Chromosomal abnormalities are present in most (if not all) patients with multiple myeloma (MM) and primary plasma cell leukemia (PCL). Furthermore, recent data have shown that numerical chromosomal changes are present in most individuals with monoclonal gammopathy of undetermined significance (MGUS). Epidemiological studies have shown that up to one third of MM may emerge from pre-existing MGUS. To clarify further possible stepwise chromosomal aberrations on a pathway between MGUS and MM, we have analyzed 158 patients with either MM or primary PCL and 19 individuals with MGUS using fluorescence in situ hybridization (FISH). Our FISH analyses were designed to detect illegitimate IGH rearrangements at 14q32 or monosomy 13. Whereas translocations involving the 14q32 region were observed with a similar incidence (60%) in both conditions, a significant difference was found in the incidence of monosomy 13 in MGUS versus MM or primary PCL. It was present in 40% of MM/PCL patients, but in only 4 of 19 MGUS individuals. Moreover, whereas monosomy 13 was found in the majority of plasma cells in MM, it was observed only in cell subpopulations in MGUS. It is noteworthy that, in a group of 20 patients with MM and a previous MGUS history, incidence of monosomy 13 was 70% versus 31% in MM patients without a known history of MGUS (P = .002). Thus, this study highlights monosomy 13 as correlated with the transformation of MGUS to overt MM and may define 2 groups of MM with possible different natural history and outcome, ie, post-MGUS MM with a very high incidence of monosomy 13 and de novo MM in which other genetic events might be involved. Serial analyses of individuals with MGUS will be needed to validate this model.
MULTIPLE MYELOMA (MM), a disorder in which malignant plasma cells (PC) accumulate in the bone marrow, is responsible for approximately 1% of all cancer-related mortality in Western countries. Specific chromosomal abnormalities have been implicated in the oncogenesis of several hematologic malignancies. Translocations t(9;22) in chronic myeloid leukemia, t(15;17) in acute promyelocytic leukemia, and t(11;14) in mantle cell lymphoma are characteristic examples. In MM, the situation is far less clear. First of all, cytogenetic analyses are hampered by the low proliferative rate of PC. Consequently, 50% to 70% of patients display a normal karyotype, because only normal metaphase bone marrow cells are in cycle and able to be analyzed.1-6 In fact, recent studies based on interphase fluorescence in situ hybridization (FISH) have shown that virtually 100% of MM patients have chromosomal abnormalities in their plasma cells.7-9 The second problem is the absence of any common specific abnormality. Many recurrent chromosomal changes have been described, including gains of odd chromosomes, rearrangements of the 14q32 region, or monosomy 13.1-6,10 11 However, none of these abnormalities is myeloma-specific or found in a large majority of MM patients.
Significant advances have been made with the recent demonstration that most human myeloma cell lines12 and 60% to 75% of MM patients13,14 have an illegitimate rearrangement involving the IGH gene at 14q32. We have recently shown that 60% of MM patients display such a rearrangement, especially through reciprocal translocations, and that the main partner chromosomal regions were 11q13 and 4p16.14,15 Moreover, we have shown that the incidence of these abnormalities was independent of the clinical stage, supporting the hypothesis that they occur early in the natural history of the disease.14 Finally, we have shown that these rearrangements were found in all of the tumor cells in a given patient, strongly implicating them as an early key event preceding clonal expansion.
MM has been clinically and pathophysiologically related to another condition termed monoclonal gammopathy of undetermined significance (MGUS). Analysis of bone marrow in MGUS is usually normal, but in a significant percentage of cases, an increased percentage of PC is found. Individuals with MGUS show a significant risk of progression to MM, with an annual actuarial risk of malignant transformation of 0.8%.16 A recent report from the Mayo Clinic has shown that 33% of newly diagnosed MM patients had a previous MGUS history.17 Analyses of chromosomal abnormalities in individuals with MGUS have shown that numerical changes were present in their PC.18-20 All of these data support the hypothesis of a possible link between MGUS and MM, with MGUS being a premalignant condition that could evolve to overt MM in some individuals with accumulation of further genetic events. A pending question remains as to whether all MM emerge from a pre-existing MGUS (ie, post-MGUS MM) or not (ie, de novo MM).
To gain insight into these questions, we have analyzed by FISH 19 individuals with MGUS, in comparison with 144 patients with MM and 15 with primary plasma cell leukemia (PCL). We searched for a specific abnormality (or abnormalities) that correlated with malignant transformation from MGUS into overt MM. In other words, do detectable genetic differences exist between these 2 conditions? To answer these questions, we looked for 3 of the 4 most common specific translocations involving IGH, ie, t(4;14),14,15,21,22t(8;14),1-6 t(11;14),1-6,14 15 and the most frequent chromosomal loss, ie, monosomy 13, using interphase FISH.
PATIENTS, MATERIALS, AND METHODS
Patients.
We analyzed 19 individuals with MGUS, according to standard criteria: less than 30 g/L of serum M-component, less than 10% PC in bone marrow, absence of lytic bone lesions on radiography, and no anemia or hypercalcemia. Two patients with AL amyloidosis and renal insufficiency, but lacking any symptom of overt MM, were included in this MGUS population. Serum monoclonal Ig levels were stable for at least 12 months. Follow-up ranged from 12 to 101 months. Age at diagnosis ranged from 48 to 86 years (median, 68 years). The M-component was IgGκ in 8 patients, IgGλ in 5 patients, and IgAκ in 6 patients. Pertinent features of these individuals are outlined in Table 1.
Characteristics of Individuals With MGUS
| Isotype . | Level of M-Component (g/L) . | Decrease of Other Ig* . | Follow-Up (mo)† . | % Plasma Cells . | IGH Configuration‡ . | Monosomy 13 (% plasma cells) . |
|---|---|---|---|---|---|---|
| IgGλ | 8.3 | No | 14 | 4 | YY | No (3) |
| IgAκ | 10.6 | No | 101 | 2 | YY | No (4) |
| IgGκ | 12 | M | 12 | 4 | t(11;14) | No (3) |
| IgAκ | 12 | No | 14 | 5 | t(11;14) | No (2) |
| IgAκ | 13.3 | No | 23 | 6 | YY | No (1) |
| IgAκ | 13.7 | No | 15 | 8 | YGO | No (2) |
| IgGκ | 14 | A | 98 | 4 | YY | Yes (18) |
| IgAκ | 14.4 | M | 19 | 8 | Y | No (2) |
| IgAκ | 14.6 | No | 17 | 2 | YGO | No (2) |
| IgGκ | 15 | No | 18 | 3 | YGO | Yes (33) |
| IgGκ | 17.5 | M | 12 | 2 | Y | No (3) |
| IgGλ | 17.8 | M | 66 | 6 | YGO | Yes (40) |
| IgGλ | 19.2 | M | 53 | 5 | YG | No (4) |
| IgGκ | 21 | No | 35 | 2 | YY | No (1) |
| IgGλ | 23 | M | 43 | 4 | t(11;14) | No (3) |
| IgGκ | 23.1 | M | 52 | 3 | YGO | No (4) |
| IgGκ | 23.5 | No | 74 | 3 | YGO | No (1) |
| IgGλ | 24.6 | No | 14 | 1 | t(4;14) | Yes (80) |
| IgGκ | 28 | No | 14 | 1 | YY | No (1) |
| Isotype . | Level of M-Component (g/L) . | Decrease of Other Ig* . | Follow-Up (mo)† . | % Plasma Cells . | IGH Configuration‡ . | Monosomy 13 (% plasma cells) . |
|---|---|---|---|---|---|---|
| IgGλ | 8.3 | No | 14 | 4 | YY | No (3) |
| IgAκ | 10.6 | No | 101 | 2 | YY | No (4) |
| IgGκ | 12 | M | 12 | 4 | t(11;14) | No (3) |
| IgAκ | 12 | No | 14 | 5 | t(11;14) | No (2) |
| IgAκ | 13.3 | No | 23 | 6 | YY | No (1) |
| IgAκ | 13.7 | No | 15 | 8 | YGO | No (2) |
| IgGκ | 14 | A | 98 | 4 | YY | Yes (18) |
| IgAκ | 14.4 | M | 19 | 8 | Y | No (2) |
| IgAκ | 14.6 | No | 17 | 2 | YGO | No (2) |
| IgGκ | 15 | No | 18 | 3 | YGO | Yes (33) |
| IgGκ | 17.5 | M | 12 | 2 | Y | No (3) |
| IgGλ | 17.8 | M | 66 | 6 | YGO | Yes (40) |
| IgGλ | 19.2 | M | 53 | 5 | YG | No (4) |
| IgGκ | 21 | No | 35 | 2 | YY | No (1) |
| IgGλ | 23 | M | 43 | 4 | t(11;14) | No (3) |
| IgGκ | 23.1 | M | 52 | 3 | YGO | No (4) |
| IgGκ | 23.5 | No | 74 | 3 | YGO | No (1) |
| IgGλ | 24.6 | No | 14 | 1 | t(4;14) | Yes (80) |
| IgGκ | 28 | No | 14 | 1 | YY | No (1) |
M means a decrease of IgM level and A a decrease of IgA level.
Follow-up from the diagnosis of MGUS.
Y means the presence of 1 yellow signal, corresponding to the colocalization of 1 green (G) and 1 orange (O) signal. Thus, YGO refers to the presence of 1 normal chromosome 14 and a second 1 bearing an illegitimate IGH rearrangement (separation of the green and orange signals).
These 19 individuals with MGUS were compared with a series of 158 patients with plasma cell disorders, including 13 indolent MM,23 17 newly diagnosed stage I MM,24 27 stage II, 58 stage III patients, 15 primary PCL patients, and 28 patients at relapse. Except for these last patients, who were studied at first relapse (after achievement of at least a partial response), all of the other patients were studied before any treatment. These patients have been previously described in detail.14 Moreover, to better evaluate the tumor mass, we recorded the serum β2-microglobulin for most and correlated it with chromosomal findings.
PC purification.
Bone marrow aspirates were collected into heparin as an anticoagulant. Mononuclear cells were separated by density gradient centrifugation (Ficoll-Hypaque). For the first 72 patients, malignant plasma cells were identified using control probes specific for previously detected numerical abnormalities.14 Because CD138 is expressed only on PC within the bone marrow, we then used the anti-CD138 B-B4 monoclonal antibody to obtain highly purified plasma cells as previously described25 in the subsequent 86 patients. Mononuclear cells were washed in phosphate-buffered saline (PBS) and then incubated with anti-CD138 antibodies for 15 minutes at 4°C. After 2 washes in PBS, magnetic beads coated with IgG1 (Miltenyi Biotec, Paris, France) were added, and PC were purified on columns using a magnet, according to the manufacturer’s instructions. After a final wash in PBS, the percentage of PC was determined on cytocentrifuge slides (median, 93%; range, 78% to 99%). Purified cells were then incubated in hypotonic KCl and fixed in methanol/acetic acid (3/1). In MGUS, PC were also purified using the anti-CD138–coated magnetic beads. Because nonclonal PC may persist in the bone marrow of MGUS patients (even if usually at a low percentage; data not shown), we performed interphase FISH with centromeric probes specific for the main additional chromosomes, ie, 3, 7, 9, 11, 15, 17, 18, and X, to evaluate the number of clonal PC (see below).
FISH.
FISH was performed as previously reported.14,15 Briefly, 50 ng of each unique sequence probe was ethanol-precipitated with 1 μg of Cot-1 DNA. After resuspension in Hybrisol VII (Oncor, Gaithersburg, MD), probes were denatured at 75°C for 10 minutes and placed at 37°C for 15 to 30 minutes for preannealing. After overnight hybridization, slides were washed and nuclei were counterstained with 4′,6-diamidino-2-phenylindole in antifade. Repetitive centromeric probes were used as previously reported.14
Probes.
The IGH locus was analyzed using 3 probes, as previously described14: a BAC probe (158A2) previously described,26 the Y6 YAC, and the Ig10 cosmid probes, kindly provided by Dr F. Matsuda (Kyoto University, Kyoto, Japan) and Dr T.H. Rabbits (Medical Research Council, Cambridge, UK), respectively. The FGFR3 locus was analyzed using the PAC probe described by Chesi et al.21 The probes were validated in the OPM2 and NCI-H929 human myeloma cell lines (HMCL), as previously reported.15 Translocation t(4;14) was shown by a fusion signal on the der(14) (Fig 1A). TheMYC locus was analyzed using 2 YAC probes (I2 and P72; generously provided by Dr M.L. Veronese, Jefferson Cancer Institute, Philadelphia, PA). These probes have been validated in 15 cases of Burkitt’s lymphoma with t(8;14). The translocation was shown as 1 or 2 fusion signals on the der(14) and sometimes on the der(8) (Fig 1B). These different probes were labeled using standard nick translation, with biotin-dUTP (Boehringer Mannheim, Mannheim, Germany), fluorescein isothiocyanate (FITC)-dUTP, Coumarin-dUTP (NEN, Postfach, Germany), Cyanin5-dUTP (Amersham, Les Ulis, France), and SpectrumOrange-dUTP (Vysis, Downers Grove, IL). TheCCND1 probe was provided by Vysis and was validated in mantle cell lymphoma cases and in the U266, XG1, and XG5 HMCL (Fig 1C). We first looked for illegitimate IGH rearrangements using the Y6 and Ig10 probes. We then looked for specific 14q32 translocation partners, even in patients without illegitimate IGHrearrangements, to assess the specificity of our FISH methodology in detecting 14q32 translocations. In this second step, the 14q32 region was analyzed using a combination of the Ig10 and 158A2 probes (thus covering the whole constant domain), labeled in green, and the partner regions were analyzed using the specific probes labeled in orange. Finally, chromosome 13 was analyzed using 2 probes: a D13S319-probe mapping at 13q14 provided by Vysis and a YAC probe mapping at 13q31. FISH results were compared with karyotype when available (clonal abnormalities in 38 of 77 patients analyzed).
(A) Photograph of a malignant PC with t(4;14) and deletion 13q. The 4p16 (FGFR3) probe is labeled in red, whereas the 14q32 probes (Ig10 and 158A2) are labeled in green and the 13q32 probe in white/grey. The t(4;14) is represented by the fusion signal and the normal chromosomes 4 and 14 by the red and green signals, respectively. Only 1 white signal is observed, reflecting deletion 13q. (B) A plasma cell of a patient with t(8;14)(q24;q32). The translocation is identified by the fusion of 1 green (14q32 probe) and 1 red (8q24) signal. The separated green and red signals correspond to the normal chromosomes 14 and 8, respectively. Finally, this patient did not display chromosome 13 monosomy, because 2 white signals (13q32 probe) were observed. (C) Translocation t(11;14)(q13;q32), not associated with monosomy 13, in a patient with multiple myeloma. The fusion of 1 green (14q32 probe) and 1 red (11q13 probe) signal corresponds to the derivative chromosome 14, whereas the separated green and red signals represent normal chromosomes 14 and 11, respectively. Two white signals (13q32 probe) are observed.
(A) Photograph of a malignant PC with t(4;14) and deletion 13q. The 4p16 (FGFR3) probe is labeled in red, whereas the 14q32 probes (Ig10 and 158A2) are labeled in green and the 13q32 probe in white/grey. The t(4;14) is represented by the fusion signal and the normal chromosomes 4 and 14 by the red and green signals, respectively. Only 1 white signal is observed, reflecting deletion 13q. (B) A plasma cell of a patient with t(8;14)(q24;q32). The translocation is identified by the fusion of 1 green (14q32 probe) and 1 red (8q24) signal. The separated green and red signals correspond to the normal chromosomes 14 and 8, respectively. Finally, this patient did not display chromosome 13 monosomy, because 2 white signals (13q32 probe) were observed. (C) Translocation t(11;14)(q13;q32), not associated with monosomy 13, in a patient with multiple myeloma. The fusion of 1 green (14q32 probe) and 1 red (11q13 probe) signal corresponds to the derivative chromosome 14, whereas the separated green and red signals represent normal chromosomes 14 and 11, respectively. Two white signals (13q32 probe) are observed.
Statistical analyses.
For statistical analyses, we used nonparametric tests, mainly the χ2 method.
RESULTS
IGH analysis and validation of probes.
Illegitimate IGH rearrangements were analyzed with the Y6 and Ig10 probes. Absence of IGH abnormality (ie, 2 normal chromosome 14) was assessed when 2 yellow signals were found (colocalization of Y6 and Ig10 probes, configuration YY). Monosomy 14 or loss of the IGH locus were defined by the presence of only 1 fusion signal (configuration Y). All other configurations corresponded to an illegitimate recombination. The most frequent abnormal figure was 1 yellow signal (ie, the normal chromosome 14), 1 green signal (ie, the derivative chromosome 14 labeled with the Ig10 probe), and 1 orange signal (ie, the derivative partner chromosome) (configuration YGO). In some instances, we found 1 yellow signal (normal chromosome 14) and 1 green signal (derivative chromosome 14) (configuration YG). This disposition was interpreted as a deletion of the IGH 5′ part or as a loss of the derivative partner chromosome. Probes were validated on bone marrow cells obtained from healthy donors. Two thousand cells were scored with the Y6 and Ig10 probes. A normal configuration (2 yellow signals) was found in 96% of nuclei. Cut-off for the assessment of an illegitimate IGH rearrangement was 8.7% (mean + 3 SD). Chromosome 13-specific probes were validated in the same way. Dual-color hybridizations were performed with the 2 probes. Based on analysis of 4,000 peripheral blood mononuclear cells from 10 healthy individuals, cut-off for deletion (mean + 3 SD) was 5.3% for the 13q14 probe and 8.6% for the 13q31 probe.
14q32 abnormalities are observed in 59% of patients with malignant plasma cell disorders, but correlate neither with disease stage nor with progression.
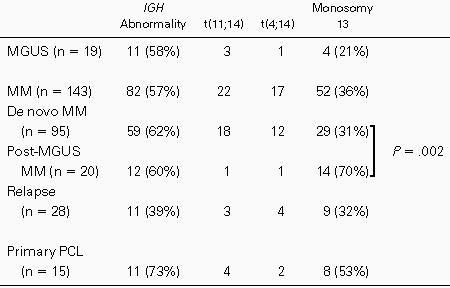
An illegitimate IGH rearrangement was found in 93 of 158 (59%) patients with MM or primary PCL (Table 2). Analysis of 14q32 abnormalities according to disease stage and status did not show any significant difference. An abnormal FISH probe disposition was found in 10 of 13 (77%), 10 of 17 (59%), 17 of 27 (63%), 34 of 58 (59%), 11 of 14 (79%), and 11 of 28 (39%) patients with indolent, stage I, stage II, stage III, primary PCL, and in first relapse, respectively. We then analyzed the 4p16 (FGFR3), 8q24 (MYC), and 11q13 (CCND1) regions. We found a t(11;14)(q13;q32) (ie, IGH-CCND1 fusion) in 26 patients, a t(4;14)(p16;q32) (ie, IGH-FGFR3 fusion) in 19 patients, and a t(8;14)(q24;q32) (ie, IGH-MYC fusion) in 3 patients. As expected, all of the cases with fusion were found in patients with an illegitimate IGH rearrangement. Comparison of FISH results with karyotype for the 38 patients with clonal abnormalities showed discrepancies in 11 patients. In 10 cases, illegitimate IGHrearrangements were observed by FISH, whereas cytogenetics did not detect any 14q32 abnormality. Of note, 5 of these 10 patients displayed a t(4;14)(p16;q32), which is not detectable by cytogenetics. In 1 patient, cytogenetics detected an add(14q32), whereas no illegitimate rearrangement was found by FISH. An extensive analysis of theIGH gene using numerous FISH probes failed to detect illegitimate recombinations. Thus, this patient had a 14q32 abnormality that did not involve the IGH gene.
No significant correlation between specific rearrangement and disease stage was found. We also correlated 14q32 abnormalities with the β2-microglobulin level. In the group of patients with β2-microglobulin less than 3 mg/L (this cut-off value has been determined as highly significant for prognosis in the IFM94 protocol; unpublished data), 24 of 36 (67%) displayed an illegitimateIGH rearrangement, as opposed to 34 of 59 (58%) in the group of patients with β2-microglobulin greater than 3 mg/L. These results confirmed the absence of any statistically significant correlation between 14q32 abnormalities and tumor mass.
Monosomy 13 is observed in 39% of patients with malignant plasma cell disorders, but correlates neither with disease stage nor with progression.
Deletion of the 13q14 region was found in 60 of 158 (38%) patients with either MM or primary PCL. Most of these deletions probably correspond to chromosome 13 monosomies (or at least large deletions), because the 13q31 region was concomitantly lost in 57 of 60 patients. Comparison of FISH and cytogenetic results was in complete agreement: the 16 patients with only one 13q14 signal by FISH displayed monosomy 13 on karyotyping. These chromosome 13 abnormalities were found in 64% to 96% of plasma cells (mean, 83%; SD, 9.94%) and, thus, are considered as secondary events. The incidences of 13q14 deletions were 23% (3/13), 35% (6/17), 44% (12/27), 38% (22/58), 53% (8/15), and 32% (9/28) in patients with indolent, stage I, stage II, stage III, primary PCL, and relapse MM, respectively (no significant difference). Correlation with β2-microglobulin gave similar results: in the low-level group, 14 of 36 patients (39%) displayed a monosomy 13, whereas in the high-level group, 24 of 59 (41%) displayed such a monosomy. Finally, we analyzed the incidence of monosomy 13 within each 14q32 subgroup, ie, patients without IGH abnormality, patients with t(4;14), patients with t(11;14), and patients with otherIGH abnormalities. Clearly, monosomy 13 was more frequent in patients with t(4;14) than in patients with either t(11;14) (P < .002) or lacking 14q32 abnormalities (P < .001).
As opposed to 14q32 translocations, monosomy 13 is less frequently observed in individuals with MGUS.
Besides these 158 patients with either MM or primary PCL, we analyzed 19 individuals with MGUS. Their main bioclinical features are described in Table 1. Illegitimate IGH rearrangements were observed in 11 of 19 cases (58%). This incidence is similar to that found in patients with MM and PCL. Partner chromosome analysis identified t(11;14)(q13;q32) in 3 cases and t(4;14)(p16;q32) in 1 case. No other specific rearrangement was found. Monosomy of chromosome 13 was observed in 4 of 19 individuals (21%), in 80%, 40%, 33%, and 18% of PC, respectively (patients no. 6, 8, 13, and 16). To know if monosomy 13 was present only in clonal PC in these 3 latter patients with low percentage of 13q− PC, we performed FISH experiments combining the D13S319 probe with centromeric probes specific for gained chromosomes or with the 14q32 probes when illegitimate IGHrearrangements were present in these patients (Fig 2A and B). In these 3 patients, we observed clonal chromosomal changes in 86%, 73%, and 82% of PC, thus confirming that monosomy 13 was only present in clonal PC subpopulations. Because the absence of illegitimate IGH rearrangement could be related to a low percentage of clonal PC, we performed in the 5 patients lacking both 14q32 and 13q14 abnormalities FISH analysis with the centromeric probes. Trisomies were detected in 3 of them (trisomies 3, 9, 11, and 15 in 2 cases, and trisomies 9 and 15 in 1 case), respectively, in 83%, 57%, and 62% of PC. Thus, PC of these 3 individuals did not display illegitimate IGH rearrangements.
(A) Analysis of a purified plasma cell of MGUS individual no. 16, using the Y6, IG10, and 13q32 probes. The yellow signal corresponds to the fusion of 1 green (IG10 probe) and 1 red (Y6 probe) on a normal chromosome 14. The second chromosome 14 is rearranged, because the Y6 and the IG10 probes are separated. This clonal plasma cell is disomic for chromosome 13 (2 white signals). (B) Another plasma cell of the same patient. In contrast to the cell shown in (A), this clonal plasma cell (with an illegitimate IGH rearrangement) displays deletion 13q (only 1 white signal).
(A) Analysis of a purified plasma cell of MGUS individual no. 16, using the Y6, IG10, and 13q32 probes. The yellow signal corresponds to the fusion of 1 green (IG10 probe) and 1 red (Y6 probe) on a normal chromosome 14. The second chromosome 14 is rearranged, because the Y6 and the IG10 probes are separated. This clonal plasma cell is disomic for chromosome 13 (2 white signals). (B) Another plasma cell of the same patient. In contrast to the cell shown in (A), this clonal plasma cell (with an illegitimate IGH rearrangement) displays deletion 13q (only 1 white signal).
Monosomy 13, but not 14q32 translocations, discriminates post-MGUS MM from de novo MM.
Recently, Kyle et al17 have shown that 33% of MM in their series occurred in patients with a previous MGUS history. In our current series, an MGUS history was documented in 20 patients with either MM or primary PCL (20/130 patients at diagnosis, ie, 15% of patients). In this subset of patients, 14q32 abnormalities were observed in 12 of 20 (60%) patients (1 case with IGH-CCND1fusion and 1 case with IGH-FGFR3 fusion), thus at a similar incidence to that found in de novo MM and primary PCL (70/110 [64%];P = .97). In contrast, incidence of deletion 13q14 was significantly different in each group: 14 of 20 (70%) in post-MGUS MM versus 29 of 95 (31%) in de novo MM at diagnosis (P = .002).
Monosomy 13 is an important event in the transition from MGUS to MM.
When MGUS and post-MGUS MM were analyzed in parallel, monosomy 13 was found in 4 of 19 of MGUS but in 14 of 20 post-MGUS MM (P = .006), underlining that monosomy 13 appears to represent an important event in the transition from MGUS to MM. In the current study, MGUS and post-MGUS MM are 2 different populations of patients. Thus, serial evaluation of individuals with MGUS will be necessary to validate this point. However, the fact that 3 patients with MGUS had monosomy 13 in subsets of their plasma cells (18%, 33%, and 40%, respectively) emphasizes the possibility of such malignant transformation at the cellular level through monosomy 13.
DISCUSSION
Chromosomal abnormalities are present in virtually 100% of MM patients.7-9 However, their characterization is often hampered by the low proliferative rate of myeloma cells. The situation is even more prominent in PC of individuals with MGUS. Cytogenetic abnormalities are very rarely detected in MGUS. Only 1 report described cytogenetic abnormalities in 11 of 44 individuals with MGUS.6 However, these abnormal karyotypes were strikingly different from those found in MM (high percentage of cytogenetically abnormal cells, but no numerical abnormality, with usually 1 single structural rearrangement). Nevertheless, a few reports based on interphase FISH clearly demonstrated that at least numerical changes are present in the majority of individuals with this indolent condition.18,19 Recently, Zandecki et al20showed that, in contrast to myeloma cells, which are usually really monoclonal, several cytogenetic subclones may coexist within these cases of MGUS. This finding might be interpreted as an ongoing tumorigenic process leading to overt MM with time. This hypothesis is corroborated by the finding that some individuals with MGUS still acquire mutations in their Ig genes, whereas patients with MM exhibit heavily mutated PC.27 This finding reflects the fact that MGUS PC are still under the exposure of the mutator.
To elucidate the oncogenetic events in MM, and especially those involved in the MGUS/MM transition, we analyzed by FISH chromosome 13 and 14q32 rearrangements in 19 individuals with MGUS and 158 patients with either MM or primary PCL. We found illegitimate IGHrecombinations in 11 of 19 individuals with MGUS (58%) and in 93 of 158 patients with plasma cell malignancies (59%). This similar incidence favors the hypothesis that these illegitimate rearrangements occur very early in the tumorigenic process leading to MM and may be not sufficient for complete malignant transformation. Moreover, in contrast to numerical changes, we demonstrated that PC of individuals with MGUS are very homogeneous regarding these structural rearrangements. These numerical chromosomal abnormalities are more likely secondary events. However, because they are not found in all the cases of MGUS or MM, illegitimate IGH recombinations are probably not the only event implicated in tumorigenesis. We were able to characterize the partner chromosome in 4 of 19 MGUS cases. The 11q13 region was involved in 3 cases, whereas the 4p16 region was involved in 1 case. These 2 chromosomal regions are also the most frequently involved in MM and primary PCL in our series.
Another chromosomal abnormality that has been associated with poor prognosis in MM is deletion or monosomy 13.9,11 28 Analysis of 130 patients with MM or primary PCL at diagnosis showed that this abnormality is found in 39% of cases. Even though this abnormality is probably a secondary event, because it is not found in all myeloma cells, it is present in the majority of PC. This finding supports the hypothesis that this chromosomal change occurs as a secondary event, but that it then confers a selective growth or survival advantage. However, this secondary event does not appear to occur during disease progression, because the analysis of patients at relapse did not show a higher incidence (9/28 [32%]). It is noteworthy that deletion/monosomy 13 appears to be associated with some particular 14q32 abnormalities. It is significantly more frequently found in association with illegitimate IGH rearrangements and especially with t(4;14). In contrast, it is less frequently found in patients with t(11;14) or lacking illegitimate IGH rearrangements. The reasons for these associations are currently unknown.
It is of major interest that the incidence of deletion/monosomy 13 was significantly lower in MGUS than in MM and primary PCL, supporting the hypothesis that loss of chromosome 13 may play a role in the MGUS/MM transition. This hypothesis is strongly reinforced by the finding that monosomy 13 incidence is much higher in post-MGUS MM than in so-called de novo MM. A careful literature analysis of large cytogenetics series enabled us to find 1 recent report in which such data were available. Smadja et al29 described 6 patients with post-MGUS MM in a cytogenetic series of 81 patients. Whereas chromosome 13 deletions were present in 33 of 75 (44%) patients without any known MGUS history, monosomy 13 was found in 5 of 6 (83%) cases of post-MGUS MM.
In this model, the loss of chromosome 13 sequences would precipitate long-lived plasma cells (possibly previously modified by other genetic events, such as 14q32 abnormalities) into myeloma cells. In this model, our 3 MGUS individuals with monosomy 13 in only PC subsets may correspond to transitional conditions. In these cases, we could hypothesize that the clone with monosomy 13 will progress and that these 3 patients will rapidly evolve to overt MM. Our model is even more supported by the fact that 4 of the 20 patients with post-MGUS MM had an indolent disease and that monosomy 13 was found in only 1 of these 4 patients. Thus, monosomy 13 would be involved in the transformation of MGUS into overt aggressive MM (Fig3).
Because deletion/monosomy 13 is found more frequently in MM patients with MGUS history, we can hypothesize the existence of 2 kinds of MM. One group would emerge as a malignant transformation of MGUS, and loss of chromosome 13 could participate in this transformation. A second group would be the de novo MM, in which chromosome 13 would have less importance in oncogenesis. In this scheme, the worse prognosis found in patients with loss of chromosome 13 would be the consequence of a different disease, more chemoresistant, as shown in another model, ie, acute myeloid leukemia with previous myelodysplasia and abnormalities of chromosomes 5 and 7. Because MGUS is an asymptomatic condition, some MM may occur in individuals with an undiagnosed MGUS. In the experience of Kyle et al,17 at least 33% of MM would emerge from MGUS. This incidence is close to that of monosomy 13 in MM (35% to 40%). An attractive hypothesis would be to associate these 2 groups of MM (post-MGUS MM and those with monosomy 13) in a single disease. In this model, the poor prognosis associated with monosomy 139,11 28 would be the consequence of the secondary character of these MM, as recently proposed in acute myeloid leukemia with deletion of chromosomes 5 and 7. To answer this important question, long-term follow-up and repetitive appropriate biological evaluations of large cohorts of individuals with MGUS will be needed. Especially serial evaluation of the same patients recognized at MGUS, followed without treatment while asymptomatic and then at the emergence of overt myeloma, should help to clarify the impact of specific chromosome abnormalities. From this point of view, careful follow-up of individuals with MGUS and monosomy 13 in PC subpopulations should be of major interest. The answer might also be given by the Mayo Clinic group by analyzing their cohort of post-MGUS versus de novo MM.
In conclusion, we demonstrated that illegitimate IGHrearrangements are present in patients with MGUS, with exactly the same incidence as in MM and primary PCL patients. In contrast, monosomy 13 seems to be rather infrequent in MGUS, whereas it is found in 40% of MM and in 70% of post-MGUS MM and thus is a good candidate event for the malignant transformation of myeloma cells. However, because most chromosome 13 abnormalities are monosomies or large deletions, many genes may be implicated in this oncogenic process.
Supported in part by the Fondation contre la Leucémie and the Comité Départemental de Loire-Atlantique de la Ligue contre le Cancer. J.-Y.L. is a grant recipient of the Conseil Régional des Pays de Loire.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Hervé Avet-Loiseau, MD, PhD, or Régis Bataille, MD, PhD, Laboratoire d’Hématologie, Institut de Biologie, 9, quai Moncousu, 44093 Nantes Cedex 1, France; e-mail: havetloiseau@chu-nantes.fr.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal