Members of the JAK family of protein tyrosine kinase (PTK) proteins are required for the transmission of signals from a variety of cell surface receptors, particularly those of the cytokine receptor family. JAK function has been implicated in hematopoiesis and regulation of the immune system, and recent data suggest that the vertebrate JAK2gene may play a role in leukemia. We have isolated and characterizedjak cDNAs from the zebrafish Danio rerio. The zebrafish genome possesses 2 jak2 genes that occupy paralogous chromosome segments in the zebrafish genome, and these segments conserve syntenic relationships with orthologous genes in mammalian genomes, suggesting an ancient duplication in the zebrafish lineage. The jak2a gene is expressed at high levels in erythroid precursors of primitive and definitive waves and at a lower level in early central nervous system and developing fin buds. jak2b is expressed in the developing lens and nephritic ducts, but not in hematopoietic tissue. The expression of jak2a was examined in hematopoietic mutants and found to be disrupted in clocheand spadetail, suggesting an early role in hematopoiesis. Taken together with recent gene knockout data in the mouse, we suggest that jak2a may be functionally equivalent to mammalianJak2, with a role in early erythropoiesis.
CYTOKINES ARE IMPORTANT regulators of proliferation, differentiation, and cell function for a wide range of cells of hematopoietic and other lineages. The JAK/STAT signaling system is required for the function of a number of cytokines, acting to transduce signals from cytokine receptors.1 2
JAK tyrosine kinases possess a distinctive structure. Amino acid sequence comparison of the 4 mammalian JAK family members (JAK1, JAK2, JAK3, and TYK2) shows the presence of 7 highly conserved domains (named JH1 through JH7).3 The N-terminal region of JAK kinases contains structures (JH7-JH3) that confer specific binding activity toward cytokine receptor cytoplasmic domains.4-9 Further C-terminal, the kinase-related domain (JH2) exhibits significant similarity to a tyrosine kinase domain yet diverges within several critical catalytic motifs.10 Indeed, this domain appears not to possess phosphotransferase activity10; rather, it is required for the stability and binding affinity of associated receptors.11,12 The C-terminal domain (JH1) contains a protein tyrosine kinase10 responsible for initiating much of the signaling activity from cytokine receptors that use this family of PTKs. Expression of kinase inactive JAK variants abrogate most if not all aspects of signaling in a dominant negative manner through receptors for cytokines such as erythropoietin (EPO),13growth hormone (GH),5 granulocyte-macrophage colony-stimulating factor (GM-CSF),14 granulocyte colony-stimulating factor (G-CSF),15 interleukin-6 (IL-6),16 interferon α (IFNα),11IFNγ,17 and IL-2.18 Analysis of the effects of dominant negative JAK variants combined with a series of somatic cell mutants deficient or defective in 1 of the JAK kinases8,19 20 demonstrates the critical role of the JAK kinase family in cell signaling.
Three lines of evidence link JAK function directly to the regulation of the growth and differentiation of hematopoietic cells in vivo. First, gene targeting of Jak loci in the mouse yields hematopoietic phenotypes: Jak1-deficient mice exhibit impaired lymphopoiesis,21,Jak2 deficiency abolishes definitive erythropoiesis,22,23 and Jak3 null mutation results in the murine equivalent of human severe combined immunodeficiency syndrome (SCID).24-27 Second, the dominant Drosophila melanogaster JAK mutant,hopscotchTum-l causes a lethal leukemia in fruit flies.28 The protein products ofhopTum-l and a recently identified allelehopT42 show increased levels of autophosphorylation and the capacity to activate Stat92E.29-31 Third, constitutive JAK2 catalytic activity can be detected in some acute lymphoblastic leukemia (ALL) cells in humans. Chromosomal translocation in a human T-cell ALL creates a TEL-JAK2 fusion protein capable of oligomerization through the TEL fusion partner, resulting in constitutive activation of JAK2 tyrosine kinase activity.32Mice expressing this fusion protein in bone marrow developed a fatal mixed myeloproliferative and T-cell lymphoproliferative disorder.33 These results indicate that JAK genes are required for normal hematopoiesis and that the deregulation of JAK catalytic activity is capable of causing hematopoietic neoplasia.
To further explore the role that the JAK family has in controlling blood cell growth, we have isolated and characterizedJAK genes from the zebrafish, Danio rerio. The zebrafish offers many technical advantages for the developmental analysis of gene function, because embryos are plentiful, develop externally, and are optically transparent. As a vertebrate experimental system, the zebrafish allows strong analogies to be drawn to human biology. For example, in contrast to Drosophila melanogasterand Caenorhabditis elegans, zebrafish has circulating blood cells of the erythroid, myeloid, and lymphoid lineages.34,35 Furthermore, a recent mutant screen performed in the zebrafish led to the isolation of mutations in more than 20 complementation groups that disrupt hematopoiesis,36,37 including a model for human congenital sideroblastic anemia, sauternes.38 Importantly, the possibility of performing modifier screens in the zebrafish offers the means of identifying other factors in these complex developmental processes by a genetic approach.
Our search using degenerate oligonucleotide polymerase chain reaction (PCR) for homologs of JAK and STAT genes in D rerio yielded several genes from both JAK (jak1, jak2a,and jak2b) and STAT (stat1 andstat3) gene families. The characterization of zebrafish members of the STAT family will be reported elsewhere.38a We report here in detail on the structure and evolutionary relationships of the zebrafish JAK2 homologs. We determined the expression of these genes in wild-type and several selected zebrafish mutants and consequently propose a role for jak2a in erythropoiesis.
MATERIALS AND METHODS
Isolation of zebrafish jak homologs.
JAK-directed degenerate oligonucleotide primers were designed after Wilks,39 based around conserved subdomains VIb and IX40 in the catalytic JH1 domain of Homo sapiens JAK1,10,Mus musculus Jak2,3,H sapiensJAK341 and TYK2,42 and D melanogaster HOP.43 Wherever complete degeneracy was required at a nucleotide position in the primers, an inosine residue was incorporated.44 The sequence of the primers is CCGAATTCCA(C/T)(C/A)GIGA(C/T)(C/T)TIGCIGCI(C/A)GIAA and CCGAATTCIACICC(A/G)(A/T)AI(G/C)(A/T)CCAIAC(G/A)TC. cDNA libraries were plated and screened at high stringency according to standard methods,45 with PCR products generated byJAK-directed degenerate oligonucleotide PCR as described above, and specific for the zebrafish jak1, jak2a, andjak2b genes. Mixed developmental stage D rerio cDNA libraries in Lambda Zap (Stratagene, La Jolla, CA), both random and poly-A primed, were a gift of J. Campos-Ortega (University of Köln, Köln, Germany). Lambda Zap cDNA libraries generated from staged embryonic mRNA populations were made by Bob Riggleman and Kathryn Helde and were a kind gift of D. Grunwald (Eccles Institute, University of Utah, Salt Lake City, UT). The “Contig Manager” application of the DNAstar suite of programs (DNAstar Inc, Madison, WI) was used to create and monitor contigs from the primary sequence data. cDNA sequences presented herein corresponding to each of the gene transcripts under study were sequenced in both directions to a minimum of 2-fold coverage.
Zebrafish care and strains.
Zebrafish were raised and maintained as described.46Zebrafish carrying the following mutant alleles of cloche(clom39),47,spadetail(sptb104),48,cabernet(cabtl236), retsina(rettr217), chianti(ciatu25f), sauternes(sauty121),38 chablis(chatu245/tu242e) (thought to be clonal alleles),weißherbst (wehth238,wehtp85c), chardonnay(cdyte216), frascati(frstm130d, frstq223),reisling (ristb237), and merlot(mottm303c)36 were studied.
Sequence analysis and evolutionary comparison.
Electronic database searches were made by submitting nucleic acid sequence and putative amino acid sequence to the public search facility at the Baylor College of Medicine (Houston, TX;http://hgsc.bcm.tmc.edu/SearchLaunches/) using “WU-BLAST.”49 To study the evolutionary relationships between the PTKs and JAKs identified, the deduced amino acid sequences of the genes in question were aligned with the “CLUSTAL” protein alignment program50 of the MegAlign application (DNAstar suite) and refined by hand using structural information, where available. These alignments were used to create maximum parsimony phylogenetic trees and distance matrices using the options of that program. The topology of the phylogenetic tree shown was insensitive to the order of sequence addition.
Whole mount in situ hybridization.
Embryos were staged according to Kimmel et al.51 Embryos raised to time points beyond 24 hours postfertilization (hpf) were transferred to E3 embryo medium with 0.003% phenylthiourea (PTU; Sigma, St Louis, MO) to prevent melanization. Riboprobe synthesis and in situ hybridization were performed essentially as described52 with the following modifications. Riboprobes were purified before use over RNA sephadex G-50 columns (Boehringer Mannheim, Indianapolis, IN). Using estimates of RNA synthesis based on 32P-CTP incorporation, probes were resuspended in HYB+52 at a concentration of 1 ng/mL for use. Embryos up to 24 hpf were not proteinase K-digested, embryos between 24 hpf and 36 hpf were digested for 10 minutes (10 μg/mL), and embryos greater than 36 hpf were digested for between 20 and 30 minutes (20 μg/mL). Hybridization and washing was performed at temperatures of 65°C to 70°C. Nonspecific antidigoxygenin Fab-AP binding (Boehringer Mannheim; used at a dilution of 1/5,000) was blocked by 2% wt/vol Blocking Reagent (Boehringer Mannheim)/10% heat-inactivated sheep serum/MABT (100 mmol/L Maelic acid, 150 mmol/L NaCl, 0.1% Tween-20, pH 7.5) for 1 hour at room temperature. Color detection reactions used BM purple substrate (Boehringer Mannheim) and were developed for up to 2 days before fixing in 4% paraformaldehyde/PBT (phosphate-buffered saline/0.1% Tween 20). Embryos were either cleared in glycerol or benzyl benzoate:benzyl alcohol (2:1) and photographed using a Leitz Wild T stereo dissection microscope (Leitz, Wetzlar, Germany) or a Nikon Microphot AX compound microscope (Nikon Inc, Melville, NY). The entire jak2b cDNA was used to generate a digoxygenin (DIG)-labeled probe, and the jak2a riboprobe contained the 5′-most 800 bp encoding the JH7 and JH6 domains.
Generation of DNA polymorphisms and genetic mapping.
A bacterial artificial chromosome (BAC) library (Genome Systems, St Louis, MO) containing large insert zebrafish genomic DNA was screened by hybridization to oligonucleotide probes derived from jak1,jak2a, and jak2b cDNA. Clones corresponding to the genomic loci were obtained for jak1 (96 E18, 100 K4, and 143 O15) and jak2a (112 K6), but not jak2b. A P1 artificial chromosome (PAC) library (C. Amemiya, Boston University, Boston, MA) was screened by hybridization to an oligonucleotide probe derived from jak2b and clones obtained (35 F6 and 58 G17). Sequence information from the ends of each of these genomic clones was determined (data not shown) and, along with the sequence of 3′ UTR regions from cDNA of the jak genes, was used to design PCR primer pairs that amplified products from genomic DNA that segregated in a C32xSJD mapping cross.53 The primers forjak1 (100 T7-1 GTAGAAGATACAGTCGCCTG, 100 T7-2 GTAAAGCAATATCAATAGAG) give a codominant size polymorphism54of 290/270 bp; the jak2a codominant size polymorphism (200/220 bp) is from the primer pair j2A.29 (GATCATCCACAGTTCAGCTCC), j2A.30 (TAATGATGAGAGAACACCCGC); jak2b was mapped with a codominant sequence polymorphism55 in the PCR product generated by the j2B.M1 (AAGAAAGTCTGTCCGCTGTCTTCACATGTC), j2B.M4 (CGCGCCAGCACTGCTAGCATAACAGAAACC) primer pair. Linkage was determined by comparison of a given marker on a C32xSJD haploid panel consisting of 96 individuals that were genotyped against approximately 600 markers for close correlation of segregation patterns53,56-59 using the program “MapMaker”60 (Massachusetts Institute of Technology, Cambridge, MA) and the program “mapmanager”61 (Roswell Park Cancer Institute, Buffalo, NY).
Assessment of linkage of jak2a to the cab andmot mutations.
Homozygous diploid embryos were generated as described46from a ABxDAR hybrid mother carrying a single mutant allele ofcabtl236 and from a ABxDAR hybrid mother heterozygous for mottm303c and scored for an erythropoietic phenotype. These embryos were typed for the segregation of the j2A.29/j2A.03 marker from the jak2a 3′ UTR with the mutant phenotype, based on a size polymorphism evident between the AB and DAR strains.
RESULTS
Cloning of zebrafish jak2 genes.
To isolate JAK homologs in zebrafish, PCR was used to amplify cDNA derived from mixed-stage embryonic mRNA with JAK-directed degenerate primer pairs, yielding 8 distinct PTK fragments corresponding to members from 3 kinase subfamilies (Fig 1a). Sequence comparison of the PCR products to known JAK genes suggested that 2 of the PTKs were closest in identity to mammalian JAK2; these were termedjak2a (HD-1) and jak2b (HD-71). A third, HD-9, was most similar to mammalian JAK1 and was designatedjak1. This manuscript will focus on the 2 jak2 genes we detected in the zebrafish. The embryonic expression of jak1will be reported elsewhere (Oates and Wilks, manuscript in preparation).
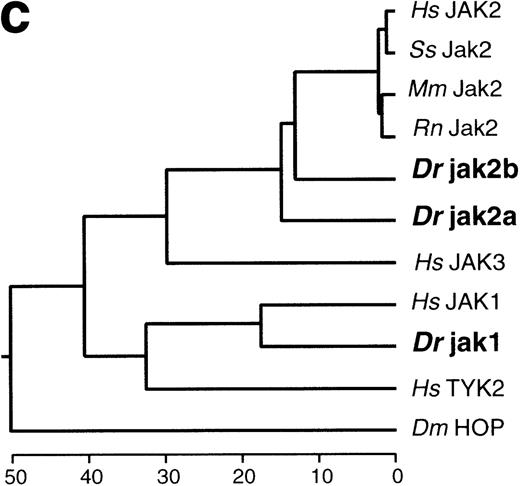
JAK genes from the zebrafish. (a) Alignment of deduced amino acid sequences for the PTKs identified by PCR from zebrafish cDNA. The alignment was generated using CLUSTAL.50 The clone name is listed as HD on the right of the sequences, followed by the name of the gene that gave the highest BLAST score in database similarity searches.49 The JAK-specific sequence elements in Hanks motif VIII are boxed. (b) Comparison of the zebrafish jak2a and human JAK2 protein. The amino acid sequence (single-letter code) of the jak2a protein from zebrafish and the JAK2 protein from human88 is numbered from the putative initiation methionine. The JAK homology (JH) domains are indicated by brackets and are labeled JH1 through JH7 according to Harpur et al.3 Conserved motifs in the catalytic JH1 domain are labeled in roman numerals. Conserved motifs in the kinase related domain (JH2) are labeled in roman numerals with the subscript a. A series of conserved residues mutated in murine Jak2 without phenotypic effect in IFN signaling8 are indicated by an asterisk above the amino acid, and are labeled according to Kohlhuber et al8 by capital letters. The site of the E665K mutation within JH2 that hyperactivates the catalytic activity31 of murine Jak2 and D melanogaster HOP is marked with a solid arrowhead. The autophosphorylation site in the JH1 domain of murine Jak2, which is required for catalytic activity,89 is marked by an open arrowhead. The structure of the variant jak2aβcDNA is marked by an arrow at the site of predicted translational termination due to alternate splicing.62 (c) Phylogeny of the JAK gene family. The amino acid sequences of zebrafish (Danio rerio, Dr) jak2a and jak2b were aligned across the known region of jak2b, consisting of most of JH2 and all of JH1, with JAK2 proteins from human (Homo sapiens, Hs),88 pig (Sus scrofa,Ss),90 mouse (Mus musculus,Mm),3 and rat (Rattus norvegicus,Rn)91; all other members of the JAK family from human (Hs),10,41,42 the zebrafish (Dr) jak1 sequence (this study), and the sequence of the D melanogaster (Dm) JAK homolog,42 hopscotch, using the CLUSTAL alignment algorithm.50This alignment was used to construct a dendrogram with the maximum parsimony options of the DNAstar MegALIGN application to infer the likely genealogy of the JAK family. The names of the sequences are displayed to the right of the dendrogram; zebrafish sequences are in bold.
JAK genes from the zebrafish. (a) Alignment of deduced amino acid sequences for the PTKs identified by PCR from zebrafish cDNA. The alignment was generated using CLUSTAL.50 The clone name is listed as HD on the right of the sequences, followed by the name of the gene that gave the highest BLAST score in database similarity searches.49 The JAK-specific sequence elements in Hanks motif VIII are boxed. (b) Comparison of the zebrafish jak2a and human JAK2 protein. The amino acid sequence (single-letter code) of the jak2a protein from zebrafish and the JAK2 protein from human88 is numbered from the putative initiation methionine. The JAK homology (JH) domains are indicated by brackets and are labeled JH1 through JH7 according to Harpur et al.3 Conserved motifs in the catalytic JH1 domain are labeled in roman numerals. Conserved motifs in the kinase related domain (JH2) are labeled in roman numerals with the subscript a. A series of conserved residues mutated in murine Jak2 without phenotypic effect in IFN signaling8 are indicated by an asterisk above the amino acid, and are labeled according to Kohlhuber et al8 by capital letters. The site of the E665K mutation within JH2 that hyperactivates the catalytic activity31 of murine Jak2 and D melanogaster HOP is marked with a solid arrowhead. The autophosphorylation site in the JH1 domain of murine Jak2, which is required for catalytic activity,89 is marked by an open arrowhead. The structure of the variant jak2aβcDNA is marked by an arrow at the site of predicted translational termination due to alternate splicing.62 (c) Phylogeny of the JAK gene family. The amino acid sequences of zebrafish (Danio rerio, Dr) jak2a and jak2b were aligned across the known region of jak2b, consisting of most of JH2 and all of JH1, with JAK2 proteins from human (Homo sapiens, Hs),88 pig (Sus scrofa,Ss),90 mouse (Mus musculus,Mm),3 and rat (Rattus norvegicus,Rn)91; all other members of the JAK family from human (Hs),10,41,42 the zebrafish (Dr) jak1 sequence (this study), and the sequence of the D melanogaster (Dm) JAK homolog,42 hopscotch, using the CLUSTAL alignment algorithm.50This alignment was used to construct a dendrogram with the maximum parsimony options of the DNAstar MegALIGN application to infer the likely genealogy of the JAK family. The names of the sequences are displayed to the right of the dendrogram; zebrafish sequences are in bold.
Multiple cDNA clones were recovered from an embryonic cDNA library by DNA hybridization using the PCR product corresponding to jak2aas a probe. Conceptual translation of the resulting sequence contig showed an open reading frame (ORF) of 1,095 amino acids with high similarity across the entire coding region to mammalian JAK2genes (65% identity to Mm JAK23; Fig1b). One cDNA possessed an internal deletion relative to all other cDNAs, consistent with the omission of an exon due to an alternate splicing event.62 The longer form of the transcript was named jak2aα and the shorter, alternately spliced form was termed jak2aβ, consistent with the nomenclature for alternately spliced forms of the mammalianSTAT1 and STAT3 genes.63 64 Multiple cDNA libraries were screened with a jak2b probe and a partial cDNA was isolated 1,967 nucleotides in length containing an ORF of 498 amino acids consisting of the C-terminal region of the protein (data not shown).
Structure of the zebrafish jak2 transcripts.
Examination of the conceptual translation of both zebrafishjak2 genes showed a high degree of sequence conservation. The jak2a protein shows approximately 65% identity to the mammalian JAK2 proteins from mouse, human, rat, and pig. As shown in Fig 1b, by comparison to human JAK2, all recognized structural elements found in mammalian JAK2 proteins are present. This high degree of sequence conservation indicated a strong likelihood of functional conservation. Comparison of the predicted amino acid sequence of the majority of the JH2 domain and the entire JH1 domain found in the jak2b cDNA with other JAK kinases indicated that it, too, is most closely related to mammalian JAK2 proteins.
The mammalian JAK kinases and Drosophila HOP were aligned with the amino acid sequence of jak2a and jak2b over the JH1 and JH2 domains, and a phylogenetic reconstruction of JAK gene evolution was derived, as shown in Fig 1c. The zebrafish jak2 proteins are approximately as different from each other (75% identity) as they are from the mammalian JAK2 proteins, indicating an ancient paralogy. jak2b is slightly more similar to the mammalian proteins (average of 78% identity) than is jak2a (75% identity). However, when nucleotide similarity is assessed across these domains, the zebrafish genes are more similar to each other (72% identity) than either is to any of the mammalian genes (jak2a v mammalian, 67% identity;jak2b v mammalian, 70% identity). Furthermore, there are 27 amino acid positions at which the zebrafish jak2 proteins are identical to each other but different from the mammalian sequence, suggesting the existence of a fish-specific substitution. A striking feature of the jak2 paralogs is their extensive divergence, suggesting that the time since paralog duplication is of similar magnitude to that since divergence of the zebrafish and mammalian lineages. This is reflected in the short distances between the nodes leading to the divergence of the zebrafish and mammalian JAK2 sequences in Fig 1c.
Genetic mapping of the jak genes.
Knowledge of the genetic position of a gene is important in assessing potential linkage to a particular mutation and the identification of candidate genes from the collection of hematopoietic mutants. To generate polymorphic markers for each of the jak genes for genetic mapping, genomic DNA or cDNA sequence was used to design PCR primers for use in single-stranded conformational polymorphism (SSCP) and simple sequence-length polymorphism (SSLP) assays.
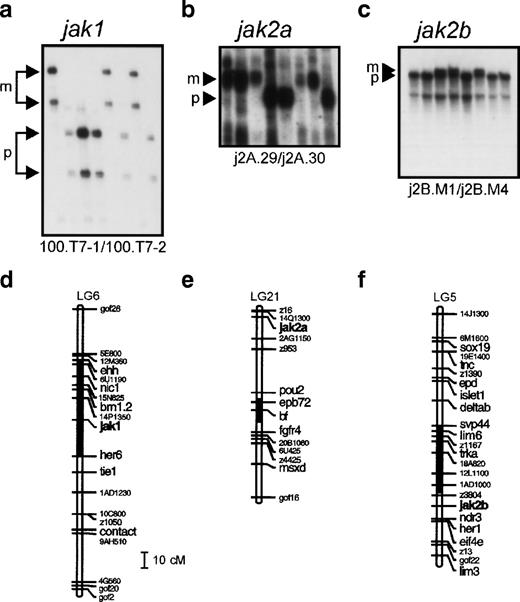
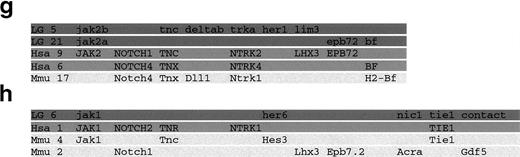
The C32xSJD mapping panel53 was typed with a polymorphic marker for each gene. Using the segregation of the 100.T7-1/100.T7-2 SSLP polymorphism derived from physically linked genomic DNA in 72 meioses (Fig 2a), jak1 was mapped to linkage group 6 (LG6) between 14P.1350.c and her6 (Fig 2d).jak2a was mapped to distal LG21, 1.9 cM proximal to z16/14Q.1300.c (Fig 2e), on the basis of the segregation of the 3′ UTR-derived j2A.29/j2A.03 SSCP polymorphism in 81 meioses (Fig2b). Segregation of an SSCP polymorphism derived from the 3′ UTR of jak 2B (j2B.M1/j2B.M4) in 48 meioses (Fig 2c) was used to place this gene on LG5 approximately equidistant between the z3804 andher1/z4299 markers (Fig 2f). The chromosome segments on which the jak genes are found were aligned with their counterparts from mouse and human, showing extensive conservation of synteny (Fig 2g and h).
Genetic mapping of the jak1,jak2a, and jak2b genes of zebrafish. (a) Segregation of the 100.T7-1/100.T7-2 SSLP jak1 polymorphism in the C32xSJD cross. The C32xSJD mapping cross was typed for a polymorphic marker derived from genomic DNA associated with the jak1 gene. PCR products amplified from genomic DNA of the haploid embryos of the C32xSJD mapping panel using the 100.T7-1/100.T7-2 primer pair were analyzed for length differences by denaturing polyacrylamide gel electrophoresis and 8 representative lanes are shown. The size variants of the products were assigned to the maternal (M) or paternal (P) genome at random, and segregation in the panel was scored. (b) Segregation of the j2A.29/j2A.30 SSLP jak2a polymorphism in the C32xSJD cross. PCR products amplified from genomic DNA of the haploid embryos of the C32xSJD mapping panel using the j2A.29/j2A.30 primer pair derived from the jak2a cDNA 3′ UTR were analyzed for length differences as described above and 8 representative lanes are shown. The size variants of the products were assigned to the maternal (M) or paternal (P) genome at random, and segregation in the panel was scored. (c) Segregation of the j2B.M1/j2B.M4 SSCP jak2bpolymorphism in the C32xSJD cross. PCR products from genomic DNA of the haploid embryos of the C32xSJD mapping panel using the j2B.M1/j2B.M4 primer pair derived from the jak2b cDNA 3′ UTR were analyzed for sequence differences by nondenaturing polyacrylamide gel electrophoresis and 8 representative lanes are shown. The size variants of the products were assigned to the maternal (M) or paternal (P) genome at random, and segregation in the panel was scored. (d) Genetic map position of the jak1 gene. Analysis of the segregation of the jak1-associated marker in 72 meiotic events of the C32xSJD mapping panel using the MapMaker and mapmanager programs59 60 and further manual refinement places the zebrafish jak1 locus on LG6. (e) Genetic map position of thejak2a gene. Analysis of the segregation of ajak2a-associated marker in 81 meiotic events of the C32xSJD mapping panel as described above places the zebrafish jak2alocus on LG21. (f) Genetic map position of the jak2b gene. Analysis of the segregation of a jak2b-associated marker in 48 meiotic events as described above places the zebrafish jak2blocus on LG5. (g and h) Synteny of the JAK family loci between the zebrafish, mouse, and human. A schematic diagram of the syntenic relationship between segments of the chromosomes of zebrafish (LG5, 6, and 21), human (Hsa 1, 9, and 6), and mouse (Mmu 2, 4, and 17) that contain members of the JAK gene family. The syntenic segments containing JAK2 homologs are indicated in (a) and those containing JAK1 homologs in (b). Note that genes have been illustrated in the same relative positions on syntenic chromosomes; however, in situ, local gene order may vary between chromosomes. The diagram is not to scale.
Genetic mapping of the jak1,jak2a, and jak2b genes of zebrafish. (a) Segregation of the 100.T7-1/100.T7-2 SSLP jak1 polymorphism in the C32xSJD cross. The C32xSJD mapping cross was typed for a polymorphic marker derived from genomic DNA associated with the jak1 gene. PCR products amplified from genomic DNA of the haploid embryos of the C32xSJD mapping panel using the 100.T7-1/100.T7-2 primer pair were analyzed for length differences by denaturing polyacrylamide gel electrophoresis and 8 representative lanes are shown. The size variants of the products were assigned to the maternal (M) or paternal (P) genome at random, and segregation in the panel was scored. (b) Segregation of the j2A.29/j2A.30 SSLP jak2a polymorphism in the C32xSJD cross. PCR products amplified from genomic DNA of the haploid embryos of the C32xSJD mapping panel using the j2A.29/j2A.30 primer pair derived from the jak2a cDNA 3′ UTR were analyzed for length differences as described above and 8 representative lanes are shown. The size variants of the products were assigned to the maternal (M) or paternal (P) genome at random, and segregation in the panel was scored. (c) Segregation of the j2B.M1/j2B.M4 SSCP jak2bpolymorphism in the C32xSJD cross. PCR products from genomic DNA of the haploid embryos of the C32xSJD mapping panel using the j2B.M1/j2B.M4 primer pair derived from the jak2b cDNA 3′ UTR were analyzed for sequence differences by nondenaturing polyacrylamide gel electrophoresis and 8 representative lanes are shown. The size variants of the products were assigned to the maternal (M) or paternal (P) genome at random, and segregation in the panel was scored. (d) Genetic map position of the jak1 gene. Analysis of the segregation of the jak1-associated marker in 72 meiotic events of the C32xSJD mapping panel using the MapMaker and mapmanager programs59 60 and further manual refinement places the zebrafish jak1 locus on LG6. (e) Genetic map position of thejak2a gene. Analysis of the segregation of ajak2a-associated marker in 81 meiotic events of the C32xSJD mapping panel as described above places the zebrafish jak2alocus on LG21. (f) Genetic map position of the jak2b gene. Analysis of the segregation of a jak2b-associated marker in 48 meiotic events as described above places the zebrafish jak2blocus on LG5. (g and h) Synteny of the JAK family loci between the zebrafish, mouse, and human. A schematic diagram of the syntenic relationship between segments of the chromosomes of zebrafish (LG5, 6, and 21), human (Hsa 1, 9, and 6), and mouse (Mmu 2, 4, and 17) that contain members of the JAK gene family. The syntenic segments containing JAK2 homologs are indicated in (a) and those containing JAK1 homologs in (b). Note that genes have been illustrated in the same relative positions on syntenic chromosomes; however, in situ, local gene order may vary between chromosomes. The diagram is not to scale.
Expression of jak2a in the developing zebrafish.
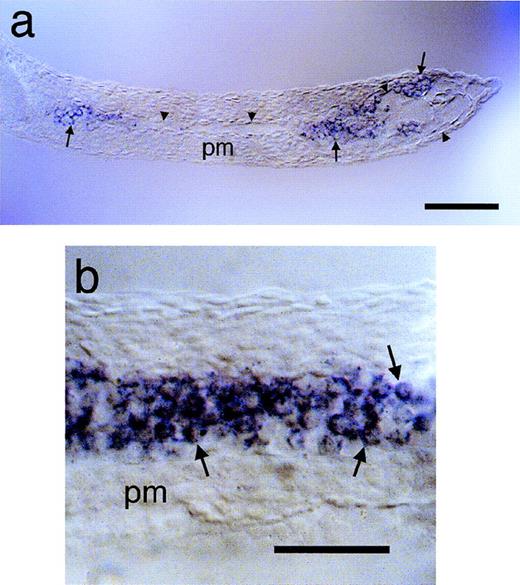
Analysis of the developmental expression pattern of jak2aindicates that it may play a role in hematopoiesis. jak2atranscripts were first detected at a low level throughout the embryo at 10 hpf (Fig3a), persisting until 14 hpf. During this period, the intensity of signal increased in the anterior part of the axis, eventually being strongest in the eyes. By 14 hpf (Fig 3b and c), cells of the medial lateral plate mesoderm expressed jak2a in a pattern consistent with the sites of earliest hematopoietic activity.65 Cells in this region have earlier expressed scl66 andgata1 and gata2 at 11 hpf.67 Costaining ofjak2a with gata1 at 14 hpf and thereafter showed that both genes were expressed in identical regions of the lateral plate mesoderm (data not shown). This suggests that the first cells of the primitive wave of erythropoiesis express jak2a and that this expression is a later event in the commitment to this lineage thangata1 expression. Cells in this region maintained jak2aexpression as they moved from a lateral position to the midline and differentiated in the intermediate cell mass (ICM; Fig 3d), consistent with jak2a expression in proliferating proerythroblasts. By 24 hpf, high level staining was restricted to cells of the anterior ICM, although a low level of expression was detected in the brain and eyes (Fig 3e). The distribution ofjak2a transcript at this stage differs from the hematopoietic expression of the vascular and stem cell marker scl in 2 important respects. As shown in Fig 3f, with white arrowheads, cells in a set of bilateral stripes located more rostrally than the ICM arescl-positive and thought to be persistent hematopoietic progenitor cells66; however, these cells did not expressjak2a. Furthermore, although both scl and jak2atranscripts were detected at high levels in the anterior ICM, onlyscl expression is detected in the posterior ICM (solid arrowhead, Fig 3f). In both of these aspects, the expression ofjak2a resembles that of gata1.67 Becausescl is expressed in both vascular and hematopoietic precursors,66 we wished to establish unambiguously the identity of the cells that expressed jak2a message. Sectioning of embryos immediately postcirculation showed that jak2aexpression was confined to cells contained within the vasculature with a large, rounded morphology (Fig4). These cells also express gata1 and hemoglobin (data not shown, see Detrich et al67) and the embryonicglobin genes (see Fig 6, below), consistent with an erythroblast identity.
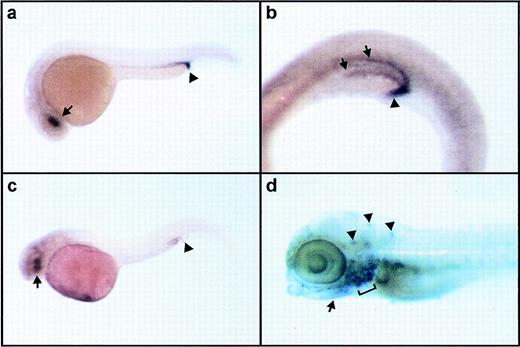
jak2a expression in the developing zebrafish. The expression of jak2a in embryos at various developmental stages was examined by whole mount in situ hybridization. (a) 10 hpf embryo, dorsal view with anterior to the top. jak2a riboprobe gives a widespread, low-level signal. Elevated expression is apparent in the dorsal axis, but this corresponds to the thickest region of the embryo and does not reflect an increase in the density of transcript. (b) 14 hpf embryo, lateral view with anterior to the top and left. Arrowheads indicate the jak2a-riboprobe labeled line of cells in the medial lateral plate mesoderm that gives rise to primitive blood lineages. (c) 14 hpf dorsal view of dissection of dorsal and posterior axial structures. Anterior (A) is to the left and posterior (P) is to the right. jak2a transcript is evident in a narrow ribbon of cells at the medial edge of the lateral plate mesoderm (arrow) that extend to a distinct anterior limit (arrowhead). (d) 20 hpf embryos, lateral view. jak2a riboprobe signal is detected at high level in the medially converging cells of the ICM (arrowheads) and in the eye. A lower level of signal is seen in the remainder of the anterior CNS, but not in the trunk or posterior body. (e) 24 hpf embryo, lateral view showing the labeling of the mature ICM (bracket) and in the eye and anterior CNS at a lower level by jak2a probe. (f) 24 hpf embryos, lateral view showing a comparison of jak2awith scl staining. Both jak2a and scl probes label the anterior ICM (see [e] for reference); however, sclis also detected in a dorso-anterior pair of bilateral stripes (open arrowhead) and in the posterior ICM (solid arrowhead), whereasjak2a is not. Expression of scl is also seen in cells of the CNS. (g) 36 hpf embryo, lateral view showing detection ofjak2a message in circulating primitive erythrocytes (ce), cells can be detected in the axial vessels of the trunk and tail, on the yolk sac, and in the heart (open arrowhead). Elevated signal is also seen in the eyes and in the midbrain (solid arrowhead). (h) 2.5 dpf embryo, lateral view showing detection of jak2a transcript in the finbud (open arrowhead), the midbrain (solid arrowhead), and the eye. Note that, in contrast to (g), there is no jak2a signal from circulating blood. (i) 3.5 dpf larva, lateral view showingjak2a message restricted to the eye and elements of the pharyngeal arches (arrowhead). (j) 8 dpf larva, lateral view.jak2a transcript can be detected in the pronephros (arrow) and in circulating cells lodged in the vasculature of the tail (bracket). A, anterior; aICM, anterior intermediate cell mass; ce, circulating erythroblasts; cv, caudal vascular plexus; e, eye; fb, fin bud; ICM, intermediate cell mass; lpm, lateral plate mesoderm; P, posterior; p, pronephros; pICM, posterior intermediate cell mass.
jak2a expression in the developing zebrafish. The expression of jak2a in embryos at various developmental stages was examined by whole mount in situ hybridization. (a) 10 hpf embryo, dorsal view with anterior to the top. jak2a riboprobe gives a widespread, low-level signal. Elevated expression is apparent in the dorsal axis, but this corresponds to the thickest region of the embryo and does not reflect an increase in the density of transcript. (b) 14 hpf embryo, lateral view with anterior to the top and left. Arrowheads indicate the jak2a-riboprobe labeled line of cells in the medial lateral plate mesoderm that gives rise to primitive blood lineages. (c) 14 hpf dorsal view of dissection of dorsal and posterior axial structures. Anterior (A) is to the left and posterior (P) is to the right. jak2a transcript is evident in a narrow ribbon of cells at the medial edge of the lateral plate mesoderm (arrow) that extend to a distinct anterior limit (arrowhead). (d) 20 hpf embryos, lateral view. jak2a riboprobe signal is detected at high level in the medially converging cells of the ICM (arrowheads) and in the eye. A lower level of signal is seen in the remainder of the anterior CNS, but not in the trunk or posterior body. (e) 24 hpf embryo, lateral view showing the labeling of the mature ICM (bracket) and in the eye and anterior CNS at a lower level by jak2a probe. (f) 24 hpf embryos, lateral view showing a comparison of jak2awith scl staining. Both jak2a and scl probes label the anterior ICM (see [e] for reference); however, sclis also detected in a dorso-anterior pair of bilateral stripes (open arrowhead) and in the posterior ICM (solid arrowhead), whereasjak2a is not. Expression of scl is also seen in cells of the CNS. (g) 36 hpf embryo, lateral view showing detection ofjak2a message in circulating primitive erythrocytes (ce), cells can be detected in the axial vessels of the trunk and tail, on the yolk sac, and in the heart (open arrowhead). Elevated signal is also seen in the eyes and in the midbrain (solid arrowhead). (h) 2.5 dpf embryo, lateral view showing detection of jak2a transcript in the finbud (open arrowhead), the midbrain (solid arrowhead), and the eye. Note that, in contrast to (g), there is no jak2a signal from circulating blood. (i) 3.5 dpf larva, lateral view showingjak2a message restricted to the eye and elements of the pharyngeal arches (arrowhead). (j) 8 dpf larva, lateral view.jak2a transcript can be detected in the pronephros (arrow) and in circulating cells lodged in the vasculature of the tail (bracket). A, anterior; aICM, anterior intermediate cell mass; ce, circulating erythroblasts; cv, caudal vascular plexus; e, eye; fb, fin bud; ICM, intermediate cell mass; lpm, lateral plate mesoderm; P, posterior; p, pronephros; pICM, posterior intermediate cell mass.
Expression of jak2a in circulating erythroblasts. The expression of jak2a in circulating cells of the primitive wave of hematopoiesis was examined in thin sections of animals after the onset of circulation (26 hpf) after whole mount in situ hybridization. Preparations are oriented with anterior to the left. (a) Transverse section of trunk and tail at the level of the dorsal aorta. Arrowheads demarcate the extent of the vasculature, indicating the anterior dorsal aorta and the posterior vascular sinus.jak2a-positive cells are confined within the vasculature (arrows). (b) Higher magnification of transverse section of the caudal vein showing large, rounded jak2a-positive cells (arrows) within the vasculature. pm, paraxial mesoderm. Scale bars: for (a), 100 μm; for (b), 50 μm.
Expression of jak2a in circulating erythroblasts. The expression of jak2a in circulating cells of the primitive wave of hematopoiesis was examined in thin sections of animals after the onset of circulation (26 hpf) after whole mount in situ hybridization. Preparations are oriented with anterior to the left. (a) Transverse section of trunk and tail at the level of the dorsal aorta. Arrowheads demarcate the extent of the vasculature, indicating the anterior dorsal aorta and the posterior vascular sinus.jak2a-positive cells are confined within the vasculature (arrows). (b) Higher magnification of transverse section of the caudal vein showing large, rounded jak2a-positive cells (arrows) within the vasculature. pm, paraxial mesoderm. Scale bars: for (a), 100 μm; for (b), 50 μm.
The primitive wave of hematopoiesis consists mainly of erythrocytes.jak2a transcripts were detected in maturing erythrocytes until 36 hpf (Fig 3g). Thereafter, jak2a expression in circulating cells decreased rapidly until 2.5 days postfertilization (dpf), at which time jak2a was not present in circulating blood or in any suspected hematopoietic site (Fig 3h). During the next 4 days of larval development, jak2a remained undetectable in blood cells, although low-level expression persisted in the dorsal midbrain, eyes, elements of the jaw, and fin buds (Fig 3h and i). At 8 days,jak2a expression was detected in the pronephros and in blood cells found lodged in the ventral tail veins (Fig 3j). Expression in the site of adult hematopoiesis, the pronephros,65 and in circulating erythrocytes indicates that definitive erythropoiesis gives rise to cells that express jak2a. Although rag1expression in the thymus68 69 was clearly detected by 5 dpf, jak2a was not expressed in the thymus (data not shown).
Thus, the timing and localization of jak2a expression in hematopoietic cells suggests that it may be involved in the transduction of signals into committed erythroblasts of both primitive and definitive lineages. Expression of jak2a in the brain and eyes suggests that intracellular signaling in these locations may also use the jak2a protein; however, the precise location of jak2a transcript in these structures was not determined.
Expression of jak2b in the developing zebrafish.
Northern blot analysis using total RNA indicated that jak2b was expressed at a very low level during embryogenesis (data not shown). Whole mount in situ hybridization demonstrated that expression at 24 hpf was restricted to the lens and the nephritic duct (Fig 5a and b), persisting until 48 hpf (Fig 5c). The rostral extent of staining ofjak2b in the nephritic duct (Fig 5b) is equivalent to that seen at 24 hpf with a probe to the ret gene.70 Low-level expression of jak2b was seen in the fin buds in embryos at 2.5 dpf, coincident with jak2a expression, but by 3.5 dpf, nojak2b transcript was detectable by this method (data not shown). At 5 dpf, low-level jak2b expression was seen in the gill arches, elements of the jaw, and the anterior and posterior lateral line, persisting until 8 dpf (Fig 5d). Thus, based on thedistribution of transcript, jak2b might play a role in signaling during embryonic lens and nephritic duct development and in signaling in the larval lateral line and gills, but not in the development of the hematopoietic system.
jak2b expression in the developing zebrafish. The expression of jak2b in embryos at various developmental stages was examined by whole mount in situ hybridization. All embryos and larvae are shown with anterior to the left and dorsal to the top of the frame. (a) 24 hpf embryo, lateral view showing detection of jak2b message restricted to the lens of the eye (arrow) and in the nephritic ducts (arrowhead). (b) 24 hpf embryo, ventro-lateral view at higher magnification, showing the region of the nephritic duct stained by the jak2b riboprobe. The bilaterally symmetric nephritic ducts are visible (arrows) as is the distal tip of the ducts at the proctodeum (arrowhead). (c) 48 hpf embryo, lateral view showing expression of jak2b persisting in the lens of the eye (arrow) and decreasing from earlier levels in the nephritic ducts (arrowhead). (d) 8 dpf larva, lateral view at higher magnification, indicating detection of a low level ofjak2b message in elements of the jaw (arrow), in the developing gills (bracket), and in the anterior lateral line (arrowheads). Expression of jak2b in the posterior lateral line is evident, but is not shown in this figure.
jak2b expression in the developing zebrafish. The expression of jak2b in embryos at various developmental stages was examined by whole mount in situ hybridization. All embryos and larvae are shown with anterior to the left and dorsal to the top of the frame. (a) 24 hpf embryo, lateral view showing detection of jak2b message restricted to the lens of the eye (arrow) and in the nephritic ducts (arrowhead). (b) 24 hpf embryo, ventro-lateral view at higher magnification, showing the region of the nephritic duct stained by the jak2b riboprobe. The bilaterally symmetric nephritic ducts are visible (arrows) as is the distal tip of the ducts at the proctodeum (arrowhead). (c) 48 hpf embryo, lateral view showing expression of jak2b persisting in the lens of the eye (arrow) and decreasing from earlier levels in the nephritic ducts (arrowhead). (d) 8 dpf larva, lateral view at higher magnification, indicating detection of a low level ofjak2b message in elements of the jaw (arrow), in the developing gills (bracket), and in the anterior lateral line (arrowheads). Expression of jak2b in the posterior lateral line is evident, but is not shown in this figure.
Analysis of jak2a expression in the zebrafish hematopoietic mutants.
Mutations that disrupt hematopoiesis have been identified in zebrafish.36,37,47,67 The majority of these mutations were discovered by screening for the presence and color of circulating erythroblasts; hence, the majority represents genes required in erythropoiesis. Because the screens were performed at developmental stages up to 5 dpf, it seems likely that the target of the screen was erythroblasts of the primitive cohort.37 Examination of mutant phenotypes allows mutant genes to be categorized into a scheme of erythroblast development as outlined by Orkin and Zon.71Embryos from selected mutant lines were examined for perturbation ofjak2a expression by in situ hybridization at various time points before and/or after the onset of a visible phenotype. A summary of the results of this investigation is presented in Table 1, and the results are described below in detail.
Expression of jak2a in Hematopoietic Mutants
| Mutant* . | Allele† . | Age of Onset‡ . | Stages Examined . | jak2a Expression1-153 . |
|---|---|---|---|---|
| cloche (clo) | m39 | 12 hpf | 13, 24, 48 hpf | Absent |
| spadetail (spt) | b104 | 12 hpf | 14, 19, 24 hpf | Absent |
| frascati (frs) | tm130d, tq223 | 2-4 dpf | 24, 36, 48 hpf, 3.5 dpf | Normal |
| chablis (cha) | tu245/tu242e | 2-3 dpf | 24, 36 hpf | Normal |
| retsina (ret) | tr217 | 3-4 dpf | 18, 24, 36, 48 hpf | Normal |
| cabernet (cab) | tl236 | 4 dpf | 24, 28 hpf | Normal |
| sauternes (sau) | ty121 | 2 dpf | 24, 26, 32, 36 hpf | Normal |
| wießherbst (weh) | th238 | 2 dpf | 24 hpf | Normal |
| chardonnay (cdy) | te216 | 2 dpf | 24, 36 hpf | Normal |
| chianti (cia) | tu25f | 3 dpf | 24, 36, 48 hpf | Normal |
| Mutant* . | Allele† . | Age of Onset‡ . | Stages Examined . | jak2a Expression1-153 . |
|---|---|---|---|---|
| cloche (clo) | m39 | 12 hpf | 13, 24, 48 hpf | Absent |
| spadetail (spt) | b104 | 12 hpf | 14, 19, 24 hpf | Absent |
| frascati (frs) | tm130d, tq223 | 2-4 dpf | 24, 36, 48 hpf, 3.5 dpf | Normal |
| chablis (cha) | tu245/tu242e | 2-3 dpf | 24, 36 hpf | Normal |
| retsina (ret) | tr217 | 3-4 dpf | 18, 24, 36, 48 hpf | Normal |
| cabernet (cab) | tl236 | 4 dpf | 24, 28 hpf | Normal |
| sauternes (sau) | ty121 | 2 dpf | 24, 26, 32, 36 hpf | Normal |
| wießherbst (weh) | th238 | 2 dpf | 24 hpf | Normal |
| chardonnay (cdy) | te216 | 2 dpf | 24, 36 hpf | Normal |
| chianti (cia) | tu25f | 3 dpf | 24, 36, 48 hpf | Normal |
The hematopoietic phenotype of the mutants examined here are described in Ransom et al,36 with the exception ofsau,38,clo,47,66,72,73 andspt.73 above).
The strongest allele was chosen for analysis of weh andret, whereas the weaker allele of sau was used. Thecha alleles tu245 and tu242e are believed to be clonal, and the frs alleles tm130d and tq223were tested both as homozygotes and trans-heterozygotes without observable differences.
The time in development when a clutch can be unambiguously scored as containing homozygous mutant embryos, taken from Ransom et al36 with the exception of clo and spt (see
Clutches of embryos from mutant incrosses were scored for disruption of jak2a expression by comparison with wild-type embryos of the same developmental stages. Absent indicates that there was nojak2a expression in any hematopoietic tissue at the stages examined. Normal indicates that there was no variation within a clutch or when compared with wild-type clutches.
Expression of jak2a in a Hemangioblast mutant,cloche.
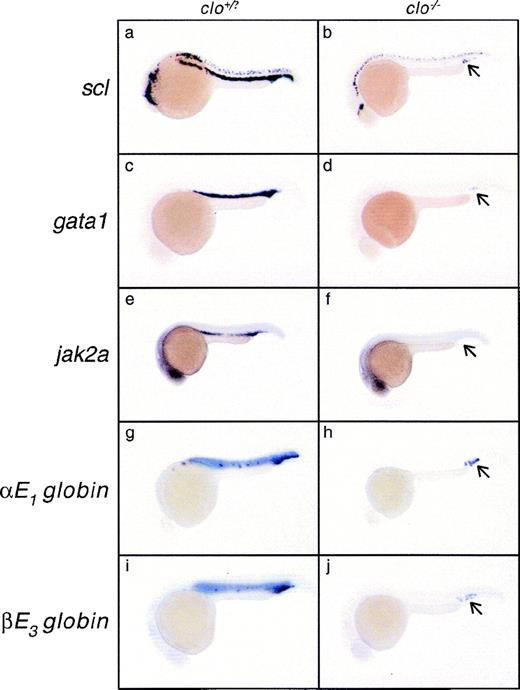
Homozygous cloche (clo) animals fail to produce blood or vasculature and die as embryos.47,66,72,73 Consistent with a general failure in clo mutants to produce cells of the hematopoietic lineage, jak2a expression in the hematopoietic lateral plate mesoderm was not initiated at 13 hpf in approximately one quarter of the embryos from a given clutch and neither was it detected in hematopoietic tissue at any stage examined thereafter (Table 1), although jak2a central nervous system (CNS) expression appears normal. The expression of jak2a in the clo mutant background was compared with the expression of other lineage and stage specific markers at 24 hpf (Fig 6). Clutches of embryos from heterozygous incrosses were examined before the onset of circulation by in situ hybridization for expression of the stem cell markerscl,66 the immature erythroblast markergata1,67 jak2a, and the embryonic α and β globins αe1 globin and βe3 globin, markers of primitive erythrocytic differentiation (Brownlie et al, manuscript in preparation). Three quarters of the embryos in a given clutch showed strong signal in the ICM from all gene probes (Fig 6a, c, e, g, and i). One quarter of embryos in a given clutch displayed a distinctive phenotype involving the dramatic reduction of signal from the ICM (Fig 6b, d, f, h, and j). In approximately 50% of the embryos that displayed a loss of expression in the ICM, persistent expression of scl, gata1, αe1 globin, and βe3 globin could be seen in 5 to 10 cells in the caudal part of the anterior ICM that appear to escape theclo−/− hematopoietic block (Fig 6b, d, h, and j; see Stainier et al47). However, ICM expression ofjak2a was not observed in any embryo that also lacked anterior ICM expression (n = 53, 4 independent experiments).
Comparison of scl, gata1, jak2a,e1 globin, and βe3 globinexpression in cloche mutant embryos. Clutches of embryos derived from heterozygous clo parents were raised for 24 hours and assayed for the expression of various hematopoietic and erythroid marker genes: scl (a and b); gata1 (c and d);jak2a (e and f); e1 globin (g and h); and βe3 globin (i and j). Embryos are displayed in a lateral position with anterior to the left and dorsal to the top of each panel. Approximately three quarters of the embryos in a given clutch had the wild-type expression pattern of the gene in question (a, c, e, g, and i); note the prominent staining of the ICM. Approximately one quarter of the embryos in a given clutch showed a near or total absence of all hematopoietic marker gene expression in the ICM (b, d, f, h, and j); presumably, these are clo homozygotes. In approximately one half of mutant embryos, from 5 to 10 cells in the ICM express thescl, gata1, e1 globin, and βe3 globin marker genes (arrow in b, d, h, and j). No jak2aexpression was observed in this area in any mutant embryo (f), ie, an embryo also lacking jak2a expression in the rostral part of the anterior ICM.
Comparison of scl, gata1, jak2a,e1 globin, and βe3 globinexpression in cloche mutant embryos. Clutches of embryos derived from heterozygous clo parents were raised for 24 hours and assayed for the expression of various hematopoietic and erythroid marker genes: scl (a and b); gata1 (c and d);jak2a (e and f); e1 globin (g and h); and βe3 globin (i and j). Embryos are displayed in a lateral position with anterior to the left and dorsal to the top of each panel. Approximately three quarters of the embryos in a given clutch had the wild-type expression pattern of the gene in question (a, c, e, g, and i); note the prominent staining of the ICM. Approximately one quarter of the embryos in a given clutch showed a near or total absence of all hematopoietic marker gene expression in the ICM (b, d, f, h, and j); presumably, these are clo homozygotes. In approximately one half of mutant embryos, from 5 to 10 cells in the ICM express thescl, gata1, e1 globin, and βe3 globin marker genes (arrow in b, d, h, and j). No jak2aexpression was observed in this area in any mutant embryo (f), ie, an embryo also lacking jak2a expression in the rostral part of the anterior ICM.
Expression of jak2a in an early onset hematopoietic mutant, spadetail.
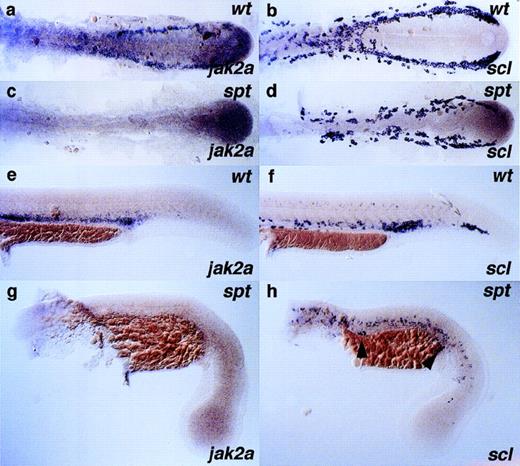
In contrast to clo, embryos homozygous for thespadetail (spt) mutation specify and differentiate vasculature correctly.73 However, the hematopoietic program is severely affected, with few, if any primitive erythroblasts reaching maturity. Clutches of embryos from spt heterozygous parents were examined for jak2a expression throughout the first 24 hours of development (Fig 7). Despite the expression of the stem cell marker scl in spt mutants in the lateral plate mesoderm at 14 somites (Fig 7d), and in contrast to wild-type embryos at this stage (Fig 7a), jak2a transcripts could not be detected in the spt homozygotes in regions of hematopoietic activity (Fig 7c). As spt homozygotes developed,scl expression in the lateral plate decreased dramatically, indicating a failure to maintain a population of early stem cells (Fig7h). Occasionally in spt homozygotes, isolated scl,gata1, and embryonic globin-positive cells were visible in the ICM, as shown by arrowheads in Fig 7h (and data not shown, see Thompson et al73), indicating the emergence of a more mature primitive erythroid cell. However, jak2a message does not accumulate in the corresponding locations or in any hematopoietic tissue at the stages examined (Fig 7g). Combined with the data fromclo mutants, this result indicates that, within the boundaries of the sensitivity of the technique, the expression of erythroid markers in the escaper cells of the caudal part of the anterior ICM is not accompanied by jak2a expression.
Expression of jak2a is perturbed in embryos with mutation in the spadetail gene. Clutches of embryos derived from heterozygous spt parents were raised for 14 and 24 hours and assayed for the expression of stem cell and erythroid marker genesscl (b, d, f, and h) and jak2a (a, c, e, and g), respectively. The spt mutant can be unambiguously scored at the developmental stages presented here by the loss of trunk somites; embryos shown in (c), (d), (g), and (h) are spt homozygotes. Note the loss of jak2a staining at all stages in thespt mutant embryos. Embryos in (a) through (d) are dissected and flat mounted, with the anterior to the left; all other embryos (e) through (h) are displayed in a lateral position with anterior to the left and dorsal to the top of each panel.
Expression of jak2a is perturbed in embryos with mutation in the spadetail gene. Clutches of embryos derived from heterozygous spt parents were raised for 14 and 24 hours and assayed for the expression of stem cell and erythroid marker genesscl (b, d, f, and h) and jak2a (a, c, e, and g), respectively. The spt mutant can be unambiguously scored at the developmental stages presented here by the loss of trunk somites; embryos shown in (c), (d), (g), and (h) are spt homozygotes. Note the loss of jak2a staining at all stages in thespt mutant embryos. Embryos in (a) through (d) are dissected and flat mounted, with the anterior to the left; all other embryos (e) through (h) are displayed in a lateral position with anterior to the left and dorsal to the top of each panel.
Expression of jak2a in late onset hematopoietic mutant zebrafish.
Expression of jak2a was examined by in situ hybridization in zebrafish embryos carrying mutations in the frascati(frs), chablis (cha), retsina(ret), weißherbst (weh), cabernet(cab), sauternes (sau), chardonnay(cdy), and chianti (cia) genes, which display a late phenotypic onset.36 In all mutants examined,jak2a expression was found in the cells of the ICM at 24 hpf and in circulating erythroblasts until 48 hpf, as seen in wild-type clutches (Table 1). In summary, jak2a expression is perturbed only in those mutants (clo, spt) that perturb erythropoiesis at early stages in development, supporting the hypothesis that jak2a is expressed in immature primitive erythroblasts in the zebrafish.
Linkage of the jak2a gene to the hematopoietic mutants.
As described above, jak2a was mapped to the distal tip of LG21 (Fig 2b and e). Comparison of the map position of jak2a with those of the mutants with known positions on the zebrafish genetic map showed that jak2a was not linked to cha, cdy,chi, clo, frs, gre, mon,pin, ris, ret, sau, spt, orweh (data not shown).Thus, jak2a is not a candidate gene for any of these mutations. Linkage to additional, currently unmapped hematopoietic mutants cab and mot was tested by typing genomic DNA from mutant embryos with thejak2a-associated marker used to map the jak2a gene, as described above. The jak2a polymorphism did not segregate with either the cabtl236 ormottm303c phenotype (data not shown), indicating that jak2a is not linked to these mutations.
DISCUSSION
Recent studies in Drosophila and mouse have shown a role for members of the JAK gene family in the control of growth and differentiation of multiple blood cell lineages and in leukemogenesis. The Jak2 gene is required in mice for successful erythropoiesis and JAK2 is implicated in ALL in human patients. We show here that the zebrafish has 2 jak2 genes that are expressed in the developing embryo and larva; however, only 1, jak2a, is expressed in the erythropoietic system. Our results in wild-type and mutant embryos suggest that jak2a may play an early role in primitive erythropoiesis in the zebrafish and, thus, is likely to represent the functional homolog of the mouse Jak2 gene with respect to hematopoiesis.
Hematological implications of zebrafish jak2a expression.
The timing of jak2a expression in wild-type embryos suggests that the presence of jak2a message defines an intermediate stage in the lineage of the primitive erythrocyte that occurs between the commitment of progenitors and the expression of the end-differentiated phenotype. Furthermore, the transience ofjak2a expression indicates that any signaling into primitive erythroblasts via receptors that use jak2a occurs in a window of time as the erythroblasts mature from 14 hpf to approximately 2 dpf. Expression of jak2a in the developing erythrocytes of wild-type and mutant zebrafish has strong implications for the involvement of jak2a in cytokine signaling in hematopoiesis.
Analysis of jak2a expression in clom39 andsptb104 homozygotes showed that jak2atranscription is not initiated at the normal time, and neither is it present in hematopoietic tissue at subsequent stages. This result is consistent with jak2a expression in cells of the hematopoietic lineage and allows inferences about jak2a function to be made. In some clo mutant embryos, a restricted number of primitive erythrocytes (5 to 10) form in a remnant tail vasculature; these cells are heme-reactive, indicating that a terminally differentiated state has been reached. Likewise, in spt mutants, isolated mature primitive erythroblasts can be detected. In contrast to wild-type erythroblasts, jak2a is not coexpressed with gata1 or the embryonic globins in these cells (Figs 6 and 7), being completely absent from the ICM in every embryo examined, indicating that jak2a expression is not required for the completion of differentiation to a globin/heme-expressing stage in these cells. It may instead be required for an aspect of survival or proliferation, as is the case in the mouse.12,13,22,23 74 Becausejak2b is not expressed in erythropoietic tissue, cells without jak2a are likely to be without JAK2 function. Thus, the surviving primitive erythrocytes of clo and spt mutant embryos would not be expected to be receptive to cytokine or other signals that require jak2. Because it is presently unclear whether these cells represent a normal stage of erythropoiesis that has been unmasked by the absence of clo or spt or whether these cells are a peculiarity of clo and spt mutant embryos, we are unable to make strong conclusions about the requirement for jak2a function in the differentiation of wild-type erythrocytes.
Examination of jak2a expression in embryos from late onset hematopoietic mutants showed that the genetic deficiencies in these fish did not disrupt erythropoietic development before the putative cytokine-receptive stage defined by expression of jak2a. This result is not surprising, because their cell morphology, by analogy to the mouse, suggests a mature post–EPO-dependent stage.71The correlation of a failure to generate or maintain blood cell number with absence or loss of jak2a expression in mutants is consistent with a potential role for this gene in the proliferation or survival of polychromatic erythroblasts.
It is of interest to compare these results with recent findings obtained from gene targeting experiments in the mouse. Mice lacking functional Jak2 exhibit significant defects in primitive and definitive erythropoiesis, although their development is otherwise overtly normal.22,23 This is consistent with the high-level expression of jak2a seen in the erythroid cells of the zebrafish embryo and larva. The erythropenic phenotype of theJak2−/− mice is more severe than mice carrying an Epo or EpoR null mutation,75,76with fewer circulating yolk sac-derived primitive erythrocytes and an earlier block in the progression of definitive erythropoiesis. Committed erythroblasts are present in these animals, but they do not expand and differentiate. Hemoglobinization of definitive erythroid cells is almost abolished in Jak2−/−fetal liver cells,23 but the expression of embryonic globins specific for the primitive cohort appears less affected.22 This phenotype is consistent with the expression and regulation of jak2a in wild-type and mutant zebrafish presented here; thus, the jak2a gene of zebrafish likely represents the functional homolog of the mouse Jak2 gene with respect to its role in hematopoiesis.
Evolutionary relationships among the jak genes.
The presence of 4 JAK genes per mammalian genome fits well with current theories about tetraploidization events early in the vertebrate lineage that suggest 2 successive duplications giving rise to 4 copies of an ancestral chromosome complement.77 Genes of theJAK family in zebrafish map to separate chromosomes, indicating that tandem duplication is not the cause of the extra jak2genes in the zebrafish. Instead, they map to regions in which syntenies are conserved compared with their homologs in mouse and human, a region known as the Katsanis paralogy group.78 This finding extends the observation that large portions of the chromosomes of early vertebrates remain intact, with disturbance mainly from local rearrangement,59,79 and indicates that the cause of the initial JAK2 duplication seems to have been a large-scale event, possibly involving 1 or more chromosomes (Fig 2g and h). If the paralogous duplication took place before the lineage of ray fin and lobe fin fish (ie, tetrapods) diverged, there must have existed a second JAK2 paralog in the genome of both lineages. In this case, there may still be a second JAK2 in existing tetrapod genomes. However, examination of mammalian JAK2 cDNA sequences and ESTs in the databases indicates that all JAK2 proteins from different mammals reported are more than 95% identical to each other and that all cDNAs or ESTs from any given species are, in fact, from the same gene (data not shown). Consideration of JAK1,JAK3, and TYK2 database entries in the same manner indicates that all listed sequences, ignoring splice variants, are orthologous or identical. In conclusion, mounting evidence of the existence of higher numbers of gene family members in zebrafish and other ray finned fish than in mammalian genomes80 81combined with the chromosomal localization data presented above favors the scenario in which duplicate jak2 genes are an innovation specific to ray finned fish.
Comparison and implications of jak2 expression patterns in the zebrafish.
The jak genes of the zebrafish are expressed at a high level in restricted groups of cells in the developing embryo and larva. Of particular interest is the divergence in expression patterns of thejak2a and jak2b genes. The only site of coexpression during development was in the lens of the eye at 18 to 36 hpf (Figs 3e and g and 5a and c). Thus, the regulatory regions of the genes have diverged profoundly in activity, whereas the coding sequence has maintained a high conservation in sequence identity, consistent with the generation of genetic novelty by the alteration of gene expression patterns without radical changes to the biochemical activity of the protein product.82
Widespread expression of mammalian JAK mRNA detected in cell lines and adult tissues by Northern blotting3,10,37,83-85and the propensity of cytokine receptors to use multiple JAK proteins in signaling1 suggested a near ubiquitous expression of these intracellular signaling components. In this view, developmental timing and positional cues would be supplied by the restricted expression of both extracellular signaling molecules and their cognate receptors. However, it is clear from this study that the expression of the appropriate signal transduction components could equally serve these timing and positional functions. Potentially, a closer examination of JAK expression in mammals may show a similar distribution and restriction of transcripts.
Prospects for genetic analysis of vertebrate JAK function using the jak2a gene in zebrafish.
The ability to screen a vertebrate genome for mutations that modifyJAK function, a task that would not be practical in the mouse, requires the identification or production of a mutation in aJAK gene. Linkage analysis presented above indicates thatjak2a is not a candidate gene for the 17 hematopoietic mutants examined. The data on the timing of jak2a expression in wild-type and mutant zebrafish suggest that any role for jak2in erythropoiesis is confined to stages after the onset ofgata1 expression, implying possible jak2a function in cells equivalent to progenitor or proerythroblasts. This, along with the phenotype of a Jak2-deficient mouse, suggests that animals with a jak2a mutation would initially express markers for hematopoietic stem cells (HSCs) and progenitor cells such as scl, lmo2,73 and gata2 in an equivalent manner to wild-type.
Other hematopoietic mutants have not been tested for linkage withjak2a, namely, bloodless, vlt, vmp,tbr, stb, paw, andclb.37,67 Of these, bloodless, vlt, and vmp show a hematopoietic phenotype that appears to be too early for the expected role of jak2a, whereas tbr,stb, paw, and clb appear to act too late. However, because these phenotypes are incompletely characterized, and it is not known whether any of these mutations is a complete loss of function in the gene in question, they cannot be ruled out as presenting potential jak2a mutant phenotypes. Because calculations presented at the conclusion of the large-scale screens indicate that approximately 50% saturation was approached,86 a mutation in the jak2a gene may not have been isolated to date. Of course, it is formally possible thatjak2a does not perform an essential function in the development of the erythrocyte lineage in the zebrafish.
An alternative strategy for the generation of a JAK phenotype would be to use 1 of the leukemogenic JAK alleles known from mammals and flies29-33 to induce a neoplastic state in the zebrafish hematopoietic system. Screening the zebrafish genome for enhancer and suppressor loci of such a phenotype should yield information on genes controlling the initiation and progression of vertebrate hematopoietic neoplasia.
Given the parallels between mouse Jak2 and zebrafishjak2a, it is interesting to speculate on the consequences of the potential subfunctionalization87 of jak2 in the zebrafish, a scenario for gene duplication in which complementary functions (eg, expression domains) can be lost by 2 gene duplicates, making both essential to survival. The expression of jak2b is not consistent with any role for this paralog in erythropoiesis; thus,jak2a appears likely to be the functional homolog of the mammalian JAK2. Mice that can be rescued from their requirement for Jak2 function in erythropoiesis by reconstituting with wild-type hematopoietic stem cells may yield interesting additional phenotypes. One prediction of the studies presented here is that lens and kidneys of the rescued mice may show defects. To generalize from this case, the presence of extra gene copies in the zebrafish would not necessarily prove a hindrance to the analysis of gene function; rather, it may allow access to restricted or late onset phenotypes that might not be observable in the mouse.
In conclusion, the findings of this study underscore the potential use of the zebrafish to model the cytokine functions of mammals. Furthermore, the ability to search the genome of the zebrafish for loci that modify the severity of a hematopoietic phenotype would be invaluable in the analysis of the complex signaling events underlying the regulation of blood growth.
ACKNOWLEDGMENT
The authors thank Jana Stickland for her invaluable help with figures. Thanks are extended also to Cuong Do and to the members of the Growth Regulation and Cytokine Biology Laboratories for many discussions. Many thanks to the members of the Zon lab fish collective for providing mutant embryos and for their support and encouragement. Thanks to Ashley Bruce, Graham Lieschke, and Jensen Hjorth for constructive criticism of the manuscript and to Robert Ho, in whose lab this work was completed.
A.C.O. was supported by an Anti Cancer Council of Victoria Postgraduate Research Scholarship. D.V.I. was supported by an Anti Cancer Council of Victoria Summer Scholarship. E.C.L. is a Pre-Doctoral Fellow of the Howard Hughes Medical Institute. B.H.P. was supported in part by a H.H.M.I. Postdoctoral Fellowship for Physicians. J.H.P. was supported by Grants No. RO1RR10715 and PHS PO1HD22486.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Andrew C. Oates, PhD, Department of Molecular Biology, Princeton University, Washington Road, Princeton, NJ 08544; e-mail: aoates@molbio.princeton.edu.



![Fig. 3. jak2a expression in the developing zebrafish. The expression of jak2a in embryos at various developmental stages was examined by whole mount in situ hybridization. (a) 10 hpf embryo, dorsal view with anterior to the top. jak2a riboprobe gives a widespread, low-level signal. Elevated expression is apparent in the dorsal axis, but this corresponds to the thickest region of the embryo and does not reflect an increase in the density of transcript. (b) 14 hpf embryo, lateral view with anterior to the top and left. Arrowheads indicate the jak2a-riboprobe labeled line of cells in the medial lateral plate mesoderm that gives rise to primitive blood lineages. (c) 14 hpf dorsal view of dissection of dorsal and posterior axial structures. Anterior (A) is to the left and posterior (P) is to the right. jak2a transcript is evident in a narrow ribbon of cells at the medial edge of the lateral plate mesoderm (arrow) that extend to a distinct anterior limit (arrowhead). (d) 20 hpf embryos, lateral view. jak2a riboprobe signal is detected at high level in the medially converging cells of the ICM (arrowheads) and in the eye. A lower level of signal is seen in the remainder of the anterior CNS, but not in the trunk or posterior body. (e) 24 hpf embryo, lateral view showing the labeling of the mature ICM (bracket) and in the eye and anterior CNS at a lower level by jak2a probe. (f) 24 hpf embryos, lateral view showing a comparison of jak2awith scl staining. Both jak2a and scl probes label the anterior ICM (see [e] for reference); however, sclis also detected in a dorso-anterior pair of bilateral stripes (open arrowhead) and in the posterior ICM (solid arrowhead), whereasjak2a is not. Expression of scl is also seen in cells of the CNS. (g) 36 hpf embryo, lateral view showing detection ofjak2a message in circulating primitive erythrocytes (ce), cells can be detected in the axial vessels of the trunk and tail, on the yolk sac, and in the heart (open arrowhead). Elevated signal is also seen in the eyes and in the midbrain (solid arrowhead). (h) 2.5 dpf embryo, lateral view showing detection of jak2a transcript in the finbud (open arrowhead), the midbrain (solid arrowhead), and the eye. Note that, in contrast to (g), there is no jak2a signal from circulating blood. (i) 3.5 dpf larva, lateral view showingjak2a message restricted to the eye and elements of the pharyngeal arches (arrowhead). (j) 8 dpf larva, lateral view.jak2a transcript can be detected in the pronephros (arrow) and in circulating cells lodged in the vasculature of the tail (bracket). A, anterior; aICM, anterior intermediate cell mass; ce, circulating erythroblasts; cv, caudal vascular plexus; e, eye; fb, fin bud; ICM, intermediate cell mass; lpm, lateral plate mesoderm; P, posterior; p, pronephros; pICM, posterior intermediate cell mass.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/8/10.1182_blood.v94.8.2622.420k39_2622_2636/7/m_blod420oate03z.jpeg?Expires=1764997330&Signature=PfqHRJe21CI1U7C8yzU-DHpAs86JBKz3-SYmW7AlBnJLMjtICrcdncyWMc7ViuEE~VsGKjRRi-zvXQsu7zHYs1Y6oHdevG5r~xFXCKlwCZehBDXNkZ2Rx90X8nY6Vi78gTd~WsrlSSXXxYcWxHV4wLeWwCxFxZKo5mNwraBkjmmUE6Fca82AwQFecquBqBKWKe5HSHpBndrgklU-37njOo-FR6m17~7EogiEP0ik~o8In1uRjkEG7fxUsU4yJw6yTmybC9k7mEevXZLZfQe2AQqvF1ZfZJNUPJxufOQ60h3k4jevIYMJrIngh5Cy0Q5kqxUnYVai4YKlM1i-U~zubA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal