Feline leukemia virus-C (FeLV-C) causes red cell aplasia in cats, likely through its interaction with its cell surface receptor. We identified this receptor by the functional screening of a library of complementary DNAs (cDNA) from feline T cells. The library, which was cloned into a retroviral vector, was introduced into FeLV-C–resistant murine (NIH 3T3) cells. The gene conferring susceptibility to FeLV-C was isolated and reintroduced into the same cell type, as well as into FeLV-C–resistant rat (NRK 52E) cells, to verify its role in viral infection. The receptor cDNA is predicted to encode a protein of 560 amino acids with 12 membrane-spanning domains, termed FLVCR. FLVCR has significant amino acid sequence homology with members of the major facilitator superfamily and especially D-glucarate transporters described in bacteria and in C. elegans. As FeLV-C impairs the in vivo differentiation of burst-forming unit–erythroid to colony-forming unit–erythroid, we hypothesize that this transporter system could have an essential role in early erythropoiesis. In further studies, a 6-kb fragment of the human FLVCR gene was amplified by polymerase chain reaction from genomic DNA, using homologous cDNA sequences identified in the human Expressed Sequence Tags database. By radiation hybrid mapping, the human gene was localized to a 0.5-centiMorgan region on the long arm of chromosome 1 at q31.3.
Cats naturally or experimentally infected with feline leukemia virus-C (FeLV-C) develop profound anemia (hematocrit = 4%-15%).1-5 There is an absence of reticulocytes in the circulation, and the marrow lacks hemoglobinized cells. As the maturation and number of granulocytic cells and platelets are normal, the presentation satisfies the clinical definition of pure red cell aplasia (PRCA).6 Burst-forming unit–erythroid (BFU-E) can be detected in the marrow of anemic animals (and may be increased in frequency), but colony-forming unit–erythroid (CFU-E) is absent.4 This finding has lead to the hypothesis that the PRCA is due to an impairment of BFU-E to CFU-E differentiation.
The exact mechanism by which FeLV-C impedes early erythropoiesis is not known. All cats chronically viremic with FeLV-C develop PRCA. This uniform response in an outbred population suggests that an immunologic pathogenesis (the most common mechanism of acquired human PRCA7) is unlikely. Similarly, in vitro studies confirm that erythropoiesis is not suppressed by an aberrant population of T cells or an autoantibody to erythroid progenitor cells or to erythropoietin.8 Also, neonatal cats (which have immature immune systems) develop PRCA more rapidly than adolescent animals,3 9 implying that viral burden, and not immunologic response, is the major correlate of disease onset.
By 4-6 weeks after cats are inoculated with virus derived from a molecular clone of FeLV-C (FeLV-C/Sarma), all hematopoietic cells, including granulocytes, monocytes, lymphocytes, BFU-E, and colony-forming unit–granulocyte-macrophage, as well as fibroblasts are infected.9-11 The consequence of FeLV-C/Sarma infection therefore differs among infected cell types. Thus, the pathogenesis of FeLV-C-induced anemia differs from the pathogenesis of anemia in human parvovirus B19 infection. As human parvovirus B19 uniquely infects and lyses erythroid precursor cells (the cell surface receptor is the P antigen),12 cell entry alone determines clinical phenotype.
Three subgroups (A, B, and C) of FeLV have been defined by host cell range, neutralization, and interference assays,13-15studies which reflect properties of the surface unit (SU) of the retrovirus envelope protein. FeLV-A has the most restrictive host cell range and only infects cat and some dog cells. FeLV-B and FeLV-C can infect cells of many species, including cat, dog, monkey, human, cow, and pig. Only FeLV-C viruses can infect guinea pig cells. Also, although the host cell range of FeLV-C is broad, this subgroup of virus cannot infect most rodent (hamster, mouse, and rat) cells. Receptor specificity has been inferred by the use of interference assays. The principle of an interference assay is that a permissive cell (eg, a cat cell) that is infected with one type of FeLV (eg, C) cannot be infected by additional viruses of the same subgroup but can be super-infected by viruses of other subgroups (eg, FeLV-A) that use different cell surface receptors.16 Two mechanisms likely contribute to retroviral interference: (1) the entrapment of the receptor within the endoplasmic reticulum by interaction with SU protein derived from integrated proviral DNA; and (2) the competitive inhibition by SU protein of the binding of additional virus to residual cell surface receptors.16 Interference assays (with human cells) have shown that FeLV-B shares a receptor with the gibbon ape leukemia virus.14,17 No other retrovirus interferes with the ability of FeLV-C to infect human cells, making its receptor of interest from a virologic, as well as a hematologic, perspective.14
FeLV-A, -B, and -C have been molecularly cloned.5,18-21Predicted amino acid sequences differ predominately in the SU protein, and variable region 1 of 5 discrete variable regions is most divergent. Both the genetic determinants of the anemia and subgroup C phenotype map to variable region 1 in studies of chimeric viruses constructed from FeLV-A/61E (a nonpathogenic subgroup A virus) and FeLV-C/Sarma.22 23 Thus, it appears that the 30 amino acid region of the SU protein that is required for anemia is also required for the binding of FeLV-C to its cell surface receptor. This observation led to the hypothesis that the cell surface receptor for FeLV-C has an important physiologic role in the normal maturation of BFU-E to CFU-E but is redundant or nonessential for granulocytic differentiation. When cells are infected by FeLV-C and receptor function or expression is impaired (via the mechanism of envelope-mediated interference), PRCA results.
To gain insights into the mechanisms controlling early erythropoiesis, we have cloned the feline complementary DNA (cDNA) encoding the FeLV-C cell surface receptor (FLVCR). The experimental strategy employed a retroviral vector cDNA library, an approach critical to the successful cloning of cDNAs for simian immunodeficiency virus co-receptors, and the human receptors for xenotropic (and polytropic) murine leukemia viruses (MuLV), and the feline endogenous virus RD114.24-27The protein that confers susceptibility to FeLV-C infection appears to be a D-glucarate transporter and member of the major facilitator superfamily (MFS) of transporter proteins.
Sequences homologous to the FLVCR cDNA were identified in the human Expressed Sequence Tags (EST) database and were utilized to amplify by polymerase chain reaction (PCR), a 6-kb fragment of the human gene. The human FLVCR (huFLVCR) was mapped, using radiation hybrid mapping, to chromosome 1q31.3. Of interest, rearrangement of the distal region of chromosome 1q has been described in a patient with Diamond-Blackfan anemia, a congenital PRCA.28
Materials and methods
Plasmids, viruses, and cell lines
The retroviral vector LAPSN (derived from Moloney MuLV,29 a gift from A. D. Miller, Fred Hutchinson Cancer Research Center, Seattle, WA) expresses human alkaline phosphatase (AP) and neomycin phosphotransferase (neo); the MSCV retroviral vectors30 (available from Clontech, Palo Alto, CA) are derived from myeloproliferative sarcoma virus, and express neo (MSCVneo), or the puromycin N-acetyl transferase gene (MSCVpuro). The retrovirus vector pMX (a gift from T. Kitamura, University of Tokyo, Japan) used for feline cDNA library expression, is derived from pBabe-puro (with deletion of the selectable marker).31
To pseudotype retroviral vectors with the FeLV-C/Sarma envelope, we subcloned a 2.0-kb XhoI/RsRII fragment of pFSC5 (a gift from J. Mullins, University of Washington, Seattle, WA) into the replication defective FeLV-A/61E-based retroviral packaging vector p61EΔΨ-A32digested with the same enzymes to generate p61EΔΨ-Cenv. Sequence analysis of p61EΔΨ-Cenv confirmed replacement of the FeLV-A env gene by the complete coding region of FeLV-Cenv. The host cell range of the virus derived from p61EΔΨ-Cenv was identical to that of FeLV-C/Sarma as assayed by transfer of the MSCVneo vector (data not shown).
The plasmid pSV-MLV-env33 (a gift from M. Emerman, Fred Hutchinson Cancer Research Center, Seattle, WA) was used to pseudotype retroviral vectors with the amphotropic envelope. The plasmid p61E-LTR-ΔΨ-gp is an FeLV-A/61E-based env-deleted packaging vector that expresses FeLV-A/61E gag and polgenes; this construct can be complemented in trans with expression vectors for FeLV or MuLV envelope to generate infectious particles (C. Meiring and J. Overbaugh, unpublished). The plasmid pCMVhph34 (a gift from M. Linial, Fred Hutchinson Cancer Research Center, Seattle, WA) contains the hygromycin B phosphotransferase gene under the control of a CMV promoter.
Replication defective FeLV-C particles carrying MSCV vector RNA, MSCVneo(61EΔΨ-Cenv), and MSCVpuro(61EΔΨ-Cenv) were produced by cotransfection [using the calcium-phosphate technique (CalPhos Mammalian Transfection Kit, Clontech)] of 293T human embryonic kidney cells35 with p61EΔΨ-Cenv, and pMSCVneo or pMSCVpuro, respectively. A stable packaging cell line able to produce replication-defective LAPSN(61EΔΨ-Cenv) particles was generated by first transfecting D17 canine osteosarcoma cells (ATCC CRL 6248) with pLAPSN followed by selection (× 14d) in G418 (1000 μg/mL active, Gibco, Gaithersburg, MD). Pools of resistant colonies were then transfected with p61EΔΨ-Cenv and pCMVHygro at a molar ratio of 20:1 and selected (× 14d) in hygromycin B (300 μg/mL, Sigma, St Louis, MO). Replication-competent FeLV-B subgroup virus (FeLV-B-90Z36) and replication-defective amphotropic pseudotype retroviral vectors were produced by cotransfection of 293T cells with pLAPSN, and pFeLV-B-90Z or p61E-LTRΔΨ-gp/pSV-MLV-env to generate LAPSN(FeLV-B-90Z) and LAPSN(61E-LTR-ΔΨ-gp/Ampho env), respectively. (The nomenclature used lists the viral vector followed by the viral genome employed to express virion proteins in parentheses.) All transient retroviral vector supernatants were collected 48 hours posttransfection, filtered, and applied to target cells or frozen at −80°C.
Mammalian cells, including mouse embryo fibroblasts, NIH 3T3 (ATCC CRL 1658); rat kidney cells, NRK 52E (ATCC CRL 1571); feline embryonic fibroblasts (a gift of E. Hoover, Ohio State University, Columbus, OH); guinea pig transformed fetal cells, GP104C1 (ATCC CRL 1405); canine D17 cells; human 293T cells; and the BOSC23 packaging cell line35 were grown in DMEM high glucose (Gibco) supplemented with 10% fetal calf serum (Summit, Fort Collins, CO).
Feline embryonic fibroblast cells were used to determine the infectious titer of FeLV pseudotype vectors. The cells, plated at a density of 2 × 105 per 60-mm tissue culture dish, were exposed after 16 hours to a dilution (10−4 to 1 mL) of viral supernatant made up to 2 mL with media and polybrene (8 μg/mL, Sigma). Two days later, 10% and 90% of the total cells from each dish were replated and selected in G418 (700 μg/mL active). Seven to 10 days later, colonies were stained with Coomassie Blue and counted. Colony-forming units (cfu) for FeLV pseudotype vectors encoding puromycin resistance were determined using puromycin selection (2.0 μg/mL, Sigma).
Expression cloning of FLVCR cDNA
A feline cDNA library derived from 3201B cells, a feline T cell line,37 was subcloned into the retroviral expression vector pMX, using the protocol of Kitamura et al31 (M. Anderson and J. Overbaugh, unpublished).
The retroviral cDNA library (complexity > 3 × 106individual clones) was pseudotyped with ecotropic MuLV envelope proteins with the use of BOSC23 packaging cells. NIH 3T3 cells (in 10 plates at a density of 2 × 105 per 100-mm dish) were transduced with the cDNA library at an estimated multiplicity of infection (MOI) of 0.1 virus per cell with the use of polybrene at 8 μg/mL (an MOI of <1 was used to decrease the number of proviral inserts per infectant26). Forty-eight hours after transduction, NIH 3T3 cells were exposed to MSCVneo(61EΔΨ-Cenv), at an MOI of 5:1. The next day, the cells were transferred to 150-mm tissue culture dishes and then were selected in G418 (600 μg/mL active) for 10 days. Pools of neomycin-resistant colonies were replated, exposed to the FeLV-C pseudotype retroviral vector MSCVpuro(61EΔΨ-Cenv) at an MOI of 0.1, and selected in puromycin (1.5 μg/mL). Distinct colonies were isolated at this stage.
Isolation of FeLV receptor DNA and expression in mammalian cells
DNA was extracted from reinfectable colonies with the use of the Puregene DNA isolation kit (Gentra, Minneapolis, MN). PCR was performed with the use of the Expand Long Template PCR kit (Boehringer Mannheim, Indianapolis, IN) in a GeneAmp 9700 thermocycler for 35 cycles each of 94°C for 30 seconds, 65°C for 30 seconds, 68°C for 4 minutes, with a final extension at 68°C for 7 minutes. The primer pair used (upstream, pMX#11, 5′-GTGGACCATCCTCTAGACTGC-3′ and downstream, pMX#14, 5′-GAAAATAAAATAGCAGCTGGTGACACG-3′) bind within the polylinker of pMX, and pMX#14 includes 3′ sequences that overlap the BstXI site used to clone the cDNA inserts into pMX; thus, these primers are designed to specifically amplify the library cDNA insert. Blunt-ended PCR products were created with the use of T4 polymerase (Gibco).
pMSCV(1.8 kb)neo (also termed pMSCV(FLVCR)neo) and pMCSV(1.6 kb)neo were constructed by ligating the 1.8-kb or 1.6-kb PCR product intoHpaI-digested pMSCVneo. These plasmids or pMSCVneo alone was introduced into BOSC23 packaging cells, and the resulting retroviral vector supernatant was used to transduce naive NIH 3T3 and NRK 52E cells. Cells were selected in G418 (600 μg/mL active) for 10 days. Pools of neomycin-resistant cells were tested for susceptibility to LAPSN(61EΔΨ-Cenv). Various dilutions of vector (10−4-1 mL in 2 mL total volume) were used in these assays so that the infectious titers (number of AP positive cells or clusters [focus-forming units (ffu)/mL] could be reliably determined. AP staining was performed as described.38
The 1.8-kb cDNA, FLVCR, was sequenced twice in both directions on a PE/ABI 373 DNA Sequencer (Applied Biosystems, Foster City, CA) with the use of Big Dye terminator sequencing chemistry (Molecular Pharmacology Facility, Department of Pharmacology, University of Washington). The GenBank accession number is AF192387. Databases39-41 were searched for homologous sequences. Predicted amino acid sequences were aligned with the use of ClustalW (MacVector, Oxford Molecular). Hydrophobic regions of the predicted FLVCR protein were inferred,42 and the presence of Prosite patterns were determined with the use of the PROSITE database.43 Exon prediction and identification of putative extended open reading frames for homologous human genomic sequences in GenBank were performed with the use of Genscan software.44
Chromosomal localization of the human FLVCR gene
PCR was performed on human genomic DNA in a GeneAmp 9700 thermocycler with the use of PCR conditions recommended in the Expand Long Template PCR kit (with an annealing temperature of 60°C) and primers (forward, Cacof255, 5′-TGTTCACATTGGCTCAAGGA-3′; reverse, w07167r144, 5′-AATTGCCGATTCTGACTGCTTGGACA-3′) with sequence common to feline FLVCR and two human ESTs (GenBank accession nos. AA305281 and W07167). The resulting 6-kb fragment of the 3′ end of huFLVCR was subcloned into the pGEM vector (Promega, Madison, WI) and partially sequenced. The chromosomal location of huFLVCR was determined with the use of the Stanford Radiation Hybrid Mapping Panel G3.45 PCR was performed on DNA from each of the 83 G3 hybrid clones with the use of primers derived from the 6-kb fragment partial sequence (forward, w07167f16, 5′-CTTGGTCTGTGGGACTGTCA-3′, and reverse, w07167r273, 5′-GCCCCTCTGTTTCAGCATTA-3′), to amplify a 277-base pair (bp) product (details are provided at reference 46). The resulting vector was submitted to the Stanford Human Genome Center RH Server for chromosomal localization.
Results
Expression library screen for the FeLV-C receptor
To use a gene transfer approach to identify the receptor, it was necessary to first identify a cell line resistant to FeLV-C infection. A number of cell lines known to be poorly infectable by FeLV-C (eg, from rodent, canine, bovine, primate, and human species13) were studied. Candidate cells were infected with high-titer FeLV-C/Sarma pseudotype viruses carrying MSCVneo and screened for neomycin resistance. Rodent cell lines, including NIH 3T3 (< 3 cfu/mL) and NRK 52E cells (< 10 cfu/mL) were identified as least susceptible to FeLV-C infection.
NIH 3T3 cells were transduced with a retroviral vector cDNA library generated from the feline T cell line, 3201B. 3201B cells are highly infectable by FeLV-C37 (J. Abkowitz, unpublished observations) and thus were predicted to encode the receptor of interest. Transduced NIH 3T3 cells were then screened to determine whether they had acquired the ability to be sequentially infected by retroviral vectors that were pseudotyped with FeLV-C envelope protein and that carried selectable markers. We used a retroviral genome that was deleted in its packaging sequence to express gag, pol, and env proteins so that the FeLV-C/Sarma env gene would not be transferred to NIH 3T3 cells during infection. Thus, NIH 3T3 cells expressing the receptor would not become resistant to reinfection as a result of interference.
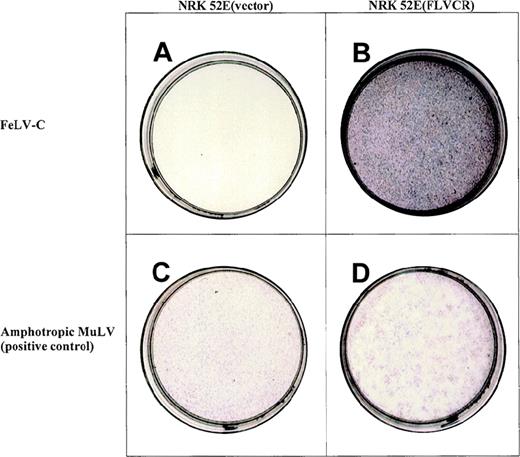
NIH 3T3 cells transduced with the library were first challenged with MSCVneo (61EΔΨ-Cenv). Cell pools identified as resistant to neomycin were challenged with the FeLV-C pseudotype vector carrying the puromycin resistance gene, at a MOI of 0.1. A low MOI was employed in the secondary screen to reduce the number of false-positive events. Of a total of 150 puromycin-resistant colonies isolated, genomic DNA from 15 colonies was subjected to PCR with the use of primers specific to the pMX vector used to generate the cDNA library. PCR products of 1.2, 1.6, and 1.8 kb were amplified in each case, suggesting that the colonies derived from a single-cell clone. The 1.6- and 1.8-kb PCR fragments were selected for further study as their sizes were more consistent with cDNAs of previously isolated retroviral receptors.25-27 47 The fragments were each subcloned into the retroviral vector MSCVneo [denoted MSCV(1.8 kb)neo and MSCV(1.6 kb)neo, respectively]. These vectors were then overexpressed in naive NIH 3T3 and NRK 52E cells. Cells expressing the 1.8-kb PCR fragment were susceptible to infection by a FeLV-C pseudotype vector (see Table1 and Figure1). Cells transduced in parallel with the MSCV(1.6 kb)neo (data not shown) or MSCVneo (vector alone) remained resistant to infection. Expression of the 1.8-kb PCR fragment had no effect either on the susceptibility of NIH 3T3 or NRK 52E cells to infection by FeLV subgroups A or B or to infection by viruses pseudotyped with amphotropic envelope, demonstrating that the 1.8-kb feline cDNA specifically confers susceptibility to FeLV-C infection and thus encodes the putative FLVCR. NRK 52E cells expressing the receptor [NRK 52E(FLVCR)] were significantly more infectable with FeLV-C than were NIH 3T3(FLVCR) cells. This finding suggests that either receptor expression was suboptimal in NIH 3T3 cells or that other cofactors not present in this cell type are required for efficient FeLV-C infection.
Infection of NIH 3T3 and NRK 52E cells expressing feline leukemia virus-C cell surface receptor (FLVCR)
| Target Cell . | Titer (ffu/mL) . | |||
|---|---|---|---|---|
| Amphotropic MuLV . | FeLV-A . | FeLV-B . | FeLV-C . | |
| FEA | 2.6 × 105 | 1.4 × 107 | 1.8 × 104 | 2.4 × 105 |
| GP104C1 | 2.0 × 105 | 1 | 0 | 9.2 × 106 |
| NIH 3T3 | 2.9 × 105 | 0 | 0 | 0 |
| NIH 3T3(FLVCR) | 3.4 × 105 | 0 | 3 | 47 |
| NRK 52E | 6.8 × 105 | <10 | 65 | <10 |
| NRK 52E(vector) | 7.2 × 105 | <10 | 48 | <10 |
| NRK 52E(FLVCR) | 4.8 × 105 | <10 | <102 | 1.4 × 105 |
| Target Cell . | Titer (ffu/mL) . | |||
|---|---|---|---|---|
| Amphotropic MuLV . | FeLV-A . | FeLV-B . | FeLV-C . | |
| FEA | 2.6 × 105 | 1.4 × 107 | 1.8 × 104 | 2.4 × 105 |
| GP104C1 | 2.0 × 105 | 1 | 0 | 9.2 × 106 |
| NIH 3T3 | 2.9 × 105 | 0 | 0 | 0 |
| NIH 3T3(FLVCR) | 3.4 × 105 | 0 | 3 | 47 |
| NRK 52E | 6.8 × 105 | <10 | 65 | <10 |
| NRK 52E(vector) | 7.2 × 105 | <10 | 48 | <10 |
| NRK 52E(FLVCR) | 4.8 × 105 | <10 | <102 | 1.4 × 105 |
FEA = feline embryonic fibroblasts; FeLV = feline leukemia virus; ffu = focus-forming unit; MuLV = murine leukemia virus. Titers were determined by alkaline phosphatase staining, which was performed 72 hours after exposure to LAPSN pseudotyped with amphotropic MuLV, FeLV-A, FeLV-B, or FeLV-C envelope proteins. Titers were averaged from three plates, from two infection studies. NIH 3T3(FLVCR) and NRK 52E(FLVCR) are cells that were transduced with MSCV(1.8 kb)neo then selected (×10d) in G418. NRK 52E(vector) are control cells that were transduced with MSCVneo then selected (×10d) in G418. The species of origin of FEA, GP104C1, NIH 3T3, and NRK 52E cells are cat, guinea pig, mouse, and rat, respectively.
Expression of the 1.8-kb cDNA allows NRK 52E to become susceptible to infection by a feline leukemia virus-C (FeLV-C) pseudotype LAPSN vector.
The NRK 52E cells in panel a contain the control vector MSCVneo, cannot be infected by the FeLV-C pseudotype LAPSN vector, thus fail to express human alkaline phosphatase, and are white. The NRK 52E cells in panel b contain MSCV (1.8 kb)neo, express FeLV-C cell surface receptor, and are readily infectable by the FeLV-C pseudotype LAPSN vector, as evidenced by their purple stain. Cells infected with the amphotropic pseudotype LAPSN vector stain purple (panels c and d). NRK-52E cells not exposed to virus do not express human alkaline phosphatase and are white (data not shown). The concentration of the FeLV-C pseudotype vector was 10 × that of the amphotropic murine leukemia virus pseudotype vector in this study. Formal titers are shown in Table 1.
Expression of the 1.8-kb cDNA allows NRK 52E to become susceptible to infection by a feline leukemia virus-C (FeLV-C) pseudotype LAPSN vector.
The NRK 52E cells in panel a contain the control vector MSCVneo, cannot be infected by the FeLV-C pseudotype LAPSN vector, thus fail to express human alkaline phosphatase, and are white. The NRK 52E cells in panel b contain MSCV (1.8 kb)neo, express FeLV-C cell surface receptor, and are readily infectable by the FeLV-C pseudotype LAPSN vector, as evidenced by their purple stain. Cells infected with the amphotropic pseudotype LAPSN vector stain purple (panels c and d). NRK-52E cells not exposed to virus do not express human alkaline phosphatase and are white (data not shown). The concentration of the FeLV-C pseudotype vector was 10 × that of the amphotropic murine leukemia virus pseudotype vector in this study. Formal titers are shown in Table 1.
Identity of the FeLV-C receptor
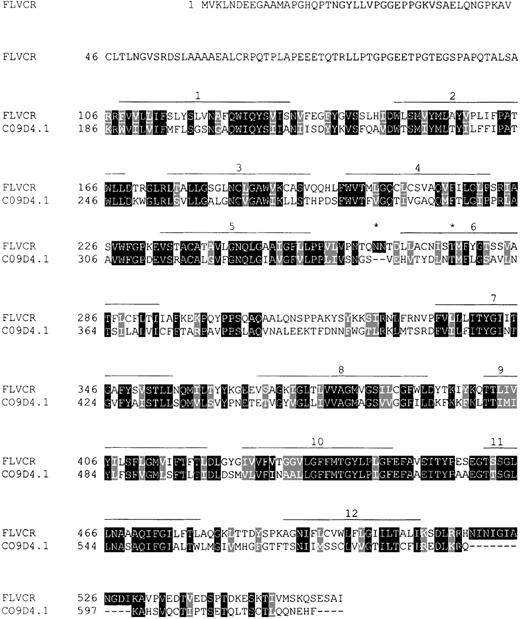
The open reading frame between bases 42 and 1724 of the FLVCR cDNA is predicted to encode a protein of 560 amino acids (Figure2) with a molecular mass of 60 kDa. The hydrophobicity plot suggests the presence of 12 hydrophobic membrane-spanning domains with the N-terminus in the cytosol.42 Canonical N-glycosylation sites occur in the third predicted extracellular and in the sixth predicted transmembrane domains. Prosite patterns43 compatible with sites of protein kinase C phosphorylation, casein kinase II phosphorylation, and N-myristoylation were identified.
Sequence comparison of the feline leukemia virus-C (FeLV-C) receptor (560 amino acids) and a putative D-glucarate transporter of C. elegans, the 623 amino acid C09D4.1 gene product (GenBank accession no. AF002196).
The receptor (aa 106-550) shares 44% sequence identity with the putative transporter (aa 186-617). Amino acid identities are marked with dark shading, and similarities are indicated with light shading. Predicted transmembrane regions are shown as lines over the amino acid sequence, and two potential N-glycosylation sites are marked by an asterisk.
Sequence comparison of the feline leukemia virus-C (FeLV-C) receptor (560 amino acids) and a putative D-glucarate transporter of C. elegans, the 623 amino acid C09D4.1 gene product (GenBank accession no. AF002196).
The receptor (aa 106-550) shares 44% sequence identity with the putative transporter (aa 186-617). Amino acid identities are marked with dark shading, and similarities are indicated with light shading. Predicted transmembrane regions are shown as lines over the amino acid sequence, and two potential N-glycosylation sites are marked by an asterisk.
BlastP40 comparisons with sequences available in the GenBank databases revealed that the predicted protein has highest homology (P < 1 × 10−106) to a hypothetical protein encoded by the gene locus C09D4.1 in C. elegans—a putative D-glucarate transporter, originally identified in bacteria (Bacillus subtilis andPseudomonas spp.). It also has high homology to a number of other putative glucarate transporter proteins in C. elegans[eg, YT45_CAEEL (GenBank accession no. Q11073) and a protein predicted for gene locus C05G5.1 (GenBank accession no. CAA94104) (P < 1 × 10−27)]. These amino acid homologies (23%-44% identities, extending across the bulk of the proteins) and the predicted topology of 12 membrane-spanning domains (Figure 2) suggest that FLVCR is a member of the anion : cation symporter family48 of the MFS (see reference 49 for review). MFS permeases are single polypeptide secondary carriers, capable of transporting small solutes across membranes (eg, sugars, Krebs cycle metabolites, inorganic phosphates) in response to chemico-osmotic gradients.48,49 They occur ubiquitously in all classifications of living organisms and comprise 3% of proteins predicted for the S. cerevisiae (yeast) genome.50 Characteristic of the anion : cation symporter family48 is the presence of numerous gene paralogs (at least 15 in C. elegans). Consistent with this description, we have noted that a portion of a bacterial artificial chromosome sequence (200 kb in length) located on human chromosome 14q23.2 may encode a protein 357 amino acids in length (predicted with the use of Genscan software44) that is highly homologous to the feline FLVCR amino acid sequence (P < 10−93). However, the DNA sequence match of this hypothetical protein to the FLVCR cDNA is imperfect (only 74%), suggesting that the bacterial artificial chromosome (GenBank accession no. AC007182.2) contains a member of this gene family.
BlastN analyses41 of the FLVCR cDNA sequence against the human EST database identified portions of the candidate huFLVCR gene. One EST (GenBank accession no. AA305281) is an unmapped 396-bp sequence derived from a colonic adenocarcinoma cell line (Caco 2) cDNA library and has a portion with 92% contiguous sequence identity (bp 137-396) to the feline FLVCR cDNA (bp 1350-1610) (P = 10−102). The second EST (GenBank accession no. W07 167), an unmapped 656-bp sequence derived from a fetal lung cDNA library, has a region with 90% homology to the 3′ end of FLVCR.
The gene encoding huFLVCR maps to human chromosome 1q31.3
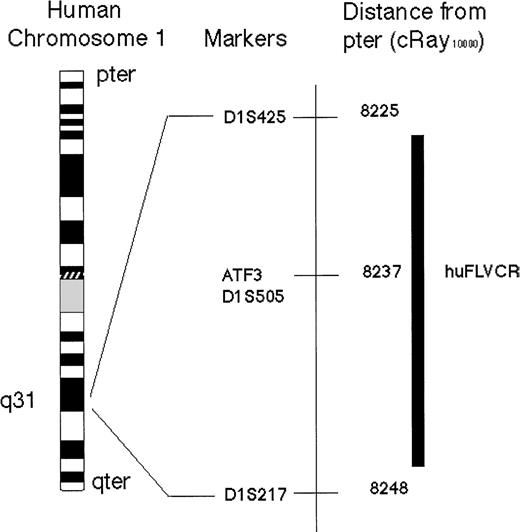
A 6-kb fragment of the 3′ end of the huFLVCR gene was amplified by PCR from human genomic DNA, using primers (Cacof255, and w07167r144) derived from sequence common to FLVCR cDNA and the two identified homologous human ESTs (GenBank accession nos. AA305281 andW07167). The fragment was partially sequenced, allowing the identification of intron-exon borders and the subsequent design of further primers (w07167f16, and w07167r273). Analysis of the Stanford Generation 3 Radiation Hybrid (G3 RH) Mapping Panel (which consists of a set of 83 hamster/human cell lines containing radiation-fragmented human DNA such that each clonal cell line contains random fragments of ∼18% of the human genome) was performed using the latter PCR primers. These primers were designed to amplify a 277-bp fragment near the 3′ end of the gene with one primer based in an intron and the second present in an exon. These primers were specific to human DNA and yielded no PCR product with hamster DNA alone. Screening of the RH DNA pools with these primers yielded the following vector 1 000 0 001 0 000 0 000 0 000 1 00 0 011 1 000 0 000 0 000 0 010 0 000 0 000 0 001 0 000 1 000 0 00 0 000 0 100 0 100 001 in which hybrids containing (1) or not containing (0) the huFLVCR sequence are indicated. This mapping vector was analyzed with the use of the radiation hybrid mapping server,51 which placed the huFLVCR gene within 11centiRay10 000 (cR) of marker D1S505 (SHGC 1592, AFMa127wb5) with a logarithm of odds score of 11.12 (Figure3). D1S505 has previously been mapped to the 6-centiMorgan (cM) region on chromosome 1q31.3 flanked by D1S491 and D1S474. The closest markers 11cRay10 000 or greater from D1S505 (8237cR) are D1S425 (8225cR) and D1S217 (8248cR). The only known gene mapping to this 0.5-cM interval is Activating Transcription Factor 3 (ATF3), and no additional ESTs have been mapped to this region (NCBI GeneMap'99).
Schematic representation of the physical location of human FeLV-C cell surface receptor (huFLVCR) gene on human Chromosome 1.
The chromosomal location of huFLVCR was determined, by polymerase chain reaction analysis of the Stanford Radiation hybrid panel G3, to lie within 11 cRay10 000 of D1S505 with a logarithm of odds score of 11.12. The only previously identified gene mapping to this interval is the transcription factor Activating Transcription Factor 3. For Chromosome 1 with the G3 RH panel, 1cRay10 000 is ∼26 kb.
Schematic representation of the physical location of human FeLV-C cell surface receptor (huFLVCR) gene on human Chromosome 1.
The chromosomal location of huFLVCR was determined, by polymerase chain reaction analysis of the Stanford Radiation hybrid panel G3, to lie within 11 cRay10 000 of D1S505 with a logarithm of odds score of 11.12. The only previously identified gene mapping to this interval is the transcription factor Activating Transcription Factor 3. For Chromosome 1 with the G3 RH panel, 1cRay10 000 is ∼26 kb.
Discussion
There are 8 interference groups of simple retroviruses (as defined by assays of human cells14), suggesting that 8 distinct receptors are responsible for cell entry. The cell surface receptors for the amphotropic MuLV, gibbon ape leukemia virus (and FeLV-B), and the feline endogenous retrovirus RD114 have been identified and are predicted to be multiple membrane-spanning proteins that function as transporter molecules, transporting phosphate ions or neutral amino acids.16,26,27 The receptor for the xenotropic MuLV is also a multiple membrane-spanning protein and may have a role in G-protein mediated signal transduction.25 Thus, these receptors are integral membrane proteins with basic physiologic functions. We have cloned the cDNA for the putative cell surface receptor for FeLV-C on the basis of its ability to confer susceptibility to FeLV-C infection when expressed in resistant rodent cells. The sequence of FLVCR suggests that it too is a transporter molecule and a member of the MFS (reviewed in references 48 and 49). Through sequence homology, it appears that FLVCR is an organic anion transporter and specifically a D-glucarate transporter molecule.
The consequence of FeLV-C infection differs from that of other simple retroviruses. Biological data suggest that the FeLV-C envelope SU protein may act as a dominant negative protein, inhibiting the function or cell surface expression of FLVCR (which is the mechanism of retroviral interference) to result in PRCA. A corollary of this hypothesis is that the FeLV-C receptor plays a crucial role in normal erythropoiesis. Thus, it is intriguing that FLVCR shares homology with D-glucarate transporters of both prokaryotes and eukaryotes.
How the transport of D-glucarate, a diacid sugar derived from glucose, may relate to erythropoiesis is unknown. D-glucarate circulates at a high level in the serum,52 and its principal metabolite inhibits beta glucuronidase activity in hepatocytes, resulting in the stabilization of bilirubin.53 A comparable role in erythroid cells could impact heme metabolism. At the stage of differentiation between BFU-E and CFU-E, globin transcription increases, iron absorption increases (via the increased cell surface expression of the transferrin receptor), and the subsequent maintenance of heme is also critical for initiation of hemoglobin synthesis.54 The identification of the FeLV-C receptor should allow the direct study of novel physiologic mechanisms that regulate early erythropoiesis.
In additional studies, the gene encoding the human ortholog of FLVCR was localized to chromosome 1q31.3 with the use of radiation hybrid mapping. Apart from a transcription factor (ATF3, Figure 3), no other genes have been mapped to this region to date. The clinical similarity between FeLV-C-induced PRCA in cats and human PRCA, including congenital PRCA (Diamond-Blackfan anemia), raises the possibility that an abnormality in the expression or structure of the huFLVCR gene could result in erythroid marrow failure. It is, therefore, of interest that a case report describes a patient with Diamond-Blackfan anemia and a rearrangement of the distal region of 1q.28 Although cytogenetic abnormalities are rare in children with Diamond-Blackfan anemia, 10%-20% of patients have a family history of anemia, implying a genetic basis for their disease.28,55 Recent studies have demonstrated that the gene on chromosome 19q13.2 that encodes ribosomal protein S19, is deleted in some,56,57 but not all,58 families with Diamond-Blackfan anemia. The identification of the huFLVCR gene should allow us to determine its relevance to this disease.
The first FeLV receptor to be identified was the receptor for FeLV-B, Pit 1.17 59 As FLVCR shares minimal structural similarity but no sequence homology with Pit 1, the relationship of the structure of these retroviral receptor proteins to their function can also be studied. In addition, the mechanisms by which the closely related viruses FeLV-C and FeLV-B have evolved to use distinct cell surface receptors can be explored.
Note: Since the submission of this manuscript, the cDNA and predicted amino acid sequences of huFLVCR have been published (Tailor et al, J Virol. 1999;73:6500). There is 88% identity among base pairs of the coding regions of the feline and human cDNAs. Feline FLVCR has 83% amino acid identity and 89% similarity to the huFLVCR protein. The proteins are most disparate (57% identity) in the predicted N terminal intracellular domain (aa1-106), a region that has no homology to CO9D4.1 (see Figure 1). HuFLVCR messenger RNA is expressed in multiple tissues (eg, pancreas, kidney) as well as marrow, consistent with FeLV-C infectivity data in cats.10 The ESTs we used to construct a huFLVCR genomic probe are 99% identical to huFLVCR cDNA, confirming the appropriateness of this probe for the chromosomal localization studies. The structural similarity of feline FLVCR and huFLVCR thus supports the contention that these proteins have an equivalent function in the regulation of erythropoiesis as well as in the regulation of retroviral entry.
Acknowledgments
The authors would like to thank Allan Dimaunahan and Zenaida Sisk for help in preparation of the manuscript.
Supported by grants R01 HL31823 and CA51080 from the National Institutes of Health. Dr Abkowitz is recipient of a Faculty Research Award from the American Cancer Society.
Reprints:Janis L. Abkowitz, Professor of Medicine, Division of Hematology, University of Washington, Box 357710, Seattle, WA 98195-7710.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal