERK1 (recognizes ERK1 and ERK2), anti-p38, or anti-JNK1 (SAPK) (all from Santa Cruz Biotechnology) overnight at 4°C; 1250 μg of total cell lysate was incubated with the bead complex for 2 hours at 4°C. Immunoprecipitates were split into equal aliquots. One aliquot was washed 4 times with kinase buffer and subjected to in vitro kinase assay. The other aliquot was subjected to standard Western blotting procedure, probing the membranes with anti-FGFR3, ERK1, p38, or JNK1 (SAPK).
Results
Human FGFR3 is functional in transduced B9 cells
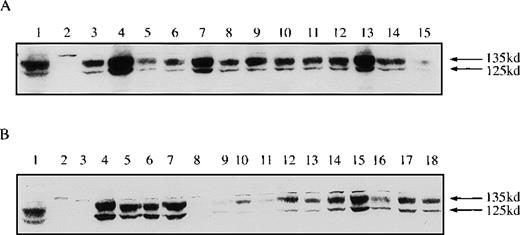
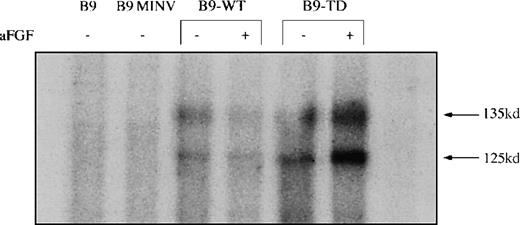
Western blots confirmed varying levels of expression of FGFR3 in the transduced B9-WT (Figure 1A) and B9-TD (Figure 1B) lines and subclones. An in vitro kinase assay demonstrated that both wild-type and FGFR3-TD lines were capable of autophosphorylation (Figure2). Phosphotyrosine Western blots further demonstrated that FGFR3 was phosphorylated only in the absence of ligand within the B9-TD line, thus indicating that the mutant receptor possessed constitutive tyrosine kinase activity. On addition of ligand, aFGF, and heparin, phosphorylation of wild-type FGFR3 was induced in B9-WT (not shown).
Expression of fibroblast growth factor receptor 3 (FGFR3) is achieved at variable levels in B9-WT and B9-TD cells.
(A) Western blot analysis, probing with anti-FGFR3, shows the expression of FGFR3 in B9 clones: KMS11 myeloma cell line positive control for FGFR3 (lane 1), parental B9 (lane 2), and B9-WT subclones (lanes 3-15). Lane 4 (clone #3) and lane 7 (clone #6) represent clones selected for further study. (B) KMS11 positive control for FGFR3 (lane 1), U266 myeloma cell line negative control (lane 2), parental B9 (lane 3), and B9-TD subclones (lanes 4-18). Lane 17 represents clone #14 selected for further study.
Expression of fibroblast growth factor receptor 3 (FGFR3) is achieved at variable levels in B9-WT and B9-TD cells.
(A) Western blot analysis, probing with anti-FGFR3, shows the expression of FGFR3 in B9 clones: KMS11 myeloma cell line positive control for FGFR3 (lane 1), parental B9 (lane 2), and B9-WT subclones (lanes 3-15). Lane 4 (clone #3) and lane 7 (clone #6) represent clones selected for further study. (B) KMS11 positive control for FGFR3 (lane 1), U266 myeloma cell line negative control (lane 2), parental B9 (lane 3), and B9-TD subclones (lanes 4-18). Lane 17 represents clone #14 selected for further study.
Fibroblast growth factor receptor 3 (FGFR3) is functional in B9-WT and B9-TD Cells.
Parental B9, B9 MINV, B9-WT, and B9-TD cells were subjected to an in vitro kinase assay with the use of anti-FGFR3 to initially immunoprecipitate FGFR3. Presence or absence of the ligand acidic fibroblast growth factor is indicated. Autophosphorylation of FGFR3 is apparent in B9-WT and B9-TD cells, demonstrating functionality of the expressed protein.
Fibroblast growth factor receptor 3 (FGFR3) is functional in B9-WT and B9-TD Cells.
Parental B9, B9 MINV, B9-WT, and B9-TD cells were subjected to an in vitro kinase assay with the use of anti-FGFR3 to initially immunoprecipitate FGFR3. Presence or absence of the ligand acidic fibroblast growth factor is indicated. Autophosphorylation of FGFR3 is apparent in B9-WT and B9-TD cells, demonstrating functionality of the expressed protein.
FGFR3 overexpression can substitute for IL-6 receptor signaling
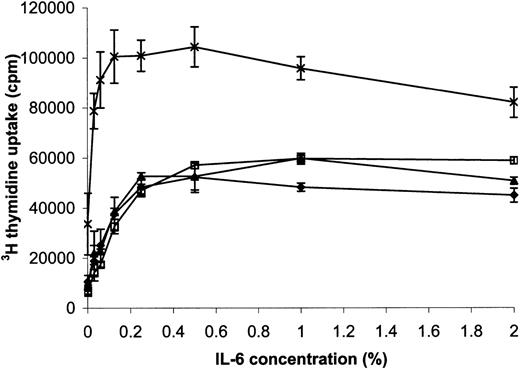
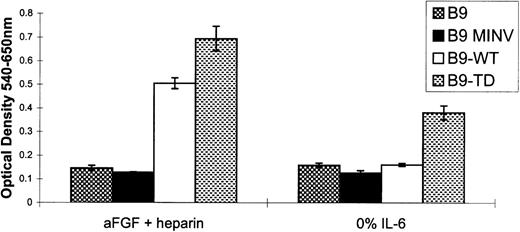
We first asked if overexpression of FGFR3 would influence IL-6– induced proliferation of B9 cells. Parental B9, B9-MINV, and pooled B9-WT cells had a similar rate of proliferation at all concentrations of IL-6 as assessed by 3H-thymidine incorporation. However, the proliferative rate of pooled B9-TD cells expressing mutant FGFR3 was higher than that for the other 3 cell lines at all concentrations of IL-6 (Figure 3). Two days after withdrawal of IL-6, B9, B9-MINV, B9-WT, and B9-TD, cells were 30%, 16%, 44%, and 75% viable, respectively. After 4 days of cytokine starvation, there were 6 times more viable cells in B9-TD culture than for the other lines, and, unlike the parental B9 cell line, B9-TD cells could be maintained indefinitely in the absence of IL-6. Thus, activated FGFR3 enhanced cell proliferation in response to IL-6 and could substitute for the absolute dependence of B9 cells on IL-6. Because many myelomas do not immediately acquire FGFR3 mutation, we next examined the response of cells expressing the wild-type or mutant protein to the addition of the aFGF ligand. On ligand stimulation of IL-6–starved cells, B9-WT proliferation was greater than that seen for parental B9 and B9-MINV cells, and the already increased proliferation of B9-TD cells was further enhanced (Figure4). Some B9-WT clones expressing the highest levels of FGFR3 (eg, clone #3 and clone #6) also exhibited IL-6 independence. These data together demonstrate an enhanced responsiveness of FGFR3-expressing cells to IL-6 stimulation and the acquisition of IL-6 independence proportional to the degree of activation of FGFR3.
Cells expressing activated fibroblast growth factor receptor 3 (FGFR3) have an enhanced proliferative rate in the presence and absence of interleukin-6 (IL-6).
Parental B9 (♦), B9-MINV (□), pooled B9-WT (▴), and B9-TD (×) cell lines were incubated in the presence of increasing concentrations of IL-6, and a 3H-thymidine proliferation assay was performed. Pooled B9-TD cells exhibit enhanced proliferation at all concentrations of IL-6.
Cells expressing activated fibroblast growth factor receptor 3 (FGFR3) have an enhanced proliferative rate in the presence and absence of interleukin-6 (IL-6).
Parental B9 (♦), B9-MINV (□), pooled B9-WT (▴), and B9-TD (×) cell lines were incubated in the presence of increasing concentrations of IL-6, and a 3H-thymidine proliferation assay was performed. Pooled B9-TD cells exhibit enhanced proliferation at all concentrations of IL-6.
Ligand-stimulated cells expressing wild-type fibroblast growth factor receptor 3 (FGFR3) proliferate in the absence of interleukin-6 (IL-6).
A chromogenic dye 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was performed on IL-6–starved FGFR3-expressing cell lines and controls in the presence or absence of ligand (acidic fibroblast growth factor and heparin). Pooled B9-WT cells exhibit enhanced proliferation in comparison to cytokine-starved controls only in the presence of ligand. Pooled B9-TD cells demonstrate increased proliferation in the absence of IL-6 and further enhancement of proliferation in response to ligand.
Ligand-stimulated cells expressing wild-type fibroblast growth factor receptor 3 (FGFR3) proliferate in the absence of interleukin-6 (IL-6).
A chromogenic dye 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was performed on IL-6–starved FGFR3-expressing cell lines and controls in the presence or absence of ligand (acidic fibroblast growth factor and heparin). Pooled B9-WT cells exhibit enhanced proliferation in comparison to cytokine-starved controls only in the presence of ligand. Pooled B9-TD cells demonstrate increased proliferation in the absence of IL-6 and further enhancement of proliferation in response to ligand.
Signaling by FGFR3 decreases apoptosis in the absence of IL-6
The mechanism of enhanced survival of B9-TD cells in the absence of IL-6 and ligand was next examined. There was little difference in the cell-cycle status of B9, B9-WT, and B9-TD in the presence or absence of IL-6. It was noted, however, that B9-TD cells had fewer sub-G0 apoptotic cells in the absence of IL-6 and ligand than did either controls or B9-WT. Indeed, with the use of a Tunel assay, the vast majority of parental B9 cells were apoptotic 5 days after withdrawal of IL-6, whereas only 53% of pooled B9-TD cells were undergoing apoptosis (Table 1). B9-MINV and pooled B9-WT had between 85% and 88% apoptotic cells in the absence of IL-6 and ligand. These data demonstrate that activated FGFR3 prevents apoptosis of IL-6–dependent cells in the absence of IL-6.
FGFR3 signaling induces phosphorylation of STAT3
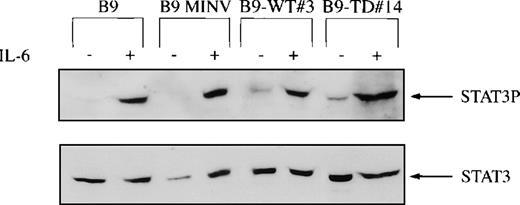
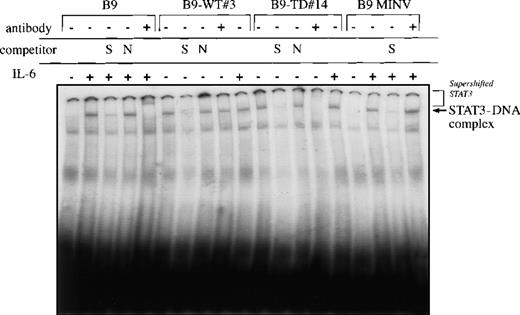
Because the STATs are a key mediator of cytokine signaling, Western blot analysis was performed with the use of antibodies that bind to tyrosine phosphorylated STAT1, STAT3, or STAT5. STAT5 was phosphorylated in all cell lines independent of the presence or absence of IL-6 and was not further activated by FGFR3. STAT1 and STAT3 were phosphorylated in all of the cell lines after IL-6 stimulation. However, STAT3 was also constitutively phosphorylated (in the absence of IL-6) in both B9-WT clones expressing high levels of FGFR3 and in B9-TD cells (Figure 5). To confirm that STAT3 was constitutively activated in cells expressing high levels of wild-type FGFR3 or mutant FGFR3, EMSAs were performed. In the absence of IL-6 stimulation, both B9-WT and B9-TD cells displayed a protein-DNA complex that was only present in parental B9 cells in the presence of IL-6 (Figure 6). This complex could be competed with an excess of unlabeled IRF-1 but was not affected by a molar excess of nonspecific oligonucleotide. It was demonstrated that the complex contained STAT3 because a peptide-specific STAT3 antibody could supershift the complex in B9-TD cells.
STAT3 phosphorylation in fibroblast growth factor receptor 3 (FGFR3)-expressing cells.
FGFR3-expressing and control cell lines were depleted of cytokine and then stimulated with or without interleukin-6 for 10 minutes at 37°C. B9-WT clones selected for high expression of FGFR3 were used. Total cell lysates were resolved via SDS-PAGE, and the membrane was probed with a tyrosine phospho-specific STAT3 antibody (upper panel). The membrane was reprobed for loading equivalence with a peptide-specific antibody that recognized total STAT3 (lower panel).
STAT3 phosphorylation in fibroblast growth factor receptor 3 (FGFR3)-expressing cells.
FGFR3-expressing and control cell lines were depleted of cytokine and then stimulated with or without interleukin-6 for 10 minutes at 37°C. B9-WT clones selected for high expression of FGFR3 were used. Total cell lysates were resolved via SDS-PAGE, and the membrane was probed with a tyrosine phospho-specific STAT3 antibody (upper panel). The membrane was reprobed for loading equivalence with a peptide-specific antibody that recognized total STAT3 (lower panel).
STAT3 DNA binding is constitutively activated in B9-WT and B9-TD cells.
Nuclear extracts were prepared from B9, B9 MINV, B9-WT#3, and B9-TD#14 stimulated with and without interleukin-6 for 10 minutes at 37°C. Complexes were resolved on a 5% native polyacrylamide gel. The specificity of DNA binding was determined by the addition of unlabeled IRF-1 oligonucleotide (S) or a nonspecific oligonucleotide from the T14 promoter (N) and by incubation with a peptide-specific STAT3 antibody. (The absence of supershifting in B9-MINV and B9-WT cells is likely technical in nature.)
STAT3 DNA binding is constitutively activated in B9-WT and B9-TD cells.
Nuclear extracts were prepared from B9, B9 MINV, B9-WT#3, and B9-TD#14 stimulated with and without interleukin-6 for 10 minutes at 37°C. Complexes were resolved on a 5% native polyacrylamide gel. The specificity of DNA binding was determined by the addition of unlabeled IRF-1 oligonucleotide (S) or a nonspecific oligonucleotide from the T14 promoter (N) and by incubation with a peptide-specific STAT3 antibody. (The absence of supershifting in B9-MINV and B9-WT cells is likely technical in nature.)
FGFR3 survival signal is mediated by up-regulation of bcl-xL
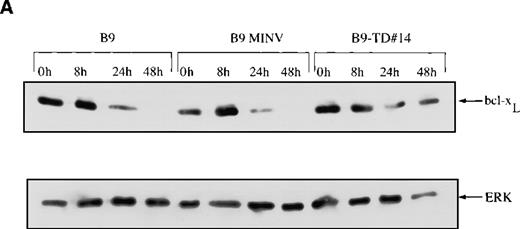
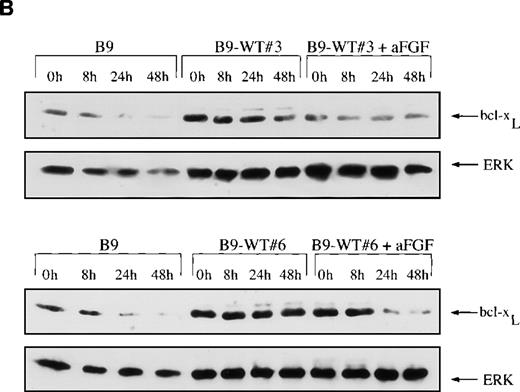
Because we have previously shown that the IL-6 survival signal in myeloma is mediated by up-regulation of bcl-xL,8 we next examined the expression of this protein by Western blots and demonstrated that the baseline level of bcl-xL is higher in B9-TD cells expressing constitutively activated FGFR3 than in B9 controls (Figure7A). With the withdrawal of IL-6 from B9 or B9-MINV control cells, there is a decrease in the amount of bcl-xL protein over 48 hours. In contrast, the expression of bcl-xL in B9-TD clones remained relatively constant after the withdrawal of IL-6 from the culture medium. Some B9-WT clones expressing high levels of FGFR3 also exhibit a marked increase in baseline expression of bcl-xL (Figure 7B). Of interest, the addition of ligand actually decreases bcl-xL expression in B9-WT clones, perhaps because of down-regulation of the receptor.
Expression of bcl-XL protein is elevated in fibroblast growth factor receptor 3 (FGFR3)-overexpressing cells.
Total cell lysates were prepared at varying time points from FGFR3-expressing clones or from control cells grown in the absence of interleukin-6 (IL-6) over a 48-hour period. The lysates were resolved via SDS-PAGE, and the membrane was probed with an anti–bcl-xS/L antibody. Membranes were reprobed with ERK as a protein-loading control. (A) bcl-xL is overexpressed at baseline in cells expressing mutant FGFR3 (B9-TD#14) and remains high after IL-6 withdrawal (top panel). (B) bcl-xL is also overexpressed at baseline in cells expressing high levels of wild-type FGFR3 (B9-WT#3 and B9-WT#6) and remains high after IL-6 withdrawal (first and third panel). With the addition of fibroblast growth factor ligand, down-regulation of bcl-xL is noted.
Expression of bcl-XL protein is elevated in fibroblast growth factor receptor 3 (FGFR3)-overexpressing cells.
Total cell lysates were prepared at varying time points from FGFR3-expressing clones or from control cells grown in the absence of interleukin-6 (IL-6) over a 48-hour period. The lysates were resolved via SDS-PAGE, and the membrane was probed with an anti–bcl-xS/L antibody. Membranes were reprobed with ERK as a protein-loading control. (A) bcl-xL is overexpressed at baseline in cells expressing mutant FGFR3 (B9-TD#14) and remains high after IL-6 withdrawal (top panel). (B) bcl-xL is also overexpressed at baseline in cells expressing high levels of wild-type FGFR3 (B9-WT#3 and B9-WT#6) and remains high after IL-6 withdrawal (first and third panel). With the addition of fibroblast growth factor ligand, down-regulation of bcl-xL is noted.
Western blots were also used to examine the expression levels of bcl-xS, bax, and bcl-2. There was no change in the expression of bax because of the presence of FGFR3 either in the presence or absence of IL-6. Levels remained similar to those expressed by parental B9 cells. Expression of bcl-2 and bcl-xS was not detected in B9 cells or in the FGFR3-expressing derivatives (data not shown).
Enhanced proliferative response to IL-6 is not mediated by MAP kinases
Although overexpression of bcl-xL may explain the enhanced survival of B9 cells overexpressing FGFR3, it does not explain the enhanced proliferative rate of the transduced cell lines in response to IL-6. Because cytokines not only activate the JAK-STAT signaling pathway but also function to phosphorylate members of the MAPK family, we next examined ERK-, p38-, and SAPK-induced activation. Although p38 was induced by IL-6, no change in SAPK, ERK1, ERK2, or p38 activity was noted in the FGFR3-expressing cell lines (not shown). Thus, the mechanism of enhanced IL-6 responsiveness in FGFR3-expressing cells remains unknown.
Discussion
The t(4;14) translocation occurs in approximately 25% of MM cell lines and patient samples and results in the ectopic expression ofFGFR3 from the der1414 and IgH-MMSET hybrid messenger RNA transcripts from the der4.18 Furthermore, activating mutations of the translocated FGFR3 have been identified in some myeloma samples, indicating an important role for the FGFR3 signaling pathway in these cases.14 One such activating mutant of FGFR3 is known to cause TD II, a severe form of human dwarfism, when the mutation arises in the germline.21,22 Prior investigation of the role of FGFR3in dwarfism has demonstrated that such mutations result in constitutive activation of the receptor independent of ligand.23,24 This activation leads to phosphorylation and translocation of STAT1 to the nucleus followed by up-regulation of the cell cycle regulator p21, resulting in growth arrest in chondrocytes.25 In a related TD I mutation ofFGFR3, STAT1 is also activated, along with the MAPK pathway, with a resulting increase in apoptosis secondary to increased levels of bax and decreased bcl-2.26 The net effect of these mutations in human chondrocytes is, therefore, growth arrest and cell apoptosis, explaining the chondrocyte growth arrest viewed clinically. Clearly, such changes would not explain why activation of FGFR3 promotes the growth or development of MM cells.
Clues to the latter come from the observation that signaling through gp130 by LIF in cardiac myocytes up-regulates bcl-xL via STAT1.27 Because the myeloma-promoting cytokine IL-6 also signals through gp130 to up-regulate bcl-xL in myeloma cells8 and because FGFR3 phosphorylates STAT1,25,26 we hypothesized that FGFR3 could enable cells to bypass gp130 signaling by constitutively up-regulating STATs and thus bcl-xL in myeloma cells. To examine the consequences of FGFR3 overexpression, IL-6–dependent murine B9 myeloma cells were engineered to express either wild-type human FGFR3 or FGFR3-TD containing the K650E-activating mutation. We observed an increased level of responsiveness to IL-6 in the resultant cell lines that correlated with the expression level of FGFR3. Indeed, some cells expressing the constitutively active FGFR3-TD or high levels of wild-type FGFR3 exhibited complete independence from IL-6. Consistent with these results, others have shown that BaF3 cells overexpressing FGFR3 also exhibit sustained proliferation in the absence of ligand,23 suggesting that, in cytokine-dependent hematopoietic cells, the net effect of FGFR3 overexpression is to promote rather than to inhibit cell growth.
The concept that FGFR3 can produce seemingly opposing signals in chondrocytes and cytokine-dependent BaF3 cells (or as shown here in IL-6–dependent B9 cells) has precedence, as cytokine signaling via gp130 may also simultaneously induce contradictory intracellular signaling pathways.36 The central orchestrator of this gp130-signaling dichotomy appears to be STAT3 in that phosphorylation of STAT3 in response to IL-6 up-regulates cyclins and down-regulates p21, resulting in cell cycle transition.36 In contrast, when STAT3 is suppressed, signaling via gp130 can induce up-regulation of p21 and growth arrest, as viewed in human dwarfism.25
We initially hypothesized that the effect of FGFR3 in conferring IL-6 independence occurred via enhanced phosphorylation of STAT1 as previously observed in chondrocytes; however, we found that expression of FGFR3 in B9 cells did not alter the phosphorylation of STAT1 but did constitutively phosphorylate STAT3. Because STAT3 is implicated in gp130 signaling6 and because gp130 signaling also up-regulates bcl-xL,8,37 38 we next examined bcl-xL expression. Indeed, B9-TD clones and clones expressing high levels of wild-type FGFR3 also express a relatively constant amount of bcl-xL protein regardless of whether IL-6 is present or absent, and this baseline level is higher than that expressed by parental B9 cells.
It has previously been reported that the activating TD II mutation of FGFR3 results in a level of kinase activity of the receptor higher than can be achieved by maximally stimulating wild-type FGFR3 with ligand.24 From this observation, we theorize that the wild-type receptor would activate the same pathways as FGFR3-TD, but the magnitude of activation is expected to be less. This theory would appear to be true with our cell lines because there is a greater proliferative effect of expressing mutant FGFR3 than when the wild-type receptor is maximally stimulated with aFGF. Indeed, a dose effect of FGFR3 was observed for STAT3 phosphorylation and for IL-6 independence in high- and low-expressing FGFR3 clones.
Whereas phosphorylation of STAT3 and resulting overexpression of bcl-xL likely explains the reduction in apoptosis of FGFR3-expressing cells after IL-6 withdrawal, it does not account for the enhanced proliferative response to IL-6 that was observed. In this regard, activated FGFR3-expressing cells exhibit an enhanced proliferative response to IL-6 at all concentrations of cytokine in comparison with controls, and cells expressing the wild-type protein demonstrate an enhanced proliferative response when ligand is present. Cells expressing high levels of wild-type FGFR3 may also show growth in the complete absence of IL-6. Although ERKs are thought to be involved in the enhanced proliferative response of IL-6–stimulated myeloma cells6,7 and FGFR3 has been shown to be capable of phosphorylating ERK1 and ERK2 in other systems,26,39 we were unable to demonstrate enhanced ERK phosphorylation secondary to FGFR3 overexpression. Similarly, activity of the other MAP kinase family members SAPK9 and p3840 was not altered by FGFR3 overexpression. The mechanism of cytokine-enhanced proliferation induced by FGFR3, therefore, remains unexplained.
Around 25% of myelomas translocate the FGFR3 gene. This fact alone may confer a survival advantage to B cells, as cells overexpressing FGFR3 proliferate at a higher rate than do controls in the presence of ligand. It can be further hypothesized that selective pressures subsequently result in the acquisition of activating FGFR3 mutations. A report by Fracchiolla et al41suggests that FGFR3 mutations occur rarely in MM. However, the authors chose to analyze DNA rather than cDNA by SSCP, which is likely to increase the background and decrease the sensitivity of this assay. Furthermore, DNA analysis for detection of mutations would be more sensitive if tumor cells were selected or if only samples containing at least 30% tumor cells were analyzed. Because serial dilutions of DNA from a cell line known to have an FGFR3 mutation, such as KMS11, and of DNA from a cell line with nonmutated FGFR3 were not performed to determine the minimal concentration of tumor DNA required to detected the abnormal migrating band by SSCP, the true incidence of FGFR3 mutation remains unknown.
In conclusion, FGFR3-overexpressing myeloma cells proliferate in the absence of IL-6, exhibit an enhanced proliferative response in the presence of IL-6, and exhibit decreased apoptosis on IL-6 withdrawal. Cells overexpressing FGFR3 phosphorylate STAT3 and up-regulate bcl-xL expression, thus inhibiting apoptosis following cytokine withdrawal. The mechanism of FGFR3-enhanced proliferation is not yet elucidated.
We speculate that an initial translocation of FGFR3 into the IgH chain switch region in a germinal center B cell likely provides the affected B cells with enhanced growth potential, with signaling occurring through both the IL-6 receptor and FGFR3. Further, activating mutations of FGFR3 would enable the cells to entirely escape the need for ligand stimulation, whether by IL-6 or aFGF, and would result in enhanced proliferation and a competitive advantage over other B cells. These findings provide a plausible, albeit partial, explanation for the myeloma-promoting properties of FGFR3 and provide a basis for the development of therapies targeting this pathway.
Acknowledgments
We are grateful to Dr Brent Zanke and Dr Ian Dubé for advice, as well as to Nathan Faliconi, Fawzy Saad, Darrin Cappe, Meenakshi Bali, and Christine Dodgson for technical assistance.
Supported by funds from MRC of Canada and the Nelson Arthur Hyland Foundation.
Reprints:A. Keith Stewart, Department of Medical Hematology-Oncology, 5th Floor Room 126, The Princess Margaret Hospital, 610 University Ave, Toronto, Ontario M5G 2M9; e-mail:k.stewart@utoronto.ca.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal