The promoter region of the cyclin-dependent kinase inhibitorp15INK4Bcontains a CpG island that is hypermethylated in many hematologic malignancies. To explore the relationship between patterns of methylation and gene transcription, we used bisulfite genomic sequencing to obtain a detailed analysis of methylation in acute leukemia, leukemia cell lines, and normal lymphocytes. The entire CpG island region of p15 was largely devoid of methylation in normal lymphocytes, but methylation of varying density was found in primary acute leukemia. Methylation density was generally conserved between the alleles from each sample, but marked heterogeneity for the specific CpG sites methylated was observed. Patterns of methylation were compared and expression assessed with reverse-transcriptase polymerase chain reaction (RT-PCR). The density of methylation within the CpG island, and not any specific location, correlates best with transcriptional loss. Leukemias with methylation of approximately 40% of the CpG dinucleotides on each allele had complete gene silencing, with variable, but diminished expression with less dense CpG island methylation. Our results suggest that the transcriptional silencing of p15 in conjunction with aberrant hypermethylation is best understood as an evolutionary process that involves progressively increasing methylation of the entire p15CpG island.
PROMOTER REGION CpG island hypermethylation has been reported for tumor-suppressor genes, including both p15 and its functional homolog p16. While p16 is inactivated by this mechanism in many epithelial-derived cancers,1-5 CpG island hypermethylation of p15 occurs almost exclusively in the hematologic malignancies.6-9 This division is most striking in acute myelogenous leukemia (AML), where p15 is frequently inactivated by hypermethylation, but p16 is not inactivated by either homozygous deletion or methylation.7 Hypermethylation ofp15 is also observed in myelodysplasia, and this process appears to be involved in disease progression.10,11 The conservation of p15 hypermethylation in a murine model of T-cell malignancy12 further supports the functional significance of this neoplastic change.
CpG island methylation associated with transcriptional silencing is an important alternative mechanism of silencing genes involved in the pathogenesis of neoplasia.1,2 13-16 However, despite the importance and prevalence of this process in neoplasia, little detail of specific patterns of methylation has been reported. Most analyses of CpG island hypermethylation employ techniques capable of studying only 2 to 4 CpG sites within an island region. CpG island hypermethylation assayed by Southern blot analysis using methylation-sensitive restriction enzymes or, more recently, with polymerase chain reaction (PCR)-based approaches based on restriction enzyme cleavage are examples of this. These techniques depend on the presence or absence of methylation at a small number of CpG sites relative to the total number found across the entire island region. With this approach, CpG islands are often described as either “unmethylated” or “hypermethylated.” Thus, the importance of hypermethylation at 1 or 2 CpG sites and their location relative to transcription start sites remain to be determined. The density and placement of methylation necessary for gene silencing and the issue of allelic heterogeneity of methylation and its relationship to transcription remain unresolved.
The relatively recent addition of bisulfite genomic sequencing17 has markedly improved our ability to assess the methylation status of CpG islands in detail. Such analysis provides the methylation status for each CpG site within a given region, and cloning can provide this information for individual alleles of a given gene. Recent studies have used this technique to demonstrate methylation patterns for critical genes involved in neoplasia, including Rb18, p1619,MGMT,20,21 and BRCA1.22 While each of these studies contributes to our understanding of the methylation-associated silencing of tumor-suppressor genes, the analysis of p16 and MGMT includes only cell lines and the transcriptional consequences of hypermethylation were not studied for BRCA1 or Rb.
In this study, we chose p15 as a model for an analysis of allelic patterns of CpG island methylation for a number of reasons. We had previously observed7 that not all primary acute leukemias were as completely methylated at this CpG island as one would have expected based on the percentage of blast cells within the sample. Because a detailed analysis of the relationship of transcriptional silencing to the methylation pattern of this island has not been reported, such a study might clarify functional questions concerning hypermethylation of p15. Primary leukemia samples provide an especially pure neoplastic sample, minimizing the influence of normal cell contamination. This allowed us to address the transcriptional consequences of specific patterns of promoter region hypermethylation in a primary malignancy, rather than in an established cell culture system.
MATERIALS AND METHODS
Sodium bisulfite DNA treatment.
DNA was extracted from frozen leukemia cell pellets using standard phenol chloroform extraction with ethanol precipitation. In the case of normal control individuals, peripheral blood lymphocytes were extracted from human blood using gradient centrifugation on Histopaque (Sigma Chemical, St Louis, MO). One to 2 μg of DNA was then treated with sodium bisulfite17 according to established methods.23 DNA was resuspended in 15 to 24 μL of distilled water before PCR.
PCR, cloning and sequencing of individual alleles.
Using 4 μL of resuspended, sodium bisulfite–treated DNA, PCR was performed in a 50-μL reaction using 400 nmol/L each primer (GIBCO/BRL, Rockville, MD), 1X PCR Buffer-Mg++ (GIBCO/BRL), 1.5 mmol/L MgCl2(GIBCO/BRL), and 200 μmol/L dATP, 200 μmol/L dCTP, 200 μmol/L dGTP, and 200 μmol/L dTTP (Pharmacia, Piscataway, NJ). Reactions were hot-started at 96°C for 3 minutes and held at 80°C before addition of 1.25 U of Taq (GIBCO/BRL). Temperature conditions for PCR were as follows: 38 cycles of 96°C for 20 seconds, 56°C for 20 seconds, and 72°C for 1 minute followed by 1 cycle of 72°C for 5 minutes. Primers used were 5′-TGAAGGAATAGAAATTTTTTGTTT-3′ and 5′-AAGCAAGCTTAAACCCTAAAACCCCAACTACCTAA-3′.
After this PCR, products were diluted 100-fold and a nested PCR was performed as follows. Either 5 or 10 μL of 100-fold dilution of initial PCR was used in a PCR reaction of 50 μL total volume. Reactions were performed exactly as above except either 2.0 mmol/L or 2.5 mmol/L MgCl2 (GIBCO/BRL) was used. Temperature conditions for PCR were as follows: 30 cycles of 96°C for 20 seconds, 56°C for 20 seconds, and 72°C for 1 minute followed by 1 cycle of 72°C for 5 minutes. Primers used were 5′-GGGGATTAGGAGTTGAG-3′ and 5′-ACCCTAAAACCCCAACTACC-3′.
A 1-μL quantity of this PCR reaction was ligated into pCR 2.1 or pCR 2.1-TOPO (Invitrogen, Carlsbad, CA) vector and cloned exactly according to protocol. Clones were single-colony–purified and cloned. Plasmid was purified using Wizard mini-prep protocol (Promega, Madison, WI); approximately 1 μg of cloned DNA was subjected to automated sequence analysis. The sequencing primer used was 5′-CACCTTCTCCACTAATCCC-3′ and is located at position +424 relative to the p15 transcription start.24 Our sequenced products include 63 CpG sites between positions −215 and +421 relative to the p15 transcription start site.
RNA extraction and reverse-transcriptase PCR of p15.
Total cellular RNA was extracted from pelleted leukemia blastic cells, normal peripheral blood lymphocytes, and cell lines using standard acid guanidium thiocyanate-phenol-chloroform extraction.25 A 3-μg quantity of total RNA was used for reverse-transcriptase (RT) reactions, which were performed in a 15.75-μL final volume. Conditions for RT reactions were as follows: 10.4 μL of a mix containing 6 μL 5X first-strand buffer (Gibco/BRL), 3 μL of 0.1-mol/L DTT (GIBCO/BRL), 0.6 μL of a 25-mmol/L dNTPS mix (≈500 μmol/L each dNTP) (Pharmacia), 0.5 μL random hexamer mix (GIBCO/BRL), and 12 U RNasin (Promega) was added to 18.1 μL of RNA mixed in distilled water. Reactions were mixed, incubated for 5 minutes at 65°C, and placed on ice. A total of 14.25 μL was moved to a new tube (the remainder was used for the RT minus reaction) and 300 U of Moloney Murine Leukemia Virus (MMLV) RT enzyme (GIBCO/BRL) enzyme was then added to this new tube. All reactions (both RT-positive and RT-negative) were then mixed and incubated for 1 hour at 37°C. Reactions were then heated for approximately 5 minutes at 95°C and were frozen at −20°C. In some cases, 6 μg of starting RNA was used and all reaction mixtures were doubled with the exception of a constant amount of MMLV RT enzyme (GIBCO/BRL).
RT reactions were then diluted 5-fold. PCR was performed in a 50-μL final volume. A 10-μL quantity of diluted RT reaction was added to either 400 nmol/L each primer (GIBCO/BRL), 1X PCR buffer,23 1.25 mmol/L dATP, 1.25 mmol/L dCTP, 1.25 mmol/L dGTP, 1.25 mmol/L dTTP (Pharmacia), 3% dimethyl sulfoxide (p15 conditions); or to 400 nmol/L each primer, 1X PCR buffer–Mg++ (GIBCO/BRL), 1.5 mmol/L MgCl2(GIBCO/BRL), 200 μmol/L dATP, 200 μmol/L dCTP, 200 μmol/L dGTP, and 200 μmol/L dTTP (Pharmacia) (GAPDH conditions). Reactions were hot-started at 98°C for 3 minutes and held at 80°C before addition of 1.25 U of Taq enzyme (GIBCO/BRL). Temperature conditions for p15 PCR were as follows: 35 cycles of 98°C for 30 seconds, 56°C for 30 seconds, and 72°C for 1.5 minutes followed by 1 cycle of 72°C for 10 minutes. Temperature conditions forGAPDH PCR were as follows: 23 cycles of 98°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. Primers used were, for p15, 5′-TGGGGGCGGCAGCGATGAG-3′ and 5′-AGGTGGGTGGGGGTGGGAAAT-3′; and, for GAPDH, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′ and 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′. Either 10 μL (p15) or 5 μL (GAPDH) was run on a 3% agarose gel and post-stained with ethidium bromide.
For relative expression quantitation (as shown in Table1), all RT-PCR results were taken into account. Expression was derived based on ethidium staining, as well as blotting and hybridization for most samples. For blotting experiments, 28 cycles of p15 or 20 cycles of GAPDH RT-PCR were performed. Products were blotted to Zeta-probe (Bio-Rad, Hercules, CA), hybridized with a labeledp15 or GAPDH cDNA probe, and the results quantitated via phosphorimager analysis. L117 and L157 were only assayed by ethidium bromide staining analysis due to limited sample.
Methylation Density, Expression Levels, and Leukemia Type for Each Sample Where Both Methylation and RNA Could Be Analyzed
| Sample . | p15 Expression . | % Methylation . | Classification . |
|---|---|---|---|
| KG1a | — | 97.7% | AML |
| L56 | — | 50.4% | AML |
| L63 | — | 37.9% | AML |
| L105 | +/++ | 7.8% | B-ALL |
| L41 | + | 6.0% | AML |
| L117 | +++ | 2.1% | B-ALL |
| Normal | ++++ | 1.8%-3.3% | Mononuclear cells |
| HL60 | +++/++++ | 1.5% | AML |
| L161 | + | 0.5% | B-ALL |
| L3 | ++++ | 0.4% | AML |
| L5* | ++ | 65.6% | AML |
| L224* | + | 85.5% | T-ALL |
| L157* | +/++ | 91.4% | T-ALL |
| Sample . | p15 Expression . | % Methylation . | Classification . |
|---|---|---|---|
| KG1a | — | 97.7% | AML |
| L56 | — | 50.4% | AML |
| L63 | — | 37.9% | AML |
| L105 | +/++ | 7.8% | B-ALL |
| L41 | + | 6.0% | AML |
| L117 | +++ | 2.1% | B-ALL |
| Normal | ++++ | 1.8%-3.3% | Mononuclear cells |
| HL60 | +++/++++ | 1.5% | AML |
| L161 | + | 0.5% | B-ALL |
| L3 | ++++ | 0.4% | AML |
| L5* | ++ | 65.6% | AML |
| L224* | + | 85.5% | T-ALL |
| L157* | +/++ | 91.4% | T-ALL |
Denotes densely methylated leukemia samples with at least 1 fully unmethylated allele present.
RESULTS
The p15 promoter region CpG island is normally devoid of hypermethylation.
To obtain more 5′ sequence of p15 and better characterize the full extent of the p15 CpG island, we cloned thep15 promoter region from a P1 clone24 (data not shown). Using a primer from the reported promoter region ofp15,24 we then obtained additional 5′ sequence.
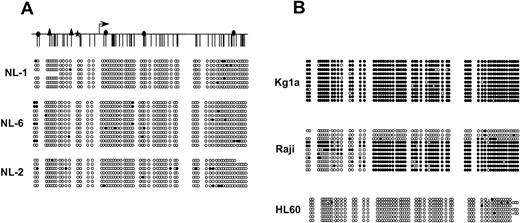
Southern blot analysis previously showed that the EagI site located +167 relative to transcription start of p15 was not methylated in normal tissues,6,8 a result confirmed by methylation-specific PCR analysis of normal bone marrow.7To further detail the patterns of methylation within this region in normal tissues, we performed bisulfite genomic sequencing of 63 CpG sites in the promoter region of p15 in lymphocytes from 3 normal individuals (Fig 1A). At least 7 individual alleles were sequenced for each normal sample studied, revealing an almost completely unmethylated CpG island. However, the existence of a few methylated CpG sites per allele in each individual was found. Interestingly, normal individuals varied slightly in the number of methylated CpG sites found within this otherwise protected region. Although there was great variation between alleles, these methylated sites tended to be more frequent in the far 5′ and 3′ portions of the CpG island.
p15 CpG island methylation state in normal individuals. Representation of the CpG island of p15 showing CpG site methylation. A schematic of the p15 CpG island is shown at the top for reference, and transcription start is denoted by the arrow. The symbols above the schematic represent potentialtrans-acting factor binding sites. (▴) Sp1 binding sites.34 Sunbursts downstream of transcription start () represent several additional potential Sp1 binding sites. (★) represents a potential G/T box. (◢) represents a purine/pyrimidine tract that binds an unidentified potentialtrans-acting factor on gel shift analysis (data not shown). For each sequenced leukemia, each row represents an individual cloned and sequenced allele following sodium bisulfite DNA modification. CpG sites are marked as circles and drawn to accurately reflect CpG density of the region. (•) Methylated CpG sites; (○) unmethylated sites. “x” represents a CpG site for which sequence data was ambiguous. (A) The p15 CpG island is largely unmethylated in normal lymphocytes taken from 3 individuals. (B) Individual cloned and sequenced alleles from 3 leukemia cell lines: KG1a, Raji, and HL60.
p15 CpG island methylation state in normal individuals. Representation of the CpG island of p15 showing CpG site methylation. A schematic of the p15 CpG island is shown at the top for reference, and transcription start is denoted by the arrow. The symbols above the schematic represent potentialtrans-acting factor binding sites. (▴) Sp1 binding sites.34 Sunbursts downstream of transcription start () represent several additional potential Sp1 binding sites. (★) represents a potential G/T box. (◢) represents a purine/pyrimidine tract that binds an unidentified potentialtrans-acting factor on gel shift analysis (data not shown). For each sequenced leukemia, each row represents an individual cloned and sequenced allele following sodium bisulfite DNA modification. CpG sites are marked as circles and drawn to accurately reflect CpG density of the region. (•) Methylated CpG sites; (○) unmethylated sites. “x” represents a CpG site for which sequence data was ambiguous. (A) The p15 CpG island is largely unmethylated in normal lymphocytes taken from 3 individuals. (B) Individual cloned and sequenced alleles from 3 leukemia cell lines: KG1a, Raji, and HL60.
p15 hypermethylation encompasses CpG sites throughout the entire promoter region in primary leukemia and leukemic cell lines.
We first examined the allelic methylation patterns of cell lines already characterized for p15 methylation by Southern analysis (Fig 1B).6 KG1a demonstrated methylation of nearly all CpG sites on every allele sequenced. Raji had dense methylation of the majority of alleles, but also maintained alleles that were nearly devoid of methylated CpG sites. HL60, like the unmethylated primary leukemias, had complete absence of p15 CpG island hypermethylation.
DNA from 13 primary acute leukemias (both AML and acute lymphoblastic leukemia [ALL]) was treated with sodium bisulfite, cloned, and multiple alleles from each sample sequenced. Although most primary AMLs and ALLs are completely methylated by Southern analysis at the EagI site located at position +167 in exon 1 ofp15,7 we chose to analyze several of the more unusual primary leukemia samples previously found to have incomplete methylation at this Eag1 site by Southern blot to investigate the consequences of methylation heterogeneity at the level of individual alleles.
The patterns of CpG island methylation in these primary leukemias were markedly different from normal lymphocytes. This methylation in primary leukemias was found to encompass the entire sequenced CpG island region, and was not restrained to the few sites previously analyzed by Southern blot (Fig 2). Patterns of the individual CpG sites methylated in each leukemia varied greatly, which suggests the absence of site-specific methylation of p15 in this disorder. Rather, the alteration occurred heterogeneously throughout the entire p15 CpG island and often involved many CpG sites.
Methylation status for individual alleles of p15from 13 primary acute leukemias, depicted as in Fig 1. Primary acute leukemias show varying degrees of methylation density, as well as allelic heterogeneity of methylation. No site-specific methylation patterns were seen.
Methylation status for individual alleles of p15from 13 primary acute leukemias, depicted as in Fig 1. Primary acute leukemias show varying degrees of methylation density, as well as allelic heterogeneity of methylation. No site-specific methylation patterns were seen.
A subset of primary leukemia samples does not have hypermethylation ofp15 based on Southern analysis.6-8 We found these to be unmethylated throughout the CpG island region by bisulfite sequencing analysis as well. In these 3 primary leukemia samples (L161, L117, and L3), all alleles are completely devoid of hypermethylation. These primary leukemia samples are hypomethylated compared with normal lymphocytes, where infrequent methylated CpG sites are observed (Fig1A).
Overall methylation density for each sample was derived by calculating the number of methylated CpG sites per sample divided by the total number of CpG sites sequenced per sample. All alleles were included in this analysis, and these data are summarized in Table1.
Allelic heterogeneity of p15 CpG methylation is pronounced in primary acute leukemia and the Raji cell line.
Although many CpG sites throughout the p15 CpG island were affected by methylation, we found that the exact location of methylated sites varied not only between samples, but also between alleles from each leukemia. Two distinct patterns of allelic heterogeneity were apparent. Frequently, we observed cases where the majority of alleles contained hypermethylation of CpG sites, but the individual CpG sites methylated varied from allele to allele. Such was the case for leukemia samples L56, L41, L63, L226, and L129 (Fig 2). This type of heterogeneity suggests instability or infidelity in the process of maintenance methylation at the p15 locus.
However, the patterns observed for leukemias L224, L5, and L157 were different. Here, individual alleles were found to have every CpG methylated on approximately 70% to 90% of alleles and every CpG unmethylated in approximately 10% to 30% of alleles. Thus, instead of showing diversity in CpG site heterogeneity within alleles, these samples show involvement of most, but not all alleles, in extremely dense hypermethylation. While one explanation for this is the presence of unmethylated alleles derived from contaminating normal cells, a similar pattern was found in our sequencing of alleles from the Raji cell line, where normal cell contamination can be excluded. Two distinct patterns of allelic methylation indicate that even within a clonal population of cells, alleles can be maintained in either methylated or unmethylated form.
Transcriptional activity of p15 correlates with overall methylation density and not site-specific methylation placement.
We next explored the relationship of methylation heterogeneity top15 expression. RNA from 10 of the sequenced primary leukemias and from normal lymphocytes, Raji, HL60, and KG1a cell lines was used for RT-PCR analysis (Fig 3). Additionally, lower cycle RT-PCR was performed for 8 of these samples, and these products were blotted, hybridized, and expression quantitated relative to GAPDH via phosphorimager analysis (data not shown). Expression levels for each sample were derived taking into account all RT-PCR data (Table 1). In the case of L157 and L117, expression levels were analyzed based on ethidium staining relative to normal lymphocyte expression and GAPDH.
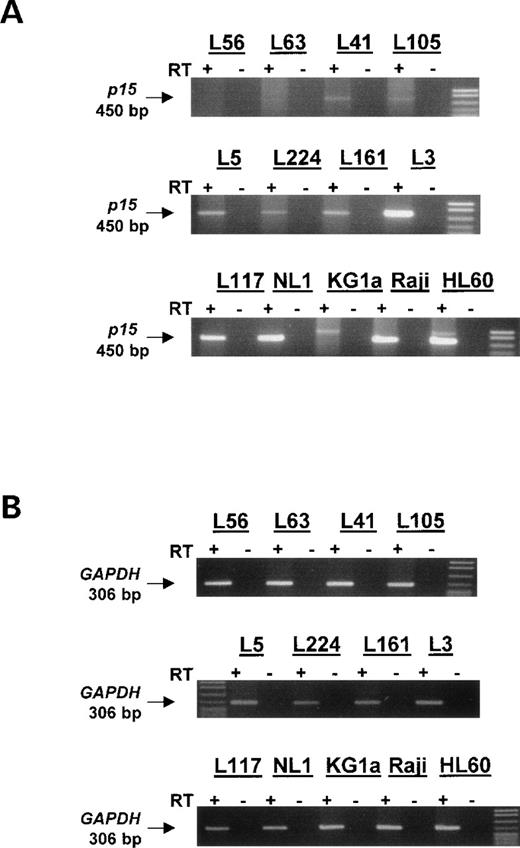
RT-PCR for primary acute leukemias, leukemia cell lines, and normal lymphocytes. Total cellular RNA was recovered from 9 of the sequenced primary acute leukemias in addition to RNA from 3 leukemia cell lines and normal lymphocytes. RT-PCR for p15 was performed to assess transcription in these samples. p15 expression was found to correlate with density of methylation and with the presence of completely unmethylated alleles in a population. KG1a, which does not express p15,6 has a nonspecific PCR product of a size not matching p15. The control GAPDH RT-PCR was done using low cycle number (20 cycles) and is shown below demonstrating that intact and relatively equal RNA was used for each sample. (+) Indicates the addition of MMLV RT enzyme; (−) indicates control RT reaction to which RT enzyme was not added. The size of p15 product is 450 bp and the size of GAPDH product is 306 bp.
RT-PCR for primary acute leukemias, leukemia cell lines, and normal lymphocytes. Total cellular RNA was recovered from 9 of the sequenced primary acute leukemias in addition to RNA from 3 leukemia cell lines and normal lymphocytes. RT-PCR for p15 was performed to assess transcription in these samples. p15 expression was found to correlate with density of methylation and with the presence of completely unmethylated alleles in a population. KG1a, which does not express p15,6 has a nonspecific PCR product of a size not matching p15. The control GAPDH RT-PCR was done using low cycle number (20 cycles) and is shown below demonstrating that intact and relatively equal RNA was used for each sample. (+) Indicates the addition of MMLV RT enzyme; (−) indicates control RT reaction to which RT enzyme was not added. The size of p15 product is 450 bp and the size of GAPDH product is 306 bp.
Normal lymphocytes from 4 sampled individuals express p15 mRNA (Fig 3, Table 1, and data not shown). Similarly, primary acute leukemias that were devoid of p15 hypermethylation (L161, L117, and L3) were shown to transcribe p15 with varying expression levels. Leukemias such as L41 and L105, which have a relatively low density of hypermethylation per allele, express p15, but at a lower level than normal lymphocytes or the unmethylated acute leukemias. Samples that were more densely methylated (>30% of total CpG sites sequenced) such as L56 and L63 did not express detectablep15 mRNA. This result indicates that methylation density, and not merely the presence of hypermethylation, may be of critical importance to transcription. Interestingly, we observed 3 leukemias (L5, L224, and L157) that were able to transcribe p15 despite having a majority of densely methylated alleles (Fig 3 and data not shown). However, as noted earlier, each of these samples also contains 1 or more completely unmethylated alleles.
DISCUSSION
Our results on the allelic methylation status of 63 CpG sites in thep15 promoter region CpG island confirm that aberrant hypermethylation found within the central region of the p15 CpG island represents a leukemic specific process. No significant methylation of CpG sites in this area could be found in normal lymphocytes. Our study also shows pronounced intra-allelic heterogeneity of methylation with varying degrees of methylation density at the p15 CpG island in the leukemias analyzed. Methylation heterogeneity, though perhaps to a smaller degree, has also been observed for Rb in sporadic retinoblastoma.18This variability for involvement of CpG sites in each leukemia suggests that the DNA methyltransferase enzyme does not faithfully maintain methylation at individual CpG sites, and could reflect a type of epigenetic instability in these cells.
In leukemic samples with abnormal methylation in each allele examined, we studied the potential relationship between CpG site methylation density and p15 mRNA. Our data suggest that methylation of 30% to 40% of CpG sites in the CpG island of this gene correlates with a state of complete gene silencing, while lower levels of hypermethylation do not. This level of methylation leading to complete gene silencing is consistent with in vitro methylation of reporter constructs, including graded loss of expression of the Rbpromoter with increasing methylation density26 and complete silencing of RSV LTR promoter activity with methylation density of 20% to 30%.27 In 2 primary leukemias (L105, L41) with methylation densities of 6% to 8%, the decreased p15expression compared with normal lymphocytes suggests that this density may be near the threshold for complete transcriptional silencing of this promoter.
Although there was little variation in p15 expression in lymphocytes taken from normal individuals, we found variable expression levels of p15 between leukemias that are unmethylated. For example, we did observe a single leukemia (L161) that had diminished expression of p15 mRNA despite its lack of methylation in thep15 CpG island. This implicates mechanisms other than CpG island methylation in the regulation of p15 expression. These may include events upstream of p15 (such as disruption of the transforming growth factor-beta [TGFβ] signaling pathway), which could result in the downregulation of p15 transcription in the absence of promoter region hypermethylation.
We found occasional leukemias that had a large proportion of completely methylated alleles and yet express p15 relatively well. Common to these was the presence of a subset of alleles with an absence of any CpG site methylation. Previous studies have not explored the presence of such unmethylated alleles within a population of largely hypermethylated alleles.18 Although these samples were predominantly leukemic blasts, the unmethylated alleles could be from normal lymphocytes and might account for the expression observed. The presence of these alleles certainly correlates with p15expression (L5, L224, and L157). However, it is important to note that in L56, a leukemia for which we show lack of transcription in the presence of a majority of heterogeneously, but densely, hypermethylated alleles, 2 alleles in this population also show a relatively normal methylation pattern. These alleles could simply reflect the randomness of the cloning process, and may not really represent 2 in 12 alleles, since this leukemia has been previously shown to be 98% methylated at the EagI site.
Similar to L5, L224, and L157, the cell line Raji also maintains a state of coexistent densely methylated and unmethylated alleles within a population of cells that are clonal and transformed. This pattern indicates the ability of 2 different allelic patterns to be maintained within a clonal population of cells, similar to the normal processes of allelic distinction such as X chromosome inactivation28 and gene imprinting.29 We have also previously documented this situation for the p16 gene in a colorectal carcinoma cell line.30
Taken together, our data suggest how hypermethylation-associated gene inactivation may occur with time during tumor progression. Recent studies suggest that aberrant promoter methylation can occur early in the progression of some tumors.31 In an experimental cell culture system, overexpression of the DNA methyltransferase results in a time-dependent hypermethylation of several CpG islands.32,33 Recent work suggests that leukemias may be evolving increasing density of methylation at the p15 CpG island over time, since the p15 methylation progresses in the transition of myelodysplasia to overt leukemia, as analyzed either by Southern analysis or methylation-specific PCR.10 11 At any point in time, there may be a wide range of density of methylation within a population of blasts. Our data suggest that once most alleles reach an average density of 30% to 40% of CpG sites methylated within this CpG island, loss of expression of p15 occurs. Such a progressive loss may be an important step in the alterations of cell cycle control characteristic of acute leukemia, and may provide a growth advantage leading to selection of those cells with the most dense p15 methylation.
ACKNOWLEDGMENT
The authors thank Dr Curt Civin for primary acute leukemia samples, Dr Jin Jen for providing us with a P1 clone containing the p15promoter region, and Dr Sanna Myohanen for helpful scientific advice and assistance.
Supported in part by Grant No. CA43318 from the National Institutes of Health.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to James G. Herman, MD, The Johns Hopkins Oncology Center, 424 N Bond St, Baltimore, MD 21231.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal