Methylation of the proximal promoter of the ABL1 oncogene is a common epigenetic alteration associated with clinical progression of chronic myeloid leukemia (CML). In this study we queried whether both the Ph′-associated and normal ABL1 alleles undergo methylation; what may be the proportion of hematopoietic progenitors bearing methylated ABL1 promoters in chronic versus acute phase disease; whether methylation affects the promoter uniformly or in patches with discrete clinical relevance; and, finally, whether methylation of ABL1 reflects a generalized process or is gene-specific. To address these issues, we adapted the techniques of methylation-specific PCR and bisulfite-sequencing to study the regulatory regions of ABL1 and other genes with a role in DNA repair or genotoxic stress response. In cell lines established from CML blast crisis, which only carry a single ABL1 allele nested within the BCR-ABL fusion gene, ABL1 promoters were universally methylated. By contrast, in clinical samples from patients at advanced stages of disease, both methylated and unmethylated promoter alleles were detectable. To distinguish between allele-specific methylation and a mixed cell population pattern, we studied the methylation status of ABL1 in colonies derived from single hematopoietic progenitors. Our results showed that both methylated and unmethylated promoter alleles coexisted in the same colony. Furthermore, ABL1 methylation was noted in the vast majority of colonies from blast crisis, but not chronic-phase CML. Both cell lines and clinical samples from acute-phase CML showed nearly uniform hypermethylation along the promoter region. Finally, we showed that ABL1 methylation does not reflect a generalized process and may be unique among DNA repair/genotoxic stress response genes. Our data suggest that specific methylation of the Ph′-associatedABL1 allele accompanies clonal evolution in CML.
CHRONIC MYELOGENOUS leukemia (CML) is a myeloproliferative disorder accounting for approximately 25% of all leukemia cases. Original pioneering work showed that the disease results from clonal expansion of a hematopoietic stem cell.1-3 The initiating molecular event is a reciprocal translocation, t(9;22), cytogenetically detectable by the presence of the Philadelphia chromosome (Ph′).4,5 This translocation results in fusion of the ABL1 gene, located on chromosome 9, to the BCR gene on chromosome 22 with formation of the BCR/ABL hybrid gene.6,7 The product of the ABL1 gene is a tyrosine kinase which, like other enzymes of this type, has been postulated to play a role in cellular growth control and response to DNA damage.8,9 Exactly how alteration of this gene is responsible for leukemogenesis is unknown, but the end result is an expansion of the stem cell pool with overproduction of mature myeloid cells with essentially normal morphology and function. After a period of 3 to 5 years of relative quiescence (“chronic phase”) characterized by functional maturation of the hematopoietic progenitors, the disease progresses to an accelerated and subsequent blastic phase that is virtually indistinguishable from acute leukemia. In these late stages of the disease, collectively termed “acute-phase CML,” a myeloid or lymphoid descendent of the originally affected stem cell loses its capacity for terminal differentiation. The early molecular events underlying disease progression to blast crisis remain largely obscure; however, a number of late cytogenetic and molecular abnormalities have been described in subsets of patients with established blastic transformation.7,10 11
Clinical diagnosis of CML is usually established in the chronic phase, with its onset, whether several days or years before detection, remaining elusive. At this stage, there is a window of opportunity for permanent cure after bone marrow transplantation (BMT); however, BMT is usually no longer effective after the disease has evolved.12 Therefore, monitoring the rate of clinical progression is of prime importance in the management of the disease. Identifying and understanding the genetic and molecular mechanisms crucial for disease evolution to the blastic phase will not only aid in determining the proper timing for BMT, but also provide greater insight into the process of malignant transformation.
The ABL1 gene is expressed in all normal human cells. This gene has 2 alternative first exons, Ia and Ib, which are differentially transcribed from their own promoters13,14(Fig 1). The proximal promoter (Pa) and a distal promoter (Pb) are 175 kb apart and direct the synthesis of 2 mRNA species of 6 and 7 kb, respectively. In approximately 90% of Ph′ translocations the proximal promoter, Pa, is nested within the BCR/ABL transcriptional unit.15 In normal cells, both ABL1 promoters are active in gene expression and the activity of the Pa promoter appears to be unaffected by the Pb promoter.
The position of the Ia CpG island within theBCR-ABL fusion gene is shown. The normal ABL1proto-oncogene (top) comprises 2 alternative promoters (Pb and Pa) each adjacent to exons Ib and Ia, respectively. Ninety percent of CML breakpoints occur in the long intron separating exons Ib and Ia. After the translocation event, the Ia promoter and its associated CpG island become nested within the fusion gene.
The position of the Ia CpG island within theBCR-ABL fusion gene is shown. The normal ABL1proto-oncogene (top) comprises 2 alternative promoters (Pb and Pa) each adjacent to exons Ib and Ia, respectively. Ninety percent of CML breakpoints occur in the long intron separating exons Ib and Ia. After the translocation event, the Ia promoter and its associated CpG island become nested within the fusion gene.
Our laboratory first reported that the CpG island associated with the proximal promoter of ABL1 undergoes methylation upon clinical progression of CML and that this epigenetic alteration may account for the loss of ABL1 expression in cell lines established from patients in the blastic phase of CML.16 Using a methylation-sensitive restriction endonuclease approach, we showed that peripheral blood and/or bone marrow samples from patients in the chronic phase of CML generally lacked methylation at this locus, whereas cell lines and clinical samples from blastic-phase CML had invariably undergone methylation. Patients in the accelerated phase of CML showed intermediate patterns of methylation but they also progressed to patterns of dense methylation upon evolution to blast crisis. We consolidated and expanded these observations in a large study of ABL1 methylation patterns in 99 CML patients at various stages of disease.17 This work demonstrated that partial methylation patterns were also observed in patients with chronic-phase disease of long duration (more than 24 months). Intriguingly, treatment with interferon-α resulted in reversal of methylation regardless of cytogenetic response; these effects were not observed with hydroxyurea treatment. Reports from other laboratories subsequently confirmed these observations.18 19
These data raise a number of important questions concerning the precise role of ABL1 promoter methylation in CML progression. Methylation may be restricted to ABL1 or represent a widespread phenomenon. Recently, reversal of the methylation status of imprinted genes was reported in blastic-phase CML, raising the possibility that maintenance of stable methylation patterns may cease upon disease evolution.20 Earlier reports on differential methylation of the calcitonin and BCR loci in acute-phase CML have also provided hints to this scenario.21-23 Additionally, a quantitative appreciation of the size of the clone bearing methylatedABL1 promoter alleles would be central to the investigation of the role of ABL1 methylation in CML. Furthermore, knowledge of the number of promoter alleles in each cell undergoing methylation in acute-phase CML has been lacking.
To address these questions we adapted the technique of methylation-specific polymerase chain reaction (MSP)24 and bisulfite sequencing to study the promoter regions of ABL1 and a number of other genes. Our results show that methylation is a highly specific process that is likely to be restricted to the ABL1 allele nested within the Ph′ fusion gene. Moreover, it appears that in blastic-phase CML, in contrast to earlier phases of the disease, the vast majority of clonogenic progenitors carry methylated ABL1 promoter alleles. Therefore, we put forward the suggestion that specific methylation of the ABL1 promoter is the mechanism disrupting the balance of BCR-ABL to ABL protein that has been postulated to underlie CML progression.25 Reversal of ABL1 hypermethylation may offer new avenues for therapeutic intervention in this common leukemia.
MATERIALS AND METHODS
Cell lines and patient samples.
Cell lines BV173, KBM5, EM2, and K562 are Philadelphia-positive CML cell lines established from patients in blast crisis (see ref 16 and references therein). The first 3 cell lines contain duplicates of a Ph′ chromosome but no normal homologue of chromosome 9; K562 contains 3 Ph′ chromosomes as well as a normal homologue of chromosome 9. The patient population used in this work has been previously studied in our laboratory using the conventional methylation-sensitive enzyme methodology.17
Cell isolation and DNA extraction from clinical samples.
Mononuclear layers of peripheral blood or bone marrow cells were isolated on a Ficoll-Hypaque density gradient (Pharmacia Biotech, Upsala, Sweden). Genomic DNA was subsequently extracted using standard organic extraction procedures.
Bisulfite modification of DNA.
Genomic DNA was modified by sodium bisulfite as follows: 5 μg DNA/reaction was denatured at 95°C for 15 minutes. Samples were subsequently chilled on ice for 2 minutes and 2N freshly prepared NaOH was added to a final concentration of 0.5N in a total volume of 30 μL. After a 15-minute incubation at 37°C, 450 μL of freshly prepared bisulfite reagent (see below) was added, and the mixtures were overlaid with mineral oil and allowed to incubate at 55°C for 4 hours. After incubation, DNA was extracted using the Wizard DNA Clean-up kit (Promega, Madison, WI). Purified DNA was recovered in water and 2N NaOH added to a final concentration of 0.3N; samples were allowed to incubate for 15 minutes at 37°C. On conclusion of this step, 1 to 1.5 μg of lambda DNA (NEB, Beverly, MA) was added and the DNA was ethanol precipitated overnight at −80°C. DNA was subsequently recovered by centrifugation and dissolved in sterile water in preparation for polymerase chain reaction (PCR). The bisulfite reagent was prepared as follows: 1.9 g of sodium bisulfite (Sigma, St Louis, MO) was slowly dissolved in sterile water on a gentle rocker; the total volume was 4 mL, which also contained 0.7 mL of a 2N NaOH solution and 0.5 mL of a saturated solution of hydroquinone (Sigma) in water.
Single-step methylation-specific PCR (ABL1, MSH2,MSH6 loci).
For the ABL1, MSH2, and MSH6 loci, a single step methylation-specific PCR approach was used. Eight hundred nanograms of genomic DNA modified as described above was included in 100 μL PCR reaction volumes containing buffer to 1X final concentration (Tris-HCl 10 mmol/L pH 9, KCl 50 mmol/L, MgCl2 1.5 mmol/L, Triton X100 0.1%, bovine serum albumin [BSA] 0.2 mg/mL), primers (see below), and 1 U Taq polymerase. dNTP concentrations were calibrated for each PCR reaction as a way to achieve gentle adjustments of the magnesium concentration. Table 1 provides primer sequences for each locus as well as the concentration of dNTPs and cycling parameters used in each PCR reaction. As a positive control, DNA from normal donor buffy coat samples was in vitro methylated at CpG sites using CpG methylase (SssI methylase) (NEB) as per the manufacturer’s recommendations.
Primers and Conditions for All Amplification Reactions Described in This Study
| Locus/Amplimer . | S/T . | 1°/2° . | M/U . | Forward Primer . | Reverse Primer . | N . | °C . | P . |
|---|---|---|---|---|---|---|---|---|
| ABL1island | S | — | — | acttctaaaataaaataaaacaaa | ttttttttggatttgtagtttatt | 3 | 50 | D |
| ABL1-I MSP | S | — | M | cgcgtttcggttaggcggagacgcggtcgc | aaacaaccccttcttaaatttacaa | 3 | 58 | A |
| ABL1-I MSP | S | — | U | tgtgttttggttaggtggagatgtggttgt | aaacaaccccttcttaaatttacaa | 3 | 58 | A |
| MSH2-I MSP | S | — | M | cgtcgtggtcggacgtcgttc | cactaaaaaaactactcac | 2 | 48 | A |
| MSH2-I MSP | S | — | U | tgttgtggttggatgttgttt | cactaaaaaaactactcac | 2 | 48 | A |
| MSH2-II MSP | S | — | M | aggacgcgttgttggtcgttc | cactaaaaaaactactcac | 2 | 48 | A |
| MSH2-II MSP | S | — | U | aggatgtgttgttggttgttt | cactaaaaaaactactcac | 2 | 48 | A |
| MSH6-I MSP | S | — | M | ttcgcgcggggtttaggcgtc | aaccttattaacatcactcaa | 3 | 49.5 | A |
| MSH6-I MSP | S | — | U | tttgtgtggggtttaggtgtt | aaccttattaacatcactcaa | 3 | 49.5 | A |
| MSH6-II MSP | S | — | M | cgcgtatcgttcgcgtacggc | aaccttattaacatcactcaa | 3 | 49.5 | A |
| MSH6-II MSP | S | — | U | tgtgtattgtttgtgtatggt | aaccttattaacatcactcaa | 3 | 49.5 | A |
| MLH1 island | T | 1° | — | tttatgtattggtatataaagttt | ttctcaactctataaattactaaa | 2 | 50 | B |
| MLH1-I MSP | T | 2° | M | acgtagacgttttattagggtcgc | ttctcaactctataaattactaaa | 2 | 50 | C |
| MLH1-I MSP | T | 2° | U | atgtagatgttttattagggttgt | ttctcaactctataaattactaaa | 2 | 50 | C |
| ATM island | T | 1° | — | taagatttaggttaaagtaaatat | atctaaaaaaaaaacaaactaaa | 2 | 50 | B |
| ATM-I MSP | T | 2° | M | gtggcggtattgaattcgc | atctaaaaaaaaaacaaactaaa | 2 | 50 | C |
| ATM-I MSP | T | 2° | U | gtggtggtattgaatttgt | atctaaaaaaaaaacaaactaaa | 2 | 50 | C |
| ATM-II MSP | T | 2° | M | cgattcgtaaacgttaagtc | atctaaaaaaaaaacaaactaaa | 2 | 50 | C |
| ATM-II MSP | T | 2° | U | tgatttgtaaatgttaagtt | atctaaaaaaaaaacaaactaaa | 2 | 50 | C |
| ATM-III MSP | T | 2° | M | cgttttcgttcgttttcggaattgtc | atctaaaaaaaaaacaaactaaa | 2 | 50 | C |
| ATM-III MSP | T | 2° | U | tgtttttgtttgtttttggaattgtt | atctaaaaaaaaaacaaactaaa | 2 | 50 | C |
| ATM-IV MSP | T | 2° | M | cgggatttgcgttgtaattatcgtcgc | atctaaaaaaaaaacaaactaaa | 2 | 50 | C |
| ATM-IV MSP | T | 2° | U | tgggatttgtgttgtaattattgttgt | atctaaaaaaaaaacaaactaaa | 2 | 50 | C |
| Locus/Amplimer . | S/T . | 1°/2° . | M/U . | Forward Primer . | Reverse Primer . | N . | °C . | P . |
|---|---|---|---|---|---|---|---|---|
| ABL1island | S | — | — | acttctaaaataaaataaaacaaa | ttttttttggatttgtagtttatt | 3 | 50 | D |
| ABL1-I MSP | S | — | M | cgcgtttcggttaggcggagacgcggtcgc | aaacaaccccttcttaaatttacaa | 3 | 58 | A |
| ABL1-I MSP | S | — | U | tgtgttttggttaggtggagatgtggttgt | aaacaaccccttcttaaatttacaa | 3 | 58 | A |
| MSH2-I MSP | S | — | M | cgtcgtggtcggacgtcgttc | cactaaaaaaactactcac | 2 | 48 | A |
| MSH2-I MSP | S | — | U | tgttgtggttggatgttgttt | cactaaaaaaactactcac | 2 | 48 | A |
| MSH2-II MSP | S | — | M | aggacgcgttgttggtcgttc | cactaaaaaaactactcac | 2 | 48 | A |
| MSH2-II MSP | S | — | U | aggatgtgttgttggttgttt | cactaaaaaaactactcac | 2 | 48 | A |
| MSH6-I MSP | S | — | M | ttcgcgcggggtttaggcgtc | aaccttattaacatcactcaa | 3 | 49.5 | A |
| MSH6-I MSP | S | — | U | tttgtgtggggtttaggtgtt | aaccttattaacatcactcaa | 3 | 49.5 | A |
| MSH6-II MSP | S | — | M | cgcgtatcgttcgcgtacggc | aaccttattaacatcactcaa | 3 | 49.5 | A |
| MSH6-II MSP | S | — | U | tgtgtattgtttgtgtatggt | aaccttattaacatcactcaa | 3 | 49.5 | A |
| MLH1 island | T | 1° | — | tttatgtattggtatataaagttt | ttctcaactctataaattactaaa | 2 | 50 | B |
| MLH1-I MSP | T | 2° | M | acgtagacgttttattagggtcgc | ttctcaactctataaattactaaa | 2 | 50 | C |
| MLH1-I MSP | T | 2° | U | atgtagatgttttattagggttgt | ttctcaactctataaattactaaa | 2 | 50 | C |
| ATM island | T | 1° | — | taagatttaggttaaagtaaatat | atctaaaaaaaaaacaaactaaa | 2 | 50 | B |
| ATM-I MSP | T | 2° | M | gtggcggtattgaattcgc | atctaaaaaaaaaacaaactaaa | 2 | 50 | C |
| ATM-I MSP | T | 2° | U | gtggtggtattgaatttgt | atctaaaaaaaaaacaaactaaa | 2 | 50 | C |
| ATM-II MSP | T | 2° | M | cgattcgtaaacgttaagtc | atctaaaaaaaaaacaaactaaa | 2 | 50 | C |
| ATM-II MSP | T | 2° | U | tgatttgtaaatgttaagtt | atctaaaaaaaaaacaaactaaa | 2 | 50 | C |
| ATM-III MSP | T | 2° | M | cgttttcgttcgttttcggaattgtc | atctaaaaaaaaaacaaactaaa | 2 | 50 | C |
| ATM-III MSP | T | 2° | U | tgtttttgtttgtttttggaattgtt | atctaaaaaaaaaacaaactaaa | 2 | 50 | C |
| ATM-IV MSP | T | 2° | M | cgggatttgcgttgtaattatcgtcgc | atctaaaaaaaaaacaaactaaa | 2 | 50 | C |
| ATM-IV MSP | T | 2° | U | tgggatttgtgttgtaattattgttgt | atctaaaaaaaaaacaaactaaa | 2 | 50 | C |
Primers for amplification of the ABL1 CpG island correspond to the antisense strand (see Materials and Methods). All other amplifications correspond to the sense strand. Primers for first-round PCR of ATM and MLH1 loci are insensitive to methylation. For each gene, a common reverse primer was used.
Abbreviations: S, single-step MSP; T, 2-step MSP; 1°, first round of 2-step MSP; 2°, second round of 2-step MSP; M, primer specific for methylated DNA; U, primer specific for unmethylated DNA; N, amount of added microliters of a standard dNTP solution (10 mmol/L of each nucleotide) per 100 μL total PCR volume; °C, annealing temperature in centrigrade; P, cycling parameters: A, 1 cycle of denaturation for 1 minute, annealing for 1 minute 30 seconds, extension for 1 minute 30 seconds followed by 39 cycles of denaturation for 1 minute, annealing for 1 minute, extension for 1 minute; B, 5 cycles of denaturation for 1 minute, annealing for 2 minutes 30 seconds, extension for 4 minutes followed by 50 cycles of denaturation for 1 minute, annealing for 2 minutes, extension for 2 minutes 30 seconds; C, 10 cycles of denaturation for 1 minute, annealing for 1 minute, extension for 1 minute; D, 5 cycles of denaturation for 1 minute, annealing for 1 minute 30 seconds, extension for 4 minutes followed by 50 cycles of denaturation for 1 minute, annealing for 1 minute, extension for 2 minutes 30 seconds.
Two-step methylation-specific PCR (ATM and MLH1loci).
For ATM and MLH1, a 2-step PCR approach was used. Two-step PCR significantly boosted sensitivity and specificity of single-step PCR and was particularly useful for islands where CpG sites were relatively sparse. In the first round of PCR, most of the CpG island in each case was amplified from modified genomic DNA using primers that were insensitive to methylation, ie, did not include any CpG sites. In the second round, a small amount of template from the first reaction was used in PCR together with primers specific to selected CpG sites. Table 1 provides the sequences for each set of primers as well as cycling parameters for each reaction.
ABL1 sequencing of bisulfite-modified DNA.
In an attempt to amplify the ABL1 CpG island, we originally designed primers flanking a 519-bp fragment corresponding to the sense strand (after bisulfite modification, opposite strands are no longer complementary and primers can be designed to amplify either strand). However, we were unable to amplify the sense strand from samples containing heavily methylated DNA, whereas any unmethylated DNA component was readily amplifiable. We speculate that this may be due to the complex conformation of heavily methylated molecules. TheABL1 promoter is very rich in guanine residues that could easily form hard-to-resolve secondary structures with cytosine residues remaining after bisulfite modification of a heavily methylated sample. This hypothesis would predict that amplification of the opposite strand should be readily achieved because the complementary cytosines should transform to thymidine after bisulfite modification. Indeed, this was found to be the case and amplification of the antisense strand was subsequently used for analysis. Because methylation of CpG sites is a symmetrical process, we were able to infer methylation patterns for the sense strand by studying the opposite strand.
DNA modified as described above was sequenced on a fluorescent sequencer, either immediately after PCR or subsequent to a cloning step using the T-vector kit (Invitrogen, Carlsbad, CA). Individual clones were screened for the correct insert by PCR before sequencing. Primers and conditions for amplifying the ABL1promoter antisense strand are given in Table 1.
Culture of colonies derived from peripheral blood hematopoietic progenitors.
Mononuclear cells were aseptically isolated from peripheral blood and cultured in semisolid medium as previously described.26
Methylation-specific PCR of colonies.
Individual colonies were harvested in 150 μL of phosphate-buffered saline (PBS), spun at maximum speed for 5 minutes, and resuspended in 22.5 μL of water containing 1.5 μg of lambda DNA per colony. Samples were heated at 95°C for 10 minutes and subsequently processed as per normal bisulfite modification protocol.
RESULTS
Methylation-specific PCR shows uniform methylation of the Ph′-associated ABL1 allele in cell lines established from blastic-phase CML.
Cell lines EM2, KBM5, BV173, and K562 were established from patients in the blastic phase of CML. The first 3 lines contain Ph′ chromosomes but lack copies of normal chromosome 9. Therefore, the onlyABL1 alleles present are the ones contained withinBCR-ABL fusion genes. Methylation-specific PCR showed complete and universal methylation of ABL1 promoters in each of these cell lines; no unmethylated promoter alleles were detected. By contrast, cell line K562 contains 3 Philadelphia chromosomes as well as a normal chromosome 9 homologue. In K562, methylation-specific PCR showed both methylated and unmethylated ABL1 sequences (Fig 2), a result which indicates thatABL1 methylation patterns in cell lines are likely to be specific and not reflect the indiscriminate spread of methylation during repeated passage. These observations suggest that methylation ofABL1 in CML is restricted to the allele nested within the Ph′ fusion gene.
Methylation-specific PCR shows ABL1 methylation patterns in CML cell lines. NBC, normal donor buffy-coat DNA; SssI-NBC, normal donor buffy-coat DNA treated in vitro with CpG methylase (SssI methylase); EM2, DNA from CML cell line EM2 containing only a duplicate of the Ph′ chromosome and lacking a normal homologue of chromosome 9; K562, DNA from cell line K562 which contains 3 copies of the Ph′ chromosome in addition to a chromosome 9 homologue; M, primers specific for methylated DNA; U, primers specific for unmethylated DNA; ◊x, ◊x/HaeIII marker.
Methylation-specific PCR shows ABL1 methylation patterns in CML cell lines. NBC, normal donor buffy-coat DNA; SssI-NBC, normal donor buffy-coat DNA treated in vitro with CpG methylase (SssI methylase); EM2, DNA from CML cell line EM2 containing only a duplicate of the Ph′ chromosome and lacking a normal homologue of chromosome 9; K562, DNA from cell line K562 which contains 3 copies of the Ph′ chromosome in addition to a chromosome 9 homologue; M, primers specific for methylated DNA; U, primers specific for unmethylated DNA; ◊x, ◊x/HaeIII marker.
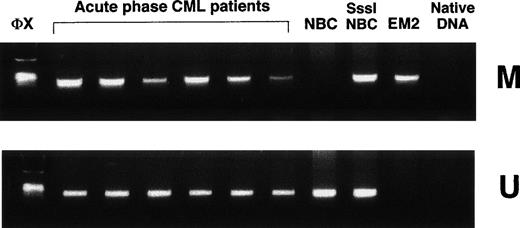
Clinical samples from patients with acute-phase CML contain both methylated and unmethylated ABL1 promoters.
In contrast to cell lines, samples from 15 patients in acute-phase CML showed both methylated and unmethylated ABL1 in every case (Fig 3). This could be accounted for by the presence of a mixed cell population consisting of CML cells with biallelic methylation of ABL1 promoters alongside residual normal cells carrying only unmethylated ABL1. Alternatively, this pattern may denote the coexistence of a methylated and an unmethylated promoter allele in each CML cell.
Methylation-specific PCR shows ABL1 methylation patterns in clinical samples from acute-phase CML. NBC, normal donor buffy-coat DNA; SssI-NBC, CpG methylase (SssI methylase)-treated normal donor buffy-coat DNA; EM2, CML cell line EM2. Native DNA lane contains DNA that was not modified by bisulfite. M, primers specific for methylated DNA; U, primers specific for unmethylated DNA; ◊x, ◊x/HaeIII marker.
Methylation-specific PCR shows ABL1 methylation patterns in clinical samples from acute-phase CML. NBC, normal donor buffy-coat DNA; SssI-NBC, CpG methylase (SssI methylase)-treated normal donor buffy-coat DNA; EM2, CML cell line EM2. Native DNA lane contains DNA that was not modified by bisulfite. M, primers specific for methylated DNA; U, primers specific for unmethylated DNA; ◊x, ◊x/HaeIII marker.
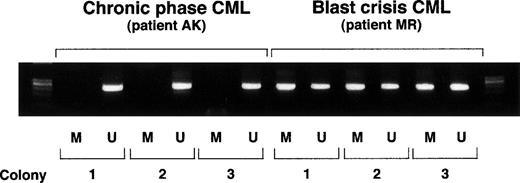
Colonies derived from single hematopoietic progenitors in acute-phase CML contain both methylated and unmethylated ABL1 promoters.
To distinguish between mixed-cell population pattern and allele-specific methylation, we examined the methylation status of individual colonies derived from hematopoietic progenitors. Peripheral blood mononuclear cells from patients at chronic phase and patients in acute-phase CML were seeded in methylcellulose-containing semisolid cultures in the presence of 10% (vol/vol) conditioned medium from cultures of the human bladder carcinoma cell line 5637. This conditioned medium contains hematopoietic growth factors that promote the growth of myeloid colonies.26 After a 14-day incubation, colonies were harvested, and their DNA extracted and subjected to modification by the bisulfite treatment protocol (see Materials and Methods). The results indicated that DNA of colonies from patients in chronic-phase CML generally lacked methylation at theABL1 promoter region, whereas colonies from patients with acute-phase disease always contained a methylated allele alongside an unmethylated allele (Fig 4).
ABL1 methylation-specific PCR on individual colonies. Each colony was split in half and used for methylation-specific PCR employing primers specific for methylated DNA (M) or for unmethylated DNA (U).
ABL1 methylation-specific PCR on individual colonies. Each colony was split in half and used for methylation-specific PCR employing primers specific for methylated DNA (M) or for unmethylated DNA (U).
The vast majority of clonogenic cells in acute-phase CML contain methylated ABL1 promoters.
The study of single-progenitor–derived colonies also gave insight into the size of the clone-bearing methylated ABL1 promoters at different phases of disease. In all patients in blastic phase studied, nearly all of clonogenic cells contained methylated ABL1promoters (Table 2). By contrast, the vast majority of patients in the chronic phase of the disease demonstrated absence of ABL1 methylation in all colonies studied. A patient recently diagnosed to be in chronic-phase CML (LR) showed ABL1promoter methylation in 6 of 12 colonies studied. Follow-up has been very short in this case, but notably this patient has maintained persistently high peripheral blood eosinophil counts and has shown resistance to hydroxyurea treatment. In accelerated-phase disease, patient DK demonstrated intermediate “methylated” clone size (2 of 10 colonies studied bore methylated ABL1). A second patient with a clinical picture of accelerated disease at the time of sample collection (patient AF) had undergone complete methylation of theABL1 promoter in all peripheral blood progenitors; this patient very promptly progressed to frank blast crisis. These findings suggest that methylation of ABL1 is tightly linked to clonal evolution of CML.
Analysis of ABL1 Methylation Patterns in Colonies Derived From Single Hematopoietic Progenitors
| Patient Identification . | Clinical Stage . | Total No. of Colonies Tested . | No. of Colonies Bearing MethylatedABL1 . |
|---|---|---|---|
| MR | Blastic | 10 | 9 |
| SS | Blastic | 4 | 4 |
| WD | Blastic | 10 | 9 |
| DK | Accelerated | 10 | 2 |
| AF | Accelerated | 12 | 12 |
| LR* | Chronic | 12 | 6 |
| AK | Chronic | 15 | 0 |
| GF | Chronic | 6 | 0 |
| GR | Chronic | 8 | 0 |
| SN | Chronic | 6 | 0 |
| JA | Chronic | 12 | 0 |
| Patient Identification . | Clinical Stage . | Total No. of Colonies Tested . | No. of Colonies Bearing MethylatedABL1 . |
|---|---|---|---|
| MR | Blastic | 10 | 9 |
| SS | Blastic | 4 | 4 |
| WD | Blastic | 10 | 9 |
| DK | Accelerated | 10 | 2 |
| AF | Accelerated | 12 | 12 |
| LR* | Chronic | 12 | 6 |
| AK | Chronic | 15 | 0 |
| GF | Chronic | 6 | 0 |
| GR | Chronic | 8 | 0 |
| SN | Chronic | 6 | 0 |
| JA | Chronic | 12 | 0 |
Samples from patient LR were collected at diagnosis. Follow-up has been very short in this case but notably this patient has shown persistently high peripheral blood eosinophil counts and resistance to hydroxyurea treatment. All other samples of chronic-phase CML have been obtained from patients with stable disease observed in follow-up periods of at least 1 year. All patients in chronic-phase CML had undergone recent cytogenetic studies while on follow-up: in all cases, 100% of metaphases carried the Ph′ chromosome.
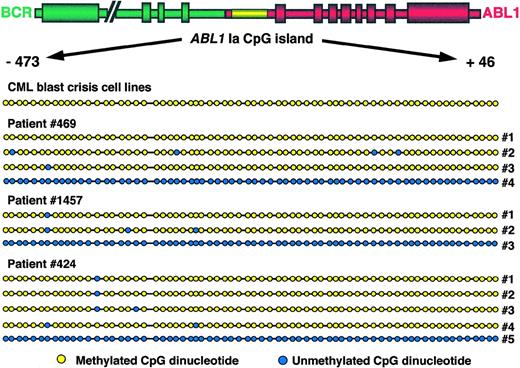
Sequencing of bisulfite-modified DNA shows methylation patterns in the ABL1 promoter region in acute-phase CML.
Traditional assays for detection of methylation using methylation-sensitive restriction enzymes allow a limited appreciation of the extent of methylation in each case because only those CpG sites that happen to be part of the recognition sequence of one or the other of these enzymes can be studied. However, sequencing of DNA after bisulfite modification has the power to reveal the methylation status of every CpG site within the region of interest. To determine the extent of methylation of the ABL1 promoter in acute-phase CML, we amplified by PCR and subsequently sequenced the ABL1promoter-associated CpG island from cell lines KBM5, BV173, and EM2 as well as 7 patients in both the accelerated and blastic phases of CML (Fig 5). Because we anticipated mixed methylation patterns in the clinical samples (see above), PCR fragments were cloned before sequencing and several clones containing full-length inserts from each patient were studied. Our results showed that in all cell lines, all CpG sites within the ABL1 promoter without exception had undergone methylation—importantly, all non-CpG cytosines had undergone conversion to thymidine after bisulfite treatment. DNA from clinical samples was also densely hypermethylated along the entire length of the CpG island. In most clones, occasional CpG sites were unmethylated in the context of a densely methylated CpG island, but they were not generally constant among patients. Clones containing unmethylated ABL1 alleles were uniformly free of methylation along the entire length of the CpG island in all cases (Fig 5).
Methylation topology of the ABL1 promoter region. The region amplified corresponds to coordinates −473 to +46 with respect to the start of exon 1a or coordinates 37352 to 37871 of GenBank sequence U07563.42 The relative position of dots on the diagram corresponds to the relative position of CpG dinucleotides along the island. Yellow dots indicate methylated CpG dinucleotides; blue dots indicate unmethylated CpG dinucleotides. Amplification products obtained from total peripheral blood and/or bone marrow mononuclear cells were cloned and individual clones were sequenced on a fluorescent sequencer. The designation “CML blast crisis cell lines” refers to cell lines EM2, BV173, and KBM5. Patients 469 and 1457 were in accelerated-phase CML when samples were collected and patient 424 was in blast crisis. For each patient, several clones of methylated DNA are depicted alongside a representative clone of unmethylated DNA.
Methylation topology of the ABL1 promoter region. The region amplified corresponds to coordinates −473 to +46 with respect to the start of exon 1a or coordinates 37352 to 37871 of GenBank sequence U07563.42 The relative position of dots on the diagram corresponds to the relative position of CpG dinucleotides along the island. Yellow dots indicate methylated CpG dinucleotides; blue dots indicate unmethylated CpG dinucleotides. Amplification products obtained from total peripheral blood and/or bone marrow mononuclear cells were cloned and individual clones were sequenced on a fluorescent sequencer. The designation “CML blast crisis cell lines” refers to cell lines EM2, BV173, and KBM5. Patients 469 and 1457 were in accelerated-phase CML when samples were collected and patient 424 was in blast crisis. For each patient, several clones of methylated DNA are depicted alongside a representative clone of unmethylated DNA.
(A) Single-step methylation-specific PCR is shown for theMSH6 locus. Lanes 1, 3, 4, 6, and 7: samples from patients in acute-phase CML; lanes 2 and 5: samples from patients in chronic phase CML; lane 8: normal donor buffy-coat DNA; lane 9: normal donor buffy-coat DNA treated in vitro with CpG methylase (SssI methylase). M, primers specific for methylated DNA; U, primers specific for unmethylated DNA; ◊x, ◊x/HaeIII marker. (B) Two-step methylation-specific PCR for the ATM promoter region. In the first round, the CpG island is amplified from bisulfite-modified DNA using primers that are insensitive to methylation (see Materials and Methods). In the second round, primers specific for methylated DNA (M) or unmethylated DNA (U) are used in PCR together with a small amount of template from the first reaction (arrow). Lane 1: normal donor buffy-coat DNA; lane 2: normal donor buffy-coat DNA treated in vitro with CpG methylase (SssI methylase); lanes 3 through 6: samples from patients in acute-phase CML. ◊x, ◊x/HaeIII marker.
(A) Single-step methylation-specific PCR is shown for theMSH6 locus. Lanes 1, 3, 4, 6, and 7: samples from patients in acute-phase CML; lanes 2 and 5: samples from patients in chronic phase CML; lane 8: normal donor buffy-coat DNA; lane 9: normal donor buffy-coat DNA treated in vitro with CpG methylase (SssI methylase). M, primers specific for methylated DNA; U, primers specific for unmethylated DNA; ◊x, ◊x/HaeIII marker. (B) Two-step methylation-specific PCR for the ATM promoter region. In the first round, the CpG island is amplified from bisulfite-modified DNA using primers that are insensitive to methylation (see Materials and Methods). In the second round, primers specific for methylated DNA (M) or unmethylated DNA (U) are used in PCR together with a small amount of template from the first reaction (arrow). Lane 1: normal donor buffy-coat DNA; lane 2: normal donor buffy-coat DNA treated in vitro with CpG methylase (SssI methylase); lanes 3 through 6: samples from patients in acute-phase CML. ◊x, ◊x/HaeIII marker.
Both cell lines and clinical samples had previously been analyzed using restriction enzyme–based methodology.17 These methods probed a solitary SacII site alongside 3 HpaII sites, all clustered at the 3′ end of the CpG island. All cell lines as well as samples from 2 patients had shown methylation at all 4 enzyme sites: bisulfite sequencing confirmed these results. The other 5 patients had shown lack of methylation at the HpaII sites while the SacII site was methylated. Bisulfite sequencing confirmed the methylation status of the SacII site in these patients; however, it also demonstrated HpaII site methylation. Therefore, HpaII analysis does not appear to be as sensitive asSacII analysis for monitoring methylation in clinical samples. Combined with the data obtained from study of progenitor-derived colonies, the enhanced resolution of sodium bisulfite sequencing showed that CML progression is characterized by an increase in the abundance of fully methylated ABL1 promoters rather than by a linear spread of methylation along the promoter region.
The ATM promoter remains free of methylation at all stages of disease.
The promoter of the ataxia telangiectasia locus (ATM) was examined by a modification of the classical methylation-specific PCR protocol, “2-step MSP” (see Materials and Methods). In essence, this method is an adaptation of nested PCR protocols, where the first amplification step encompasses most of the CpG island using primers that are insensitive to methylation; ie, do not contain CpG dinucleotides and, therefore, equally amplify methylated and unmethylated sequences. It should be emphasized that primers are still specific for bisulfite-modified DNA because thymidine is substituted for all non-CpG cytosines. In the second PCR step, methylation-specific primers are added to a small amount of template from the first step.
Using this approach, mononuclear cell samples from patients in chronic-phase CML and patients in acute-phase CML as well as CML cell lines (see above) were examined for methylation at 4 loci of theATM promoter region. The promoter was found to be free of methylation in each of the 4 regions at all stages of disease (Fig 6B, Table 3).
Methylation Status of the MSH2, MSH6, MLH1, and ATM Loci at Different Stages of CML
| Locus . | Chronic-Phase CML . | Acute-Phase CML . | ||
|---|---|---|---|---|
| Total . | Methylated . | Total . | Methylated . | |
| MSH2-I | 11 | 0 | 13 | 0 |
| MSH2-II | 13 | 0 | 15 | 0 |
| MSH6-I | 13 | 0 | 15 | 0 |
| MSH6-II | 10 | 0 | 14 | 0 |
| MLH1 | 13 | 0 | 13 | 0 |
| ATM-I | 16 | 0 | 14 | 0 |
| ATM-II | 16 | 0 | 14 | 0 |
| ATM-III | 16 | 0 | 14 | 0 |
| ATM-IV | 16 | 0 | 14 | 0 |
| Locus . | Chronic-Phase CML . | Acute-Phase CML . | ||
|---|---|---|---|---|
| Total . | Methylated . | Total . | Methylated . | |
| MSH2-I | 11 | 0 | 13 | 0 |
| MSH2-II | 13 | 0 | 15 | 0 |
| MSH6-I | 13 | 0 | 15 | 0 |
| MSH6-II | 10 | 0 | 14 | 0 |
| MLH1 | 13 | 0 | 13 | 0 |
| ATM-I | 16 | 0 | 14 | 0 |
| ATM-II | 16 | 0 | 14 | 0 |
| ATM-III | 16 | 0 | 14 | 0 |
| ATM-IV | 16 | 0 | 14 | 0 |
“Acute-phase” designation includes accelerated and blast crisis stages of CML. “Total” designates the total number of patients studied for each locus. “Methylated” designates the number of patients in whom methylation was detected at that locus in bone marrow and/or peripheral blood mononuclear cells.
Promoters of mismatch repair genes MLH1, MSH2, andMSH6 are unmethylated even in acute stages of CML.
Methylation-specific PCR was adapted to study the promoters of mismatch repair genes MSH2, MSH6, and MLH1. ForMSH2 and MSH6, a classical, single-step MSP protocol was used; to boost the specificity of detecting methylation of theMLH1 promoter, 2-step MSP was developed for this gene. Our panel of patients at chronic phase and patients in acute-phase CML as well as CML cell lines was tested for methylation at 2 loci of theMSH2 promoter, 2 loci of the MSH6 promoter, and 1 locus of the MLH1 promoter. In all cases, all these loci were found to be free of methylation in both chronic and acute stages of disease (Fig 6A, Table 3).
DISCUSSION
The presence of a pathognomonic molecular abnormality in nearly all cases as well as a rather invariable clinical course distinguish CML from other myeloproliferative disorders. Despite significant advances in the elucidation of the mechanisms responsible for disease initiation and the molecular targets of the BCR-ABL tyrosine kinase, the events underlying transition to accelerated and blastic phases remain largely unknown.
We and others have shown that epigenetic alteration of the proximal promoter of ABL1 appears to be specifically and consistently associated with progression of CML.16-19 Lack of methylation in this region appears to be restricted to the chronic phase whereas hypermethylation is observed when blast crisis has evolved.16 Furthermore, long-standing chronic disease as well as accelerated-phase disease are associated with intermediate patterns of methylation, which can be reversed after treatment with interferon-α.17
Enzyme-based approaches to study methylation patterns in disease are limited in many ways: Firstly, only a small proportion of CpG sites can be studied and the possibility of patchy methylation cannot be excluded. Secondly, the danger of false-positive results due to the inability of restriction enzymes to digest is very real, given the fact that many archival samples are rich in impurities that can inhibit these enzymes. Thirdly, enzyme-based approaches result in the inability to visualize unmethylated alleles because these are digested by the enzyme and do not contribute to the subsequent PCR amplification. To circumvent these problems, we adapted the techniques of methylation-specific PCR and sequencing of sodium bisulfite-treated DNA24 so as to study the promoter regions of ABL1and other candidate genes.
We used the ability of methylation-specific PCR to reveal unmethylated alleles to ask whether one or both ABL1 promoter alleles undergo methylation with disease progression. Whereas cell lines containing only Ph′-associated ABL1 alleles showed uniform methylation, evidence of both methylated and unmethylated promoter alleles was obtained from acute-phase CML clinical samples. This may be attributable to the presence of a mixture of cell populations: a leukemic population containing only methylated alleles alongside a population of normal hematopoietic cells containing unmethylated alleles; alternatively, it may indicate allele-specific methylation. To distinguish between these possibilities, we studied colonies derived from single hematopoietic progenitors grown in semisolid medium. The results indicated that in acute-phase CML, each and every progenitor carrying a methylated allele also carried an unmethylated allele. Thus, in acute-phase CML, methylation is likely to be an allele-specific process. Given the evidence of specific methylation of ABL1 in all 3 cell lines bearing only Ph′-associated ABL1 alleles, it would not be unreasonable to assume that in clinical samples too the proximal ABL1promoter nested within the Ph′ fusion gene specifically undergoes methylation in acute-phase CML. However, our data do not exclude the possibility that in some patients the non-Philadelphia allele may undergo methylation.
The study of colonies also enabled us to semi-quantitatively appreciate the size of the methylated ABL1-bearing clone. This is very difficult to infer from the study of uncloned populations where even a few methylated ABL1 molecules can produce strongly positive amplification results. Our data indicate that in blastic-phase CML, in contrast to the chronic phase, the vast majority of clonogenic progenitors have undergone methylation. This observation indicates thatABL1 promoter methylation is likely to be an inherent step in the evolution of the disease.
Sequencing of bisulfite-modified DNA from cell lines and patients in acute-phase CML was used to investigate the spatial degree of DNA methylation. In both cell lines and patient samples, nearly all CpG sites in the promoter region were methylated. Despite the fact that in acute-phase CML methylation appears to be a fairly uniform process, it is conceivable that study of methylation topology at earlier stages of disease may show conserved patterns by which methylation spreads to affect the whole CpG island.
Furthermore, we addressed the degree of methylation of genes other thanABL1. Among a large choice of genes to examine for methylation, we chose to concentrate on the DNA mismatch repair genes MSH2,MSH6, and MLH127-33 as well as the ataxia-telangiectasia locus, ATM.34,35 Investigation of the methylation pattern of mismatch repair genes might help to resolve the current controversy concerning the type of mutator phenotype observed in acute-phase CML36,37 because loss of MSH6expression has been linked to mutator phenotype without microsatellite instability,38 whereas loss of MSH2 expression is associated with replication errors at these repeat loci.28-31 The product of the ataxia-telangiectasia locus (ATM) was found to constitutively interact with theABL1 tyrosine kinase and activate the latter upon radiation-induced DNA damage.9,39 We reasoned that a reciprocal relationship may exist in which methylation of at least one member of the ABL1/ATM pair is required for CML evolution. After a comprehensive search in samples from patients at all stages of disease, we concluded that the promoters of mismatch repair genes as well as ATM remain free of methylation even in acute stages of disease. These findings, together with the previously reported absence of methylation of the p16 and p15 genes in acute-phase CML,40 suggest that ABL1, distinctly among DNA repair and genotoxic stress-response genes, undergoes epigenetic modification in this disorder.
Methylation of the ABL1 promoter appears to closely correlate with CML progression: both the proportion of patients with methylatedABL1 and the numbers of progenitors bearing methylated sequences in individual patients correlate with disease progression to more aggressive forms. These data are compatible with 2 models addressing the origins of ABL1 methylation. In the first, methylation may be a stochastic event leading to clonal expansion: a single CML progenitor may undergo methylation of ABL1 and/or other critical gene promoters and acquire a growth advantage, with subsequent clonal expansion and establishment of blastic transformation, either directly or following accumulation of secondary mutations. Alternatively, methylation may serve as a molecular “clock,” counting the time that has elapsed from the initiating mutation, the generation of the BCR-ABLfusion gene. In the latter model, methylation of the proximal promoter of ABL1 may be an inevitable consequence of juxtaposition to upstream Alu-rich BCR sequences, acting as “methylation centers.”41 Progenitors having survived long enough will have undergone ABL1 methylation; they will also have had the time to accumulate further mutations, thereby acquiring a more aggressive phenotype that eventually would lead to the establishment of blastic transformation.
ABL1 methylation appears to be an early and nearly universal epigenetic alteration with effects integral to CML progression. BCR-ABL and ABL appear to exert opposing and antagonistic actions at the level of apoptosis, cell-cycle progression, and genetic instability.25 BCR-ABL is the malignant counterpart, promoting cell-cycle progression and genetic instability and inhibiting apoptosis, whereas the opposite roles have been attributed to ABL. It is conceivable that a balance between the two, maintaining the clinically benign chronic phase of CML, may be dependent on continued transcription from the proximal ABL1promoter. Silencing of the internal ABL1 promoter by methylation may tip the balance in favor of BCR-ABL and provide the ground for resistance to apoptosis and genomic instability that characterize blastic transformation. Interferon therapy appears to reestablish this balance leading to prolongation of the chronic phase.17 In the future it will be fascinating to test this hypothesis in animal models and provide new avenues for gene therapy of this common and fatal form of leukemia.
ACKNOWLEDGMENT
The authors thank Aviva Simberger and Aliza Treves for valuable help with colony assays, Dr Reuven Orr for provision of clinical samples, and Dr Zahava Siegfried-Kluger for critical review of the manuscript.
Supported by grants from the Golda Meir Fellowship Fund (to F.A.A.), the Gabriella Rich Leukemia Fund, and the Office of the Chief Scientist of the State of Israel (to D.B.-Y.).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Dina Ben-Yehuda, MD, Department of Hematology, Hadassah University Hospital-Ein Karem, Jerusalem 91120, Israel; e-mail: dbyehuda@hadassah.org.il.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal