Abstract
Plasminogen activator inhibitor type-1 (PAI-1), a serine protease inhibitor, affects the processes of fibrinolysis, wound healing, and vascular remodeling. We have demonstrated that PAI-1 transcription is induced by D dimer, a plasmin proteolytic fragment of fibrin, supporting its role in negative feedback on peri-cellular proteolysis. The focus of this study was to define the mechanism of D dimer’s effects on PAI-1 transcription. D dimer increased the binding activity of the transcription factor activator protein-1 components c-fos/junD and c-fos mRNA levels in a time- and concentration-dependent manner to a greater extent than fibrinogen. Both basal and D dimer-induced PAI-1 transcriptional activity were entirely dependent on elements within the −161 to −48 bp region of the PAI-1 gene in fibroblasts. Mutations within the AP-1–like element (−59 to −52 bp) in the PAI-1 gene affected D dimer-induced transcriptional activity, c-fos/junD DNA binding, and basal and c-fos inducible PAI-1 transcriptional activity. Furthermore, expression of either wild-type or mutant c-fos proteins augmented or diminished the response of the PAI-1 promoter (−161 to +26 bp) to D dimer, respectively. D dimer-induced binding of c-fos/junD to the highly conserved and unique AP-1 like element in the PAI-1 gene provides a mechanism whereby specific fibrin fragments control fibrin persistence at sites of inflammation, fibrosis, and neoplasia.
PERSISTENCE OF CROSS-LINKED fibrin is a frequent feature of the development of atherosclerosis, fibrosis, and neoplasia. However, both the mechanism of its persistence and the nature of its effect on specific cellular events remain largely undefined.1-6 Type I plasminogen activator inhibitor (PAI-1) is a 50,000-kD glycoprotein from the serpin superfamily of genes.7,8 Like other serpins, PAI-1 forms protease inhibitor complexes by suicide inactivation, with a second order rate constant for inhibition of tissue type-plasminogen activator (t-PA) and urokinase (u-PA) of approximately 107mol/L−1s−1.9,10 Its physiologic roles include inhibition of fibrinolytic activity in thrombi and extracellular matrix, as well as regulation of other plasmin-dependent processes, including proteolytic and nonproteolytic cell migration through an extracellular matrix and modulation of plasmin-dependent transforming growth factor-β (TGF-β) activation.11-13 PAI-1 participates in the biology of wound healing, lung injury/fibrosis, cancer metastasis, and atherosclerosis through modulation of fibrinolysis or cell adhesion.14-17However, these complex in vivo processes share the common features of PAI-1–expressing mesenchymal cells that adhere to, migrate through, proliferate in, and/or actively remodel a fibrin-rich extracellular matrix.4,18 19
Cellular expression of PAI-1 in fibroblasts can be modulated by numerous inflammatory cytokines, including TGFβ, tumor necrosis factor-α (TNF-α), interleukin-1 (IL-1), and IL-6, as well as by glucocorticoids.20-25 We have recently shown that PAI-1 gene expression is upregulated in fibroblast-like cells in vivo in a rodent model of lung fibrosis, in association with fibrinous matrix deposition.4,26 We reasoned that the fibrin deposition may not only reflect the consequences of PAI-1 inhibition of PA-dependent fibrinolytic activity, but may directly participate in the regulation of PAI-1 expression through a feedback loop mechanism. To examine this possibility, terminal plasmin proteolytic fragments of fibrin were purified and tested for their ability to induce PAI-1 expression in rat lung fibroblasts.27 The carboxyterminal fragment, D dimer, induced PAI-1 transcription and expression to a greater degree than the parent molecule, fibrinogen.27 The focus of this study was to characterize the fibrin fragment-inducedtrans-acting factors and their corresponding ciselements within the regulatory region of the PAI-1 gene. Heterodimers of c-fos/JunD heterodimers were increased in response to D dimer stimulation and bound to the AP-1 like element (−59 to −52 bp) of the PAI-1 promoter in a time-and concentration-dependent manner. Moreover, this D dimer-induced c-fos expression was found to increase PAI-1 transcriptional activation through this same element. The promoter region encompassing this functional element is not present in other known promoters, yet it exhibits strict evolutionary conservation at the nucleotide level among mammalian species (97% identity).28 These findings suggest a key role for this regulatory site in the molecular pathways underlying fibrin dissolution and cell adhesion during the processes of angiogenesis, atherosclerosis, and fibrosis.
MATERIALS AND METHODS
Materials.
The 2,400-bp 5′ flanking region of the rat PAI-1 gene was kindly provided by Dr T. Gelehrter (University of Michigan, Ann Arbor, MI).28,29 Promoterless firefly and Renilla luciferase expression plasmids (pGL2 Basic, pRL-TK) and all reagents for cell lysate luciferase assay were obtained from Promega Corp (Madison, WI). The cytomegalovirus (CMV) expression vector pcDNA3 was obtained from Invitrogen (San Diego, CA). All antibodies to fos/jun family members were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). The rat lung fibroblast cell line (RLF-6) was obtained from ATCC (Bethesda, MD). Cationic lipid transfection reagent (lipofectamine; 3:1 DOSMA:DOPE), serum-free transfection media (Optimem), and HEPES (N-2-hydroxyethylpiperazine-N′-2-ethane sulfonic acid) were obtained from Life Technologies (Gaithersburg, MD). Human fibrinogen and human factor XIII were obtained from Calbiochem (San Diego, CA). Plasmin was obtained from American Diagnostica (Greenwich, CT). Lysine and gelatin sepharose were obtained from Pharmacia Biotech Inc (Piscataway, NJ). The cell propagation media (Ham’s F-12) were obtained from Mediatech (Herndon, VA). Chemical reagents were obtained from Sigma Chemical Co (St Louis, MO). Pathogen-free, male F344 rats were obtained from Charles River Labs (Wilmington, MA). Purified c-fos and JunD proteins and mammalian expression vectors encoding the acidic fos mutant (A-fos) were provided by Dr C. Vinson (Laboratory of Biochemistry, National Cancer Institute, Bethesda, MD).30
Cell isolation and propagation.
Animal protocols used for tissue harvesting were approved by the Institutional Animal Care and Use Committee (#9502067) of the University of Alabama at Birmingham (Birmingham, AL). Primary cultures of rat lung fibroblasts (F344) were established from 6-week-old F344 rats by enzymatic digestion and mechanical disaggregation as previously described.27 Fibroblasts were selected by differential adherence to tissue culture plastic and maintained at 37°C in humidified 5% CO2 in minimal essential media supplemented with 10% fetal bovine serum (FBS), 20 mmol/L HEPES, and penicillin/streptomycin/gentamycin. Cultures were determined to be free of Mycoplasma contamination by polymerase chain reaction (PCR) using Mycoplasma-specific probes as described (Stratagene, La Jolla, CA). Cell identity of primary cultures was verified by the absence of tight junctions, by production of collagen types I and III, and by typical morphology on phase contrast and electron microscopy.27Confluent primary cultures (less than passage number 5) were made quiescent by placement in 0.4% serum for 48 hours before stimulation with and without the indicated agonists in serum-free media. The rat lung fibroblast nontransformed cell line (RLF-6) was grown to confluence in Ham’s F-12 containing 20% FBS as recommended by ATCC. Preliminary experiments showed a PAI-1 induction in the RLF-6 cell line similar to that reported by our laboratory for primary cultures.27
Isolation and preparation of purified plasmin proteolytic fragments of cross-linked fibrin.
Fibrin(ogen) fragments were prepared and purified as published by the investigators as modified from previous methods.27,31-34Separation and purification steps were validated by Coomassie staining of electrophoresed pooled fractions on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and by Western blotting with thrombin, fibronectin, plasminogen, fibrin fragment E, and D domain specific antibodies, as published.31
Preparation of PAI-1 gene-Luciferase expression constructs.
Segments of the rat PAI-1 gene 5′ flanking region were cut (5′-p928-Sma I, p280-Kpn I, p161-Eco72I, 3′ +26-Xho I) and ligated to the Luciferase expression plasmid, pGL2-Basic (Promega). The 5′ deletional construct at −48 to +26 (p48) was generated by PCR using 5′ primers complementary to the PAI-1 gene sequence. Mutations at the putative AP-1 site (p161wt; −59 to −52 site; TGAGTTCA) to the nonbinding mutant site (p161mut; TGTGTTTG) or to the consensus AP-1 binding sequence (p161con; TGAGTCA) were performed by single-strand mutagenesis using commercially available reagents (QuikChange Mutagenesis kit; Stratagene, San Diego, CA) according to the manufacturer’s instructions (Fig 1). Briefly, mutated constructs were generated using the wild-type double-stranded p161-Luciferase as a template. The mutated single-stranded primers were annealed to the template, followed by first-strand synthesis using thermostable Pfu I DNA polymerase (16 cycles of 95°C, 55°C, and 68°C), digestion of the wild-type template with Dpn I, and transformation of the resultant DNA into competent cells (Epicurean Coli-XL2). All constructs were verified by automated DNA sequencing as performed by the University of Alabama at Birmingham Core DNA Sequencing Facility.
Effect of 5′ deletion of the PAI-1 gene on basal and D dimer-inducible PAI-1 transcriptional activity. Progressive deletions of the 5′ flanking region of the PAI-1 gene were generated by restriction digestion or PCR and ligated to a luciferase-encoding reporter plasmid. The plasmids were transiently transfected into rat lung fibroblasts along with the thymidine kinase promoter-Renilla luciferase encoding plasmid. For determination of promoter activity, cells were incubated for 48 hours posttransfection in 0.4% serum containing media followed by either serum-free media ([A], right, basal activity) or SFM plus D dimer (1.1 μmol/L; [B], inducible activity) for 6 hours. Raw data are tabulated as the ratio of firefly luciferase activity to that of its Renillaluciferase activity (mean ± SD) and reported relative to the value for the p928 plasmid (assigned value of 100). *P < .05 relative to the p928 values. +P < .05 relative to the p161wt values by ANOVA/Newman-Keuls test. (A) Relative basal activity. Constructs are as indicated at left. Mutations were performed at the AP-1–like site (−59 to −52) of the PAI-1 gene with otherwise intact −161 to +26. PAIcon marks the TGAGTCA element, PAIwt marks the TGAGTTCA element, and PAImut marks the TGTGTTTG element at the −59 to −52 positions of p161. Underscored letters denote deviations from the consensus AP-1 element. (B) D dimer-inducible transcriptional activity. Transfected plasmids are as indicated.
Effect of 5′ deletion of the PAI-1 gene on basal and D dimer-inducible PAI-1 transcriptional activity. Progressive deletions of the 5′ flanking region of the PAI-1 gene were generated by restriction digestion or PCR and ligated to a luciferase-encoding reporter plasmid. The plasmids were transiently transfected into rat lung fibroblasts along with the thymidine kinase promoter-Renilla luciferase encoding plasmid. For determination of promoter activity, cells were incubated for 48 hours posttransfection in 0.4% serum containing media followed by either serum-free media ([A], right, basal activity) or SFM plus D dimer (1.1 μmol/L; [B], inducible activity) for 6 hours. Raw data are tabulated as the ratio of firefly luciferase activity to that of its Renillaluciferase activity (mean ± SD) and reported relative to the value for the p928 plasmid (assigned value of 100). *P < .05 relative to the p928 values. +P < .05 relative to the p161wt values by ANOVA/Newman-Keuls test. (A) Relative basal activity. Constructs are as indicated at left. Mutations were performed at the AP-1–like site (−59 to −52) of the PAI-1 gene with otherwise intact −161 to +26. PAIcon marks the TGAGTCA element, PAIwt marks the TGAGTTCA element, and PAImut marks the TGTGTTTG element at the −59 to −52 positions of p161. Underscored letters denote deviations from the consensus AP-1 element. (B) D dimer-inducible transcriptional activity. Transfected plasmids are as indicated.
Transient transfection of rat lung fibroblasts.
Transient transfection of a nontransformed cell line of rat lung fibroblasts (RLF-6; 3.5 × 105 cells in 35-cm2 wells) was performed with cationic liposomes (6 μg/1.5 μg DNA; Lipofectamine). Cells were plated at the above-noted density in Ham’s F-12/20% FBS, washed twice with Optimem, and incubated with the lipid/DNA complexes for 6 hours in Optimem. At this point, the cells were incubated with Ham’s F12 with 0.4% FBS or Ham’s F12 containing D dimer (1 μmol/L) for 48 hours. Alternatively, the cells were transfected incubated in Ham’s F12 containing 0.4% serum for 48 hours followed by either serum-free media or D dimer (1 μmol/L) in serum-free media for 6 hours. The full coding regions of wild-type rat c-fos (c-fos) and the c-fos mutant (fosBR; basic DNA binding region deletion; kindly provided by Dr T. Townes, University of Alabama at Birmingham) were ligated into the mammalian cell expression plasmid, pcDNA3, downstream of the CMV early enhancer-promoter region and a 3′ bovine growth hormone polyadenylation signal. The indicated PAI-1 promoter-luciferase plasmids (1 μg/well) were transfected along with either c-fos, fosBR, or acidic fos (Afos; acidic amino acid substituted DNA binding region) expression plasmids (0.15 μg/well) and a Renilla luciferase encoding plasmid (pTK-RL; 0.15 μg/well) as a standard for transfection efficiency in Lipofectamine (5 μg/well). Firefly luciferase activity was assayed in cell lysates (25 mmol/L Tris-phosphate, pH 7.8, 1% TX-100, 2 mmol/L dithiothreitol [DTT], 2 mmol/L 1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid, 10% glycerol) in the presence of coenzyme A, ATP, and the substrate luciferin. Renilla luciferase activity was assayed sequentially in the same sample by quenching the firefly luciferase activity and adding the Renilla luciferase specific substrate, coelenterazine, according to the manufacturer’s instructions (Promega). Light units were measured on a Luminometer (Turner Model 20e; Promega), and data are expressed as the ratio of firefly luciferase activity to that of Renilla luciferase to account for variability in transfection efficiency among wells and conditions. Preliminary experiments using cell lysates demonstrate linearity (r2 = .998) and reproducibiity (CV = 3%) of the luciferase assays over 6 logs in the range of our measurements. Luciferase activity values of greater than 3 standard deviations above background were accepted.
Electrophoretic mobility shift assay (EMSA).
Primary rat lung fibroblasts were exposed to D dimer (at the indicated concentrations in Dulbecco’s modified Eagle’s medium [DMEM]) or serum-free DMEM for 15 to 60 minutes, and nuclear extracts were prepared, as described.35 36 Washed cells (106 to 107) were pelleted in phosphate-buffered saline (PBS; 4°C) and lysed by repeated pipetting at 4°C (in 10 mmol/L HEPES, pH 7.9, 1.5 mmol/L MgCl2, 10 mmol/L KCL, 0.5 mmol/L DTT, and 0.2 mmol/L phenylmethyl sulfonyl fluoride [PMSF]). Nuclear proteins were extracted (420 mmol/L NaCl, 20 mmol/L HEPES, pH 7.9, 1.5 mmol/L MgCl2, 0.5 mmol/L DTT, and 0.2 mmol/L PMSF) and total protein was measured by commercially available methods (BCA method; Pierce, Rockford, IL). Oligonucleotides used in EMSA assays were (1) PAIcon, a 21 mer consisting of the AP-1 consensus region (TGAGTCA) flanked by 14 bp from the PAI-1 gene (−66 to −60 and −51 to −45); (2) PAIwt, a 21 mer composed of the wild-type PAI-1 sequence (TGAGTTCA; −60 to −52) plus flanking sequence (−66 to −45); or (3) PAImut, a 21 mer composed of a mutated AP-1 site (TGTGTTTG) flanked by the PAI-1 sequence (−66 to −60 and −51 to −45; bold denotes variation from the consensus AP-1). Oligonucleotides were labeled with [α-32P]-nucleotide using the Klenow filling reaction, resulting in a specific activity of at least 106 cpm/μg DNA. The labeled oligonucleotide (20,000 cpm) was allowed to combine with 1 μg of nuclear protein for 20 minutes at 25°C (10 mmol/L HEPES, 0.1 mmol/L EDTA, 10% glycerol, 100 mmol/L KCl, 200 μg/mL bovine serum albumin [BSA], 1 mmol/L DTT, and 0.2 mmol/L PMSF) in the presence of the nonspecific competitor poly dI-dC (100 μg/mL). For specific competition experiments, a varying molar excess (up to 100-fold) of cold unlabeled oligonucleotide was added to the labeled probe before binding reaction. For immunological identification of the transcription factor(s), factor-specific antibodies (1 to 1,000 ng/reaction; Santa Cruz Biotechnologies, Santa Cruz, CA) were added to the binding reaction (30 minutes at 4°C) before the addition of the oligonucleotide. In all cases, the entire reactions (20 μL) were electrophoresed under nondenaturing conditions (4.8% to 6% acrylamide/0.5× TBE at 500 V for 1 hour at 4°C), followed by autoradiography. Band radioactivity was quantified using a phosphorimager (Molecular Dynamics, Sunnyvale, CA), and autoradiograms were made for presentation in light tight cassettes with intensifier screens at −70°C (Kodak XAR film; Eastman Kodak, Rochester, NY).
Northern blot analysis.
Total RNA was isolated from rat lung fibroblasts under the indicated conditions and analyzed by Northern blotting as previously published by our laboratory.4 27 Band density was quantified using a phosphorimager, and data are presented as the ratio of the c-fos mRNA specific band to that of the 18S rRNA band to normalize for loading and transfer. Autoradiograms were made for presentation in light tight cassettes with intensifier screens at −70°C (Kodak XAR film).
Statistical analysis.
Interval data were compared by means of an ANOVA.37 Where differences were detected, a Dunnett’s test or Newman-Keuls multiple comparisons procedures were used to test for differences in inducible firefly/Renilla luciferase activities between specific promoter constructs or in c-fos mRNA induction or AP-1 binding activity under specific conditions.37 Results are expressed as the mean ± standard deviation unless otherwise indicated. Significance was accepted at the P < .05 level for all analyses.37
RESULTS
Determination of the critical DNA regions for basal and D dimer-inducible PAI-1 transcriptional activity.
To determine the basal and D dimer-responsive elements from the 5′ flanking region of the PAI-1 gene, we performed transient transfections with several PAI-1 promoter-luciferase reporter constructs in rat lung fibroblasts. Upon progressive 5′ deletion of the PAI-1 promoter, the basal activity first increased slightly (p928 v p280) and then decreased 75-fold (p928 v p48; Fig 1A). Furthermore, upon mutation at the AP-1–like element at −59 to −52 bp (p161wt to p161mut), the basal PAI-1 promoter activity was reduced to approximately one third of its wild-type activity, whereas replacement of the same element with that of the consensus AP-1 sequence (p161con) increased the PAI-1 promoter activity by 50% greater than that of the wild-type (Fig 1A).
D dimer directly increased transcription of PAI-1 mRNA, as measured by nuclear run-off assay, and similarly enhanced PAI-1 promoter-driven luciferase activity 4.5- ± 1-fold (using p928) in rat lung fibroblasts. Relative to the largest promoter fragment tested, D dimer’s effect was preserved in the wild-type −161 to −48 region (p161) but was reduced by two thirds upon mutation of the AP-1–like element from the same region (−59 to −52 bp; p161mut) and lost upon deletion of this region (p48; Fig 1B). In support of the importance of the AP-1–like element, D dimer’s effect was slightly enhanced by the presence of the consensus AP-1–containing element (in p161con; Fig 1B). We also noted a blunting of D dimer’s effect using the p280 construct (−280 to +26 bp), but its restoration upon further 5′ deletion (to p161wt; Fig 1B). In summary, these data indicate that the precise sequence at the AP-1–like element at −59 to −52 regulates both basal and D dimer-inducible PAI-1 promoter activity.
Time and concentration dependence of D dimer-induced DNA-protein binding.
Having demonstrated the AP-1–like element at −59 to −52 of the PAI-1 gene was functionally relevant, we sought to characterize the time and dose response of nuclear protein binding to D dimer initially by an oligonucleotide containing a known AP-1 consensus element (PAIcon). Rat lung fibroblasts were incubated with highly purified fibrin D dimer, and nuclear extracts were assayed by EMSA. Band density was quantified by densitometry. D dimer exposure resulted in a rapid, transient, 5-fold increase in binding activity to PAIcon oligonucleotide (PAIcon; −66 to −46; with consensus AP-1 site at −59 to −53; TGAGTCA) that was concentration dependent (n = 5/condition; P < .01). This activity peaked at 45 minutes after exposure to 1 μmol/L D dimer and was noted with as little as as little as 50 nmol/L D dimer, with the curve plateau at 1 μmol/L D dimer after 45 minutes. Although this activity was similarly demonstrable with an oligonucleotide consisting of the wild-type PAI-1 gene sequence (PAIwt; wt −66 to −45; −59 to −52 is TGAGTTCA), it was sequence specific. The DNA-protein complex B was effectively competed by 100-fold excess of unlabeled oligonucleotide, either PAIcon or PAIwt, but not a mutated PAI gene oligonucleotide PAImut (PAImut; wt −66 to −45; −59 to −52 is TGTGTTTG; see Fig 2B), and the DNA-protein complex formed with the PAIcon or PAIwt oligonucleotides had the same migration pattern, whereas there were no retarded bands with the free probe control (Fig 2B). Because the DNA element within the PAI-1 gene (−59 to −52) varies from the AP-1 consensus by a 1-bp insert (consensus, TGAGTCA; PAI-1 gene, TGAGTTCA), we compared the in vitro affinity of the induced nuclear proteins towards the consensus AP-1 sequence, the AP-1 like sequence in the wild-type PAI-1 gene, and a triple AP-1 site mutant (TGTGTTTG; PAImut) by competition EMSA. Quantitative cross-competition analysis for binding of the D dimer-induced nuclear proteins to32P-labeled PAIcon showed a 50-fold difference in apparent affinity to the wild-type sequence relative to that of the consensus sequence (Fig 2A [• and ○] and B). In contrast, the triply mutated oligonucleotide (PAImut) did not effectively compete for binding (Fig 2A [▪] and B).
Relative affinity of D dimer-induced proteins for the PAI-1 AP-1–like sequence and the AP-1 consensus sequence. Nuclear proteins from D dimer-exposed fibroblasts (1 μmol/L for 45 minutes) were analyzed by EMSA with a 32P-PAIcon oligonucleotide (−66 to −45, del-55; TGAGTCA). The indicated molar excesses of unlabeled PAIcon, PAIwt (TGAGTTCA; PAIwt) or PAImut (TGTGTTTG) oligonucleotides were added to the binding reactions, and the reactions were analyzed by EMSA. Band densities of the retarded B complexes were quantified on a phosphorimager. (A) Quantitative competition of protein binding to32P-labeled PAIcon by unlabeled PAIcon, PAIwt, and PAImut. Data are plotted as band radioactivity (mean + SE) relative to that with no unlabeled PAIcon. (B) Representative EMSA autoradiogram. Lane 1, free labeled PAIcon probe; lanes 2 through 5, labeled PAIcon with indicated molar excesses of unlabeled PAIcon added to the protein binding reaction; lanes 6 through 9, labeled PAIcon with indicated molar excesses of unlabeled PAIwt; and lanes 10 and 11, labeled PAIcon with indicated molar excess of unlabeled PAImut.
Relative affinity of D dimer-induced proteins for the PAI-1 AP-1–like sequence and the AP-1 consensus sequence. Nuclear proteins from D dimer-exposed fibroblasts (1 μmol/L for 45 minutes) were analyzed by EMSA with a 32P-PAIcon oligonucleotide (−66 to −45, del-55; TGAGTCA). The indicated molar excesses of unlabeled PAIcon, PAIwt (TGAGTTCA; PAIwt) or PAImut (TGTGTTTG) oligonucleotides were added to the binding reactions, and the reactions were analyzed by EMSA. Band densities of the retarded B complexes were quantified on a phosphorimager. (A) Quantitative competition of protein binding to32P-labeled PAIcon by unlabeled PAIcon, PAIwt, and PAImut. Data are plotted as band radioactivity (mean + SE) relative to that with no unlabeled PAIcon. (B) Representative EMSA autoradiogram. Lane 1, free labeled PAIcon probe; lanes 2 through 5, labeled PAIcon with indicated molar excesses of unlabeled PAIcon added to the protein binding reaction; lanes 6 through 9, labeled PAIcon with indicated molar excesses of unlabeled PAIwt; and lanes 10 and 11, labeled PAIcon with indicated molar excess of unlabeled PAImut.
Identification of the components of the D dimer-inducible DNA-protein complex.
To identify the transcription factor(s) that was induced to bind to the functional AP-1–like element (−66 to −45) in the PAI-1 gene upon D dimer exposure, EMSAs were performed on the same nuclear extracts in the presence of factor-specific antibodies. Direct competition and/or supershifting of complex B was noted with antibodies to both fos and jun family proteins, but not those raised against the irrelevant antibody CD-11b (32P-PAIcon oligo; Fig 3A). This pattern of competition/supershifting strongly suggests that members of the AP-1 family are induced to bind by D dimer. A more detailed analysis using antibodies specific to individual fos/jun proteins indicates that c-fos and junD are major components of D dimer-induced fos/jun proteins in complex B, whereas fra-2 and junB appear to play a minor role (32P-PAIcon; Fig 3B). Identical fos/jun family members bind to the PAI-1 wild-type oligonucleotide (32P-PAIwt oligo; Fig 3C) as the consensus. Taken together, these observations indicate that D dimer fragment will rapidly induce the DNA binding activity of c-fos/junD heterodimers in rat lung fibroblasts.
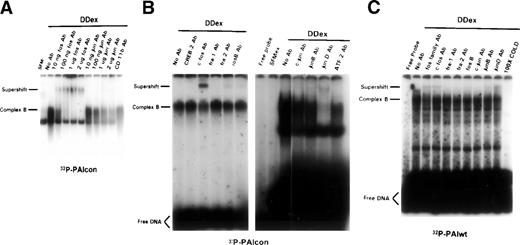
Immunologic identification of D dimer-induced DNA binding proteins. Subconfluent rat lung fibroblasts were incubated in 0.4% serum for 48 hours followed by exposure to D dimer (1 μmol/L for 45 minutes) or serum-free media controls (SFM). Nuclear proteins were extracted and analyzed by EMSA with 32P-PAIcon (A and B; PAIcon) or 32P-PAIwt (C) as indicated below the autoradiograms. The effect of antibodies that are broadly cross-reactive to fos/jun proteins and to specific fos/jun family members on the protein-DNA binding was assessed through an examination of the competition or supershifting of complex B in the EMSA autoradiogram. (A) Increasing titer of fos (lanes 3 through 6) and jun (lanes 7 through 10) family antibodies from 1 to 2,000 ng/binding reaction. Antibody to CD11b serves as an irrelevant control antibody. (B) Left autoradiogram, indicated antibodies to specific fos family members, with negative control (CREB-2 Ab). Right autoradiogram, indicated antibodies to specific fos family members, with negative control (ATF-2 Ab). (C) Indicated antibodies to specific fos and jun family members, with 32P-labeled PAIwt.
Immunologic identification of D dimer-induced DNA binding proteins. Subconfluent rat lung fibroblasts were incubated in 0.4% serum for 48 hours followed by exposure to D dimer (1 μmol/L for 45 minutes) or serum-free media controls (SFM). Nuclear proteins were extracted and analyzed by EMSA with 32P-PAIcon (A and B; PAIcon) or 32P-PAIwt (C) as indicated below the autoradiograms. The effect of antibodies that are broadly cross-reactive to fos/jun proteins and to specific fos/jun family members on the protein-DNA binding was assessed through an examination of the competition or supershifting of complex B in the EMSA autoradiogram. (A) Increasing titer of fos (lanes 3 through 6) and jun (lanes 7 through 10) family antibodies from 1 to 2,000 ng/binding reaction. Antibody to CD11b serves as an irrelevant control antibody. (B) Left autoradiogram, indicated antibodies to specific fos family members, with negative control (CREB-2 Ab). Right autoradiogram, indicated antibodies to specific fos family members, with negative control (ATF-2 Ab). (C) Indicated antibodies to specific fos and jun family members, with 32P-labeled PAIwt.
The AP-1 dependence of PAI-1 promoter activity is sequence specific and a function of c-fos/JunD availability.
To determine the functional significance of the AP-1 increases noted in response to D dimer, wild-type and altered PAI-1 promoter fragments were tested for their c-fos dependence in cotransfection assays. Expression of wild-type c-fos augmented both basal (SFM or 0.4% serum) and D dimer-inducible PAI-1 promoter activity in a sequence-specific manner. Upon triple mutation (p161mut) or deletion (p48) of the AP-1–like element, there was a 3- and 100-fold loss of basal transcriptional activity, respectively, as well as a loss of c-fos inducibility (Fig 4A). A comparison of the c-fos inducibility seen with consensus AP-1 sequence-containing promoter (p161con) with that of the wild-type (p161wt) showed an increased dependence of the consensus-containing promoter (p161con) to c-fos by 2-fold (Fig 4A).
Effect of AP-1 protein overexpression on basal and D dimer-inducible PAI-1 transcriptional activating activity. Subconfluent rat lung fibroblasts were cotransfected with plasmids expressing either wild-type c-fos (c-fos) or JunD (JunD), a nonfunctional mutant c-fos (DNA-binding basic region deleted; fos BR), or a peptide containing an acidic amino acid-substituted DNA binding region (A-fos) along with the indicated PAI-1 promoter-luciferase reporter constructs and the transfection control Renillaluciferase reporter plasmid. p161con, p161wt, p161mut, and p48 PAI-1 promoter-luciferase constructs are as defined in Fig 1. Results are plotted as the ratio of firefly/Renilla luciferase activities (mean + SE) relative to the activity ratio seen with the empty vector. (A) Sequence specificity of c-fos induction. (▨) Empty vector; (□) fos BR vector; (▪) c-fos vector. *P < .05 relative to the mutant fos values. (B) Effect of dominant negative c-fos (A-fos) on D dimer inducibility of p161wt. Fibroblasts were transfected as described above and exposed to 0.4% serum-containing media for 48 hours followed by either D dimer (1 μmol/L) or serum-free media for 6 hours. (▨) Empty vector; (□) A-fos vector; (▪) JunD or JunD/c-fos vectors. *P < .05 relative to the empty vector. +P < .05 relative to the c-fos vector by ANOVA/Newman-Keuls test.
Effect of AP-1 protein overexpression on basal and D dimer-inducible PAI-1 transcriptional activating activity. Subconfluent rat lung fibroblasts were cotransfected with plasmids expressing either wild-type c-fos (c-fos) or JunD (JunD), a nonfunctional mutant c-fos (DNA-binding basic region deleted; fos BR), or a peptide containing an acidic amino acid-substituted DNA binding region (A-fos) along with the indicated PAI-1 promoter-luciferase reporter constructs and the transfection control Renillaluciferase reporter plasmid. p161con, p161wt, p161mut, and p48 PAI-1 promoter-luciferase constructs are as defined in Fig 1. Results are plotted as the ratio of firefly/Renilla luciferase activities (mean + SE) relative to the activity ratio seen with the empty vector. (A) Sequence specificity of c-fos induction. (▨) Empty vector; (□) fos BR vector; (▪) c-fos vector. *P < .05 relative to the mutant fos values. (B) Effect of dominant negative c-fos (A-fos) on D dimer inducibility of p161wt. Fibroblasts were transfected as described above and exposed to 0.4% serum-containing media for 48 hours followed by either D dimer (1 μmol/L) or serum-free media for 6 hours. (▨) Empty vector; (□) A-fos vector; (▪) JunD or JunD/c-fos vectors. *P < .05 relative to the empty vector. +P < .05 relative to the c-fos vector by ANOVA/Newman-Keuls test.
To define the significance of c-fos and JunD to D dimer’s effect on PAI-1 promoter activity, fibroblasts were cotransfected with the p161 reporter construct and either a wild-type c-fos expressing vector, an empty vector, or an acidic mutant fos expressing vector and incubated with D dimer. Preliminary experiments documented the capacity of a 2-fold molar excess of the acidic mutant fos protein to block 99% of c-fos/JunD binding to the wild-type PAI-1 oligonucleotide (PAIwt; −66 to −45 bp) by EMSA (data not shown). Expression of wild-type c-fos (c-fos) or JunD (junD) enhanced the capacity of the cell to respond to D dimer with an increase in PAI-1–promoter activity, whereas transfection of the mutant fos (A-fos) diminished the capacity of the cell to respond to D dimer (Fig 4B). These data indicate that manipulation of the availability of c-fos and junD to bind to DNA will alter the basal and D dimer responsiveness of the PAI-1 promoter in a sequence-specific manner.
Relative induction of c-fos by D dimer and its parent molecule fibrinogen.
Because we have shown in prior work that PAI-1 expression was greater in fibroblasts incubated with D dimer than with fibrinogen, the effects of fibrinogen and D dimer on AP-1 element binding activity and mRNA levels for c-fos and Jun D were similarly compared. Both proteins induced DNA binding activity to both the wild-type oligonucleotide (PAIwt) and the AP-1 consensus-bearing oligonucleotide (PAIcon) by EMSA relative to serum-free media after 45 minutes (n = 3/condition; P = .0002, ANOVA; Fig 5, complex B: left, PAIcon; right, PAIwt). However, the increase in DNA binding activity in cells exposed to D dimer was 2.5-fold greater than that of the fibrinogen (P< .05) and equivalent to that of 10% serum-exposed cells (Fig 5). Similarly, whereas both proteins increased c-fos mRNA steady-state levels, the peak D dimer response was 4-fold greater in magnitude (n = 3/condition; P = .001, ANOVA) and tended to last longer (P = .08) than that of fibrinogen (Fig 6A and B). JunD mRNA, in contrast, was not induced by either protein (data not shown). This quantitation was performed by normalizing the c-fos or JunD mRNA band density of each lane with that of the 18S RNA to account for any variability in loading and transfer. In summary, the differences between D dimer and fibrinogen-induced AP-1 responses in the fibroblasts were largely in the magnitude of the response in c-fos mRNA.
Comparison of fibrinogen and D dimer induction of AP-1–DNA binding activity by EMSA. Fibroblasts were incubated with either fibrinogen or D dimer (1 μmol/L) for 45 minutes in serum-free media, followed by harvesting of nuclear extracts, which were analyzed by EMSA using a 32P-labeled PAIcon (A, left) or PAIwt (A, right) oligonucleotides. Complex B band density was quantified on a phosphorimager and plotted as mean +SE (n = 3/condition) in (B). Serum-free media or serum (10%) -containing media-exposed cells served as negative and positive controls, respectively. DDex, FGNex, 10%Sex, and SFMex denote nuclear extracts from D dimer, fibrinogen, 10% serum-containing media, and serum-free media-exposed cells, respectively. 100X PAIcon denotes inclusion of a 100-fold molar excess of unlabeled PAIcon oligonucleotides in the binding reaction. (A) Left, EMSA using the 32P-labeled PAIcon oligonucleotide; right, EMSA using the 32P-labeled PAIwt oligonucleotide. Complex B is indicated. (B) Quantitative presentation of complex B band density (AP-1). (□) Data from 32P-labeled PAIcon oligonucleotide; (▪) 32P-labeled PAIwt oligonucleotide.
Comparison of fibrinogen and D dimer induction of AP-1–DNA binding activity by EMSA. Fibroblasts were incubated with either fibrinogen or D dimer (1 μmol/L) for 45 minutes in serum-free media, followed by harvesting of nuclear extracts, which were analyzed by EMSA using a 32P-labeled PAIcon (A, left) or PAIwt (A, right) oligonucleotides. Complex B band density was quantified on a phosphorimager and plotted as mean +SE (n = 3/condition) in (B). Serum-free media or serum (10%) -containing media-exposed cells served as negative and positive controls, respectively. DDex, FGNex, 10%Sex, and SFMex denote nuclear extracts from D dimer, fibrinogen, 10% serum-containing media, and serum-free media-exposed cells, respectively. 100X PAIcon denotes inclusion of a 100-fold molar excess of unlabeled PAIcon oligonucleotides in the binding reaction. (A) Left, EMSA using the 32P-labeled PAIcon oligonucleotide; right, EMSA using the 32P-labeled PAIwt oligonucleotide. Complex B is indicated. (B) Quantitative presentation of complex B band density (AP-1). (□) Data from 32P-labeled PAIcon oligonucleotide; (▪) 32P-labeled PAIwt oligonucleotide.
Induction of c-fos mRNA by D dimer. Subconfluent rat lung fibroblasts were incubated in 0.4% serum for 48 hours followed by incubation with either fibrinogen or D dimer (1 μmol/L) in serum-free media for 15 to 60 minutes as indicated. Northern blotting of the RNA was performed as described in Materials and Methods. Autoradiogram bands were analyzed by phosphorimagery. Data are plotted as the mean + SE of the band density relative to that of SFM alone (n = 3/time point). (A) Representative autoradiogram. SFM, FBS, FGN, and D dimer denote the mRNA from serum-free media, 10% fetal bovine serum, fibrinogen, and D dimer-exposed cells, respectively. (B) Fold induction of c-fos/18S RNA (loading and transfer control) band intensity values relative to that of cells incubated in serum-free media. (□) Fibrinogen-exposed cells; (▪) D dimer-exposed cells.
Induction of c-fos mRNA by D dimer. Subconfluent rat lung fibroblasts were incubated in 0.4% serum for 48 hours followed by incubation with either fibrinogen or D dimer (1 μmol/L) in serum-free media for 15 to 60 minutes as indicated. Northern blotting of the RNA was performed as described in Materials and Methods. Autoradiogram bands were analyzed by phosphorimagery. Data are plotted as the mean + SE of the band density relative to that of SFM alone (n = 3/time point). (A) Representative autoradiogram. SFM, FBS, FGN, and D dimer denote the mRNA from serum-free media, 10% fetal bovine serum, fibrinogen, and D dimer-exposed cells, respectively. (B) Fold induction of c-fos/18S RNA (loading and transfer control) band intensity values relative to that of cells incubated in serum-free media. (□) Fibrinogen-exposed cells; (▪) D dimer-exposed cells.
DISCUSSION
The major finding in this study is the identification of the AP-1–like DNA element as an important transcriptional control element in the PAI-1 gene. In support of our key observation, deletion or mutation of this AP-1–like element significantly affects the level of basal transcription and D dimer responsiveness of a PAI-1 promoter-luciferase reporter construct. Furthermore, the basal transcription and D dimer responsiveness of the PAI-1 promoter is altered in a sequence-specific manner by manipulation of intracellular AP-1 levels in cotransfection assays. Our analysis of AP-1 family members in D dimer exposed cells indicates that D dimer enhances c-fos/junD DNA binding activity to the AP-1–like element (−59 to −52) in a dose- and time-dependent manner. We have demonstrated the specificity of this binding activity by competition EMSA by varying both the nucleotide sequence and by using antibodies to specific fos/jun family members. Furthermore, altering the levels of fos/jun resulted in detectable changes in PAI-1 promoter activity, which was similarly nucleotide sequence specific. Taken together, these data suggest that, in response to D dimer stimulation, AP-1 activity increases to enhance PAI-1 transcription through its unique AP-1–like element at −59 to −52 in the PAI-1 promoter.
The PAI-1 gene sequence of the rat, mouse, and human species are strikingly similar (97% homologous) in the region from −60 to the TATA box (−31), with identical AP-1–like element sequences, whereas the upstream region (−928 to −60) is significantly less homologous (70%).28 Furthermore, this proximal region appears to be entirely unique among known promoters in the NCBI database, and this same element has been implicated in the PAI-1 transcriptional response to TGF-β and to phorbol myristate acetate (PMA) in several cell types.38-40 This suggests that the elements within this proximal region of the PAI-1 promoter participate in cell responses that are both unique and critical to PAI-1 physiology. One possible function of this highly conserved proximal region may be to coordinate the AP-1–dependent expression of PAI-1 with the cell cycle.41-46 In this context, PAI-1 protein may participate in cell cycle regulation by modulating the integrity of cell-matrix contacts, either through its matrix proteolytic inhibitory action and/or through blocking of cell surface receptor-ligation sites on matrix vitronectin.14,15 47
The AP-1 response to D dimer, as characterized, is both specific and physiologically relevant. First, the D dimer fragments used were shown to be free of other PAI-1 regulatory molecules, including plasmin, thrombin, fibronectin, t-PA, and TGF-β, by immunoblotting. Second, the magnitude of the increase in AP-1 binding activity and c-fos mRNA in response to D dimer was 3 times that of equimolar amounts of fibrinogen. D dimer was also a more potent inducer (3-fold) of AP-1 binding activity than equimolar fibrinogen fragment E (1 μmol/L) or 0.4% serum (280 μg/mL protein; not shown), thereby ruling out a nonspecific protein effect. Interestingly, the relative induction of c-fos mRNA and AP-1–DNA binding activity in response to D dimer versus fibrinogen corresponds to their relative potency of induction of PAI-1 protein and mRNA.27 This suggests that D dimer possesses conformationally dependent AP-1 signaling epitopes that regulate PAI-1 expression. These epitopes may be exposed upon plasmin cleavage of cross-linked fibrin in a manner similar to that seen with thrombin-cleavage of fibrinogen. The thrombin-cleaved fibrinogen β chain possesses heparin-binding domains important for its binding to endothelial cells.48,49 Potential D dimer conformational epitopes have yet to be characterized, despite that fact that it is the major soluble fibrin-derived molecule in tissue during the matrix-remodeling process seen after inflammatory injury or thrombosis.26,50 51
Under the conditions of the competitive EMSA, it appears that the affinity of the inducible AP-1 for the PAI-1 wild-type promoter AP-1–like element (TGAGTTCA) is lower than that for the consensus element (TGAGTCA). However, it is apparent that the weak interactions at the wild-type element can result in physiologically significant transcriptional responses. In support of this contention, the level of c-fos-inducible PAI-1 promoter activity within the wild-type construct (p161wt) or the consensus construct (p161con) differed by only 2-fold, and the basal PAI-1 promoter activity was approximately equivalent (Figs 1, 2, and 4), whereas the nonbinding mutant promoter activity was one third less and was not inducible upon c-fos cotransfection. This apparent difference in relative affinity and transcriptional activating activity may reflect the importance of the state of activation of fos, of interactive effects with coactivators, of DNA conformational effects, or of redundancy among AP-1 family members.52-54
Several lines of evidence point towards c-fos/junD as the AP-1 member of greatest significance. Namely, c-fos/junD are the most prominent AP-1 family members present in D dimer exposed cells by EMSA, the effect of cotransfection of c-fos is dependent on the sequence at the AP-1–like element (−59 to −52), and cotransfection of A-fos, which will bind to Jun protein, thereby blocking Jun-related AP-1 transcriptional activity, partially abrogated the PAI-1 promoter response to D dimer. Additional indirect evidence is that the relative potency of c-fos induction by D dimer and fibrinogen parallels that of their PAI-1 inducing capacity.27 Because EMSA is a semiquantitative technique, it remains possible that quantitatively minor, or as yet unknown, AP-1 family members participate in the transcriptional response.
The persistence of extracellular matrix fibrin in disease states, including fibrosis, neoplasia, and atherosclerosis, demonstrates the consequences of blocked fibrinolysis in these disorders, and recent studies in mice that are deficient in plasminogen and/or fibrinogen illustrate the basic biological significance of fibrin/fibrinolysis to matrix remodeling.1,5,26,51 55 Our model suggests a mechanism whereby the fibroblast may control peri-cellular proteolysis of its matrix environment through AP-1–mediated PAI-1 induction (Fig 7). A PAI-1–inductive signal is initiated by fibrin fragments that are the product of plasminogen activator-dependent plasmin action and are mediated by AP-1–dependent transcriptional activation of the PAI-1 gene. The PAI-1 produced will block further fibrinolysis (D dimer formation) through PAI-1’s inhibitory action on plasminogen activation, thereby closing a negative regulatory feedback loop. This feedback of proteolytically derived matrix fragments on peri-cellular fibrinolytic activity may underlie the cellular sensing and maintenance of extracelluar matrix integrity in both normal and disease states.
Model of D dimer-initiated negative regulatory feedback loop of peri-cellular fibrinolysis. As a consequence of plasmin proteolysis of cross-linked fibrin, D dimer is generated and initiates a signal that results in AP-1–dependent induction of PAI-1 transcription using the −59 to −52 element. New PAI-1 protein is synthesized and, upon secretion, binds with high affinity to plasminogen activators, thereby blocking further activation of plasminogen to active plasmin. Upon loss of plasmin action, fibrinolysis ceases, allowing fibrin persistence and halting further generation of D dimer.
Model of D dimer-initiated negative regulatory feedback loop of peri-cellular fibrinolysis. As a consequence of plasmin proteolysis of cross-linked fibrin, D dimer is generated and initiates a signal that results in AP-1–dependent induction of PAI-1 transcription using the −59 to −52 element. New PAI-1 protein is synthesized and, upon secretion, binds with high affinity to plasminogen activators, thereby blocking further activation of plasminogen to active plasmin. Upon loss of plasmin action, fibrinolysis ceases, allowing fibrin persistence and halting further generation of D dimer.
In summary, we have demonstrated the dependence of basal, fibrin D dimer and fibrinogen-stimulated PAI-1 transcription on an AP-1–like element unique to the PAI-1 gene. At least 2 proteins that comprise the heterodimeric transcription factor, AP-1, after D dimer stimulation are c-fos and junD. Although the affinity of the induced protein complex for the AP-1–like element in the PAI-1 promoter varied considerably from its affinity to a consensus AP-1 element, dependence of PAI-1 transcription of the wild-type sequence on c-fos expression was easily demonstrated. The strict evolutionary conservation of this sequence underscores the importance of its role in regulating the expression of PAI-1, a molecule essential to the maintenance of fibrin and matrix homeostasis in disease states.
ACKNOWLEDGMENT
The authors acknowledge Patrick Miller, Dr Wenqi Jiang, and Dr James J. Marsh for their technical assistance and provision of reagents.
M.A.O. was a Parker B. Francis Research Foundation fellow during part of this work. This work was supported by grants from the American Lung Association, the American Federation of Clinical Research, the Veterans Administration MERIT Review, the National Institutes of Health to M.A.O. (HL-58655), the Alabama Chapter of the American Lung Association (to W.L.S.), and the National Institutes of Health to J.S.H. (HL-03239 and HD-28831).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Mitchell A. Olman, MA, MD, Department of Medicine, Division of Pulmonary and Critical Care Medicine, University of Alabama at Birmingham Medical Center, 1900 University Blvd, 215 THT, Birmingham, AL 35294; e-mail:Olman@pulm.dom.uab.edu.

![Fig. 1. Effect of 5′ deletion of the PAI-1 gene on basal and D dimer-inducible PAI-1 transcriptional activity. Progressive deletions of the 5′ flanking region of the PAI-1 gene were generated by restriction digestion or PCR and ligated to a luciferase-encoding reporter plasmid. The plasmids were transiently transfected into rat lung fibroblasts along with the thymidine kinase promoter-Renilla luciferase encoding plasmid. For determination of promoter activity, cells were incubated for 48 hours posttransfection in 0.4% serum containing media followed by either serum-free media ([A], right, basal activity) or SFM plus D dimer (1.1 μmol/L; [B], inducible activity) for 6 hours. Raw data are tabulated as the ratio of firefly luciferase activity to that of its Renillaluciferase activity (mean ± SD) and reported relative to the value for the p928 plasmid (assigned value of 100). *P < .05 relative to the p928 values. +P < .05 relative to the p161wt values by ANOVA/Newman-Keuls test. (A) Relative basal activity. Constructs are as indicated at left. Mutations were performed at the AP-1–like site (−59 to −52) of the PAI-1 gene with otherwise intact −161 to +26. PAIcon marks the TGAGTCA element, PAIwt marks the TGAGTTCA element, and PAImut marks the TGTGTTTG element at the −59 to −52 positions of p161. Underscored letters denote deviations from the consensus AP-1 element. (B) D dimer-inducible transcriptional activity. Transfected plasmids are as indicated.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/6/10.1182_blood.v94.6.2029/5/m_blod41825001ax.jpeg?Expires=1767725152&Signature=qi74fb6fH~jsBl5xnHf~V6sgLEN-icqauQtkjR3yDAb9qncHeZNsCeGQKXgfKAaZnhLUcuSOSq~zaYolwAi43IsitRheJYDa8eehAjkpiwwT4Mjo-DjMlu2tejP0spG-EJJamG9K0wTNCyWbMeNjUTyetp2pUjVvqo~rlx3KUmQQPHC758zAd~fL2rKQdk7UeV9CR0mgDfVq0758ez8Ddaahfempf1bbzZ9Kqdwygf3XpTl7oBX1X9NvJ8wBoTSqhqsVlUOdAHSinpPHPbag~Y-KAX6lxq6ZrkXEY10DgHubigjRYnUuhsn6--xtUb0d5aNwBwu9JGSMkoPXqpeBXA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 1. Effect of 5′ deletion of the PAI-1 gene on basal and D dimer-inducible PAI-1 transcriptional activity. Progressive deletions of the 5′ flanking region of the PAI-1 gene were generated by restriction digestion or PCR and ligated to a luciferase-encoding reporter plasmid. The plasmids were transiently transfected into rat lung fibroblasts along with the thymidine kinase promoter-Renilla luciferase encoding plasmid. For determination of promoter activity, cells were incubated for 48 hours posttransfection in 0.4% serum containing media followed by either serum-free media ([A], right, basal activity) or SFM plus D dimer (1.1 μmol/L; [B], inducible activity) for 6 hours. Raw data are tabulated as the ratio of firefly luciferase activity to that of its Renillaluciferase activity (mean ± SD) and reported relative to the value for the p928 plasmid (assigned value of 100). *P < .05 relative to the p928 values. +P < .05 relative to the p161wt values by ANOVA/Newman-Keuls test. (A) Relative basal activity. Constructs are as indicated at left. Mutations were performed at the AP-1–like site (−59 to −52) of the PAI-1 gene with otherwise intact −161 to +26. PAIcon marks the TGAGTCA element, PAIwt marks the TGAGTTCA element, and PAImut marks the TGTGTTTG element at the −59 to −52 positions of p161. Underscored letters denote deviations from the consensus AP-1 element. (B) D dimer-inducible transcriptional activity. Transfected plasmids are as indicated.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/6/10.1182_blood.v94.6.2029/5/m_blod41825001bx.jpeg?Expires=1767725152&Signature=1UNk2V3vJ7AhKHgpPo9muUEQddsGM5j4QHNpfmxoO8ONpwOc4MAN9V8hp~LKxkdL8gLgIR35JhTsgpXhKCJsvgpJ9hLHOz3UHodI11tVIT9ZiN5qfEWxyI1CRL3I-2Qa8FVF6gjjwILGrNfZy4590OoFhbwPicgnvTvkdThhpxrPQHTPpG4af0MMaa4U~NbO7wZmA6CJkeFWJ68QQAKsucJq43sfHsDEruaOLpNFm0nxDByuOsL4t2EMx8jmIoKhcA7OQLW6l-ir08IzqVu7Y4di2jaYdQ92iBK0MABio8oATHTmysimclj-SSYlrqkI8NRi~ynHsKTlkrLfqvd3mg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)







This feature is available to Subscribers Only
Sign In or Create an Account Close Modal