Abstract
Primary central nervous system lymphoma (PCNSL) represents 1% to 3% intracranial tumors. Most PCNSL are located in the brain, and 75% are large B-cell lymphomas. The largest subgroup of these tumors contains cells that resemble centroblasts and has been labelled diffuse centroblastic (polymorphous) lymphoma. To investigate the cell of origin and the clonal history of these tumors, we have analyzed VH gene of 5 cases of PCNSL, all confirmed by histological studies to be Epstein-Barr virus (EBV)-negative, high-grade diffuse B-cell lymphomas. The V4-34 gene of the VH4 family was used in 4 of 5 cases. All VHgenes were found to have accumulated very high levels of somatic mutation (14% to 25%). In 3 of 5 cases, intraclonal nucleotide heterogeneity, including codon deletion in some clones in 1 case, was observed, indicating that the VH genes were still under the influence of the somatic hypermutation mechanism. Analysis of the distribution of silent and replacement mutations showed evidence for preservation of immunoglobulin structure in all cases. These results suggest that, although there is no evidence for germinal center formation in the brain tissue, PCNSL is derived from a B cell with features associated with location in a germinal center environment.
PRIMARY CENTRAL NERVOUS system lymphoma (PCNSL) is an uncommon neoplasm, representing 1% to 2% of lymphomas and 1% to 3% of intracranial tumors.1,2 While most examples of PCNSL are sporadic, some are associated with congenital or acquired immunodeficiency. An increasing incidence of PCNSL can partly be attributed to a growing population of immunocompromised patients in which PCNSL is mainly associated with the acquired immunodeficiency syndrome, organ transplantation, and chemotherapy. PCNSL has been associated with a poor prognosis, which is worse than that for similar non-Hodgkin’s lymphomas outside the CNS.3However, recent trials of radiotherapy with various chemotherapeutic regimens have shown an improvement in survival.4,5 Most PCNSLs are located in the brain, and about 75% are large B-cell lymphomas (REAL classification). The largest subgroup of these large cell lymphomas contains cells that resemble centroblasts6 and has been labeled diffuse centroblastic (polymorphous) lymphoma (Kiel classification). Other B-cell PCNSLs are low-grade lymphocytic, lymphoplasmacytic, and plasmacytic lymphomas. T-cell PCNSLs are very rare, but large and small cell examples have been reported.1
Generation of functional antigen receptors by somatic recombination of variable, diversity, and joining segments is a unique feature of B cells and T cells. In a normal immune response, B lymphocytes stimulated by antigen can undergo somatic hypermutation in their immunoglobulin (Ig) variable region genes in a germinal center microenvironment, thus generating a more diverse repertoire of antibodies. B cells with high affinity for antigen are selectively rescued from apoptosis to differentiate into either circulating memory cells or antibody-producing plasma cells.7,8 From the developmental point of view, B-cell tumors can be regarded as B cells arrested at a certain differentiation stage when neoplastic transformation took place. Analysis of Ig V region genes has provided invaluable information on the status of tumors in B-cell differentiation and has added another dimension to classification of B-cell tumors at the molecular level.9,10 In the case of follicular center lymphoma (FCL), V genes are often somatically mutated and in some cases significant accumulation of replacement mutations is found in the complementarity-determining regions (CDRs), suggesting a selective role for antigenic stimulation. Furthermore, intraclonal nucleic acid sequence variation appears to be a common feature of FCL, an indication that tumor cells are still under the influence of a mutation mechanism.11-14 Recent studies have also shown that VH genes in primary diffuse large cell lymphomas (DLCL) are somatically mutated,15-18 with a significant level of intraclonal heterogeneity, again indicating ongoing somatic mutation.16 Interestingly, the heterogeneity is reduced after successful chemotherapy, possibly due to selective pressure on the clone.16 There have been few studies of Ig V genes on PCNSL. Southern blotting and polymerase chain reaction (PCR) amplification of the VH CDR3 sequences have demonstrated Ig V-gene rearrangement in most cases, confirming the B-cell nature of PCNSL.19-21 To gain insight into the nature of the Ig V genes in PCNSL, we cloned and sequenced the complete VHgenes of 5 cases of PCNSL. We have found frequent usage of the V4-34 gene. We have also found that the VH genes are extensively mutated and show intraclonal heterogeneity in some cases, reminiscent of alterations influenced by the germinal center environment.
MATERIALS AND METHODS
Immunohistochemistry and in situ hybridization.
Tumor biopsies were examined by standard histological techniques, including immunohistochemistry as previously described.22 A range of antibodies (DAKO, Glostrup, Denmark) to immunoglobulins, and B-cell and T-cell–specific antigens was used at the following concentrations: IgM 1:500; IgG 1:1,000; IgA 1:500; Kappa 1:500; lambda 1:500; CD20 1:400; CD79a 1:250; CD3 1:50; CD45RO 1:1,000. Sections (5 μm) were also probed for Epstein-Barr virus (EBV)-encoded RNA (EBER) transcripts using an in situ hybridization kit (Novocastra, Newcastle, UK) with appropriate positive and negative controls.
Preparation of cDNA.
Frozen tumour tissue (≈0.1 g) was cut into small pieces and ground into fine powder in liquid nitrogen in a mortar. The powder was transferred to a microcentrifuge tube, and total RNA was isolated using RNAzol according to the supplier’s instructions (Cinna Biotecxlabs Inc, Houston, TX). First-strand cDNA was synthesized from 5 μg of total RNA in a final volume of 33 μL using a cDNA synthesis kit and oligo(dT) primer (Pharmacia, Uppsala, Sweden).
Amplification, cloning, and sequencing of VH genes.
For amplification of VH genes from cDNA, 1 to 5 μL of cDNA was used as template for PCR using a mixture of oligonucleotide primers specific for each of the leader sequences of VH 1-6 families (Table 1), together with a mixture of primers complementary to the sequences of the germ line JH genes.23 In some cases, a Cμ constant region primer was also used. PCR was performed in a final volume of 50 μL containing 1 to 5 μL of cDNA, 20 pmol of each primer, 250 mmol/L of dNTPs, 2.5 U of Taq DNA polymerase with the reaction buffer supplied by the manufacturer (QIAgen, Hilden, Germany). PCR consisted of an initial denaturation step of 5 minutes at 94°C, followed by 35 cycles of 94°C for 60 seconds, 65°C for 60 seconds, and 72°C for 60 seconds, with a final extension step of 10 minutes at 72°C. PCR products (≈400 bp) were separated by agarose gel electrophoresis, purified using the GeneClean kit (Bio 101 Inc, Vista, CA), and cloned into the pGEM-T vector (Promega, Madison, WI). Randomly picked bacterial clones were sequenced on an ABI 377 automatic sequencer (Perkin-Elmer, Foster City, CA).
VH Leader and Cμ Primers for VH Gene Amplification
| VH1/7Lead | 5′-CTC ACC ATG GAC TGG ACC TGG AG-3′ |
| VH2Lead | 5′-ATG GAC ACA CTT TGC T(A/C)C AC(G/A) CTC-3′ |
| VH3Lead | 5′-CCA TGG AGT TTG GGC TGA GCT GG-3′ |
| VH4Lead | 5′-ACA TGA AAC A(C/T)C TGT GGT TCT TCC-3′ |
| VH5Lead | 5′-ATG GGG TCA ACC GCC ATC CT(C/T) G-3′ |
| VH6Lead | 5′-ATG TCT GTC TCC TTC CTC ATC TTC-3′ |
| Cμ | 5′-GGA ATT CTC ACA GGA GAC GAG G-3′ |
| VH1/7Lead | 5′-CTC ACC ATG GAC TGG ACC TGG AG-3′ |
| VH2Lead | 5′-ATG GAC ACA CTT TGC T(A/C)C AC(G/A) CTC-3′ |
| VH3Lead | 5′-CCA TGG AGT TTG GGC TGA GCT GG-3′ |
| VH4Lead | 5′-ACA TGA AAC A(C/T)C TGT GGT TCT TCC-3′ |
| VH5Lead | 5′-ATG GGG TCA ACC GCC ATC CT(C/T) G-3′ |
| VH6Lead | 5′-ATG TCT GTC TCC TTC CTC ATC TTC-3′ |
| Cμ | 5′-GGA ATT CTC ACA GGA GAC GAG G-3′ |
Analysis of VH gene usage and mutation pattern.
Tumor-related VH genes were identified as predominant repeated or similar sequences with clonally related CDR3.23Sequence alignment analysis was to the Entrez database (National Center for Biotechnology Information, Bethesda, MD) and to the VBase, which contains all known human germ line immunoglobulin V region genes24 using MacVector sequence analysis software (Oxford Molecular, Oxford, UK). The germ line genes with highest homology to the tumor-derived sequences are considered as the germ line gene donors. Statistical analysis of mutation distribution was performed according to Chang and Casali.25 In this method, each V gene is analyzed codon by codon for significance of deviation from the germ line sequence. A modified binomial distribution model is then applied to calculate whether the probability (p) of excess (in CDR) or scarcity (in framework region [FR]) of R mutations resulted from chance alone.
RESULTS
Clinical and histological data.
All 5 patients, of whom 4 were female, presented with CNS symptoms and signs. Their ages ranged from 50 to 71 years (median, 60 years). None had detectable lymphadenopathy, and none was immunocompromised. Neuroimaging showed mass lesions in the cerebrum (3 patients) or cerebellum (2 patients), which were thought to be neoplastic in nature. Biopsy of all tumors was undertaken at the Wessex Neurological Centre where concurrent radiological investigations failed to show tumor outside the CNS. In all cases, histological examination showed the typical architectural and cytological features of a high-grade (polymorphous) B-cell PCNSL (Fig 1), including the characteristic perivascular arrangement of neoplastic cells and invasion of adjacent brain tissue to form intraparenchymal masses.
PCNSL histology. (A) Brain at the periphery of a PCNSL characteristically shows accumulation of neoplastic cells in the perivascular spaces (periodic acid Schiff; original magnification [OM] × 250). (B) Invasion of surrounding brain by neoplastic cells occurs from the perivascular space (periodic acid Schiff; OM × 500). (C) A sheet of neoplastic lymphoid cells characterizes this intraparenchymal large cell PCNSL. Moderate nuclear pleomorphism, a variable number of nucleoli per cell, and mitotic figures are evident (hematoxylin and eosin; OM × 500). (D) All tumors in this study were positive for CD20 (L-26 antibody, immunoperoxidase; OM × 500).
PCNSL histology. (A) Brain at the periphery of a PCNSL characteristically shows accumulation of neoplastic cells in the perivascular spaces (periodic acid Schiff; original magnification [OM] × 250). (B) Invasion of surrounding brain by neoplastic cells occurs from the perivascular space (periodic acid Schiff; OM × 500). (C) A sheet of neoplastic lymphoid cells characterizes this intraparenchymal large cell PCNSL. Moderate nuclear pleomorphism, a variable number of nucleoli per cell, and mitotic figures are evident (hematoxylin and eosin; OM × 500). (D) All tumors in this study were positive for CD20 (L-26 antibody, immunoperoxidase; OM × 500).
Immunohistochemistry confirmed a B-cell immunophenotype for the neoplastic cells in each case (Table 2). All tumors expressed CD20 (Fig 1) and CD79a. Antibodies to CD3 and CD45RO labeled moderate numbers of reactive T lymphocytes among the neoplastic cells. Immunohistochemistry with antibodies to IgM, IgG, IgA, and kappa and lambda light chains was feasible in 3 of 5 cases. IgM and kappa light chains were detected in neoplastic cells in all 3 cases. In 1 tumor, sparse IgG-positive cells were detected in addition to more widespread IgM. No IgA was detected. All 5 tumors were EBER-negative by in situ hybridization.
Clinical, Immunophenotypic, and EBV Data of PCNSL Cases
| Patient . | Age/Sex . | Tumor Location . | CD20 . | CD79a . | CD3 . | CD45RO . | IgM . | IgG . | IgA . | κ . | λ . | EBER . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 61/F | Cerebral | + | + | − | − | NA | NA | NA | NA | NA | − |
| 2 | 71/M | Cerebella | + | + | − | − | + | ± | − | + | − | − |
| 3 | 59/F | Cerebral | + | + | − | − | + | − | − | + | − | − |
| 4 | 60/F | Cerebral | + | + | − | − | NA | NA | NA | NA | NA | − |
| 5 | 50/F | Cerebella | + | + | − | − | ± | − | − | + | − | − |
| Patient . | Age/Sex . | Tumor Location . | CD20 . | CD79a . | CD3 . | CD45RO . | IgM . | IgG . | IgA . | κ . | λ . | EBER . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 61/F | Cerebral | + | + | − | − | NA | NA | NA | NA | NA | − |
| 2 | 71/M | Cerebella | + | + | − | − | + | ± | − | + | − | − |
| 3 | 59/F | Cerebral | + | + | − | − | + | − | − | + | − | − |
| 4 | 60/F | Cerebral | + | + | − | − | NA | NA | NA | NA | NA | − |
| 5 | 50/F | Cerebella | + | + | − | − | ± | − | − | + | − | − |
Abbreviations: +, positive; −, negative; ±, a few immunopositive cells only; NA, not available.
Usage of VH, D, and JH genes in the tumors.
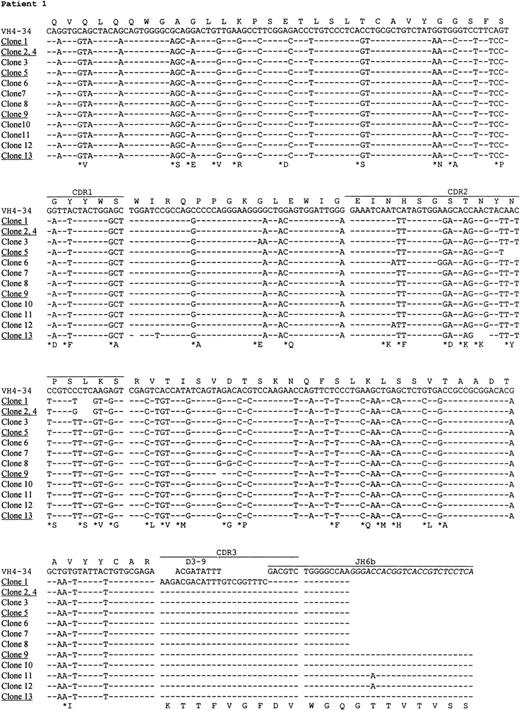
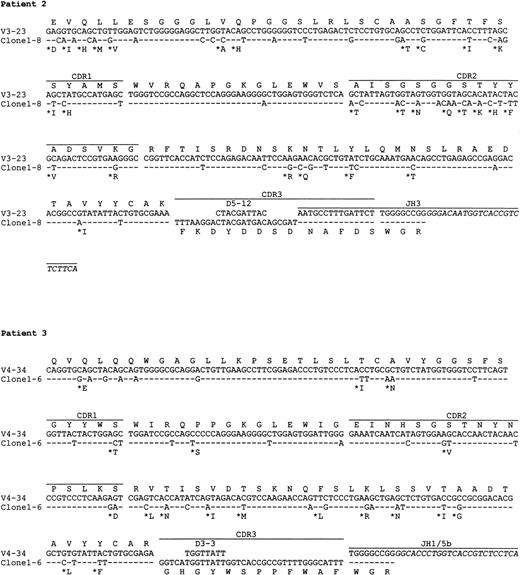
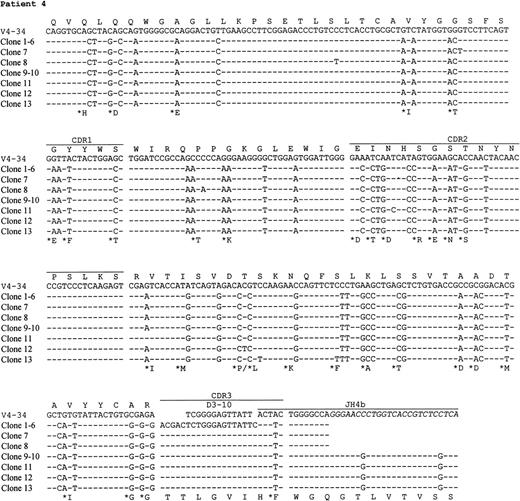
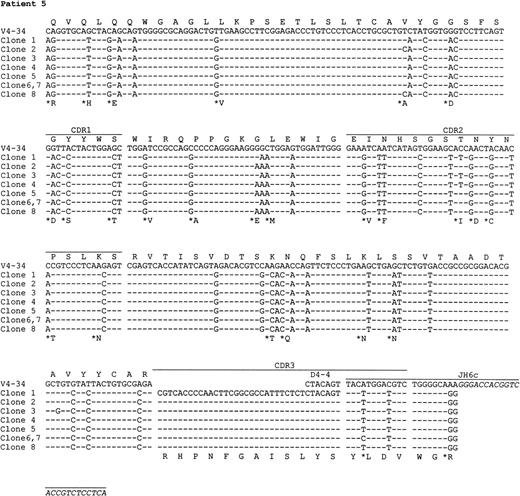
Preparations of cDNA from all 5 patients were amplified, cloned, and sequenced. In all cases, predominant repeated or similar clones with identical CDR3 sequences were obtained, consistent with tumor derivation (Table 3). The usage of VH, D, and JH genes and the homology to the corresponding germ line genes are summarized in Table 3. Sequence analysis indicates that the V4-34 gene of the VH4 family was used in 4 of 5 patients, and in the remaining case, the VH gene was derived from V3-23, a member of the VH3 family. Assignment of D gene segments was based on the homology between CDR3 sequences and the germ line D genes. According to the rule proposed by Corbett et al26 that at least 10 consecutive nucleotides of identity are required to assign a D segment, D gene can only be identified with confidence in patient 4. However, shorter D segments or D segments with nucleotide substitutions could have been used in the other cases and are also shown in Table3. There was no apparent bias in JH gene usage. JH6 gene was used in cases 1 and 5, JH3 in case 2, and JH4 in case 4. The JH gene in case 3 was extensively truncated at the 5′ end, which could be derived from either JH1 or JH5.
Analysis of Tumor-Derived VH Gene Sequences
| Patient . | VH Family . | GL Donor . | Homology (%) . | D Gene . | JH Gene . | Related Clones3-150/ Clones Sequenced . |
|---|---|---|---|---|---|---|
| 1 | VH4 | 4-34 | 75 | D3-10 | JH6b | 13/14 |
| 2 | VH3 | 3-23 | 81 | D5-12 | JH3 | 8/8 |
| 3 | VH4 | 4-34 | 86 | D3-3 | JH1/5b | 6/8 |
| 4 | VH4 | 4-34 | 82 | D3-10 | JH4b | 13/13 |
| 5 | VH4 | 4-34 | 84 | D4-11 | JH6c | 8/9 |
| Patient . | VH Family . | GL Donor . | Homology (%) . | D Gene . | JH Gene . | Related Clones3-150/ Clones Sequenced . |
|---|---|---|---|---|---|---|
| 1 | VH4 | 4-34 | 75 | D3-10 | JH6b | 13/14 |
| 2 | VH3 | 3-23 | 81 | D5-12 | JH3 | 8/8 |
| 3 | VH4 | 4-34 | 86 | D3-3 | JH1/5b | 6/8 |
| 4 | VH4 | 4-34 | 82 | D3-10 | JH4b | 13/13 |
| 5 | VH4 | 4-34 | 84 | D4-11 | JH6c | 8/9 |
Clones sharing identical or near identical CDR3 sequences.
Somatic hypermutations in tumor-derived VH genes.
The nucleotide sequences of the tumor VH genes and the deduced amino acid sequences are shown in Figs2 and 3. A large number of nucleotide substitutions was found in all tumor VH genes, ranging from 41 to 73 nucleotides (14% to 25% of the VH genes). In addition to single nucleotide change, double and triple substitution also frequently occurred, mostly within the same codon. No nucleotide insertion was observed, but triple in-frame as well as single nucleotide deletion was seen in a number of clones in case 1 (see below). Among the V4-34–derived sequences, common nucleotide substitutions leading to both silent and replacement mutations were seen in several codons, such as Gln5, Val71, and Ser35 (Fig 3). To see if there was significant clustering of replacement (R) mutations in the CDRs and silent mutations in the FRs, the distribution of somatic mutations was analyzed by the method of Chang and Casali25(Table 4). The analysis shows that the number of R mutations in FRs was significantly lower in all cases (P < .05), probably as a result of selection against R mutations within the FRs to preserve the structures of expressed immunoglobulins. In patient 2, the number of R mutations in CDRs was significantly higher than would be expected to arise solely by chance (P < .05), whereas in the other cases, the number of replacement mutations in the CDRs was not significantly higher than expected (P > .05).
Nucleotide and deduced amino acid sequences of VH genes derived from PCNSL. Comparisons were made with the most homologous germ line VH genes. Dashes represent identity with the representative germ line sequence. Replacement amino acids are starred. Blank space indicates nucleotide deletion. The binding sites for JHprimers are shown in italics. For patients 1 and 4, full JHsequences are shown for the clones obtained by PCR using the VH leader and the Cμ primers. Clones with nucleotide deletions in case 1 are underlined. The VH gene sequences have been deposited to the EMBL database with following accession numbers: patient 1, AJ235649-AJ235661; patient 2,AJ235662; patient 3, AJ235663; patient 4, AJ235664-AJ23566; patient 5,AJ235671-AJ235678.
Nucleotide and deduced amino acid sequences of VH genes derived from PCNSL. Comparisons were made with the most homologous germ line VH genes. Dashes represent identity with the representative germ line sequence. Replacement amino acids are starred. Blank space indicates nucleotide deletion. The binding sites for JHprimers are shown in italics. For patients 1 and 4, full JHsequences are shown for the clones obtained by PCR using the VH leader and the Cμ primers. Clones with nucleotide deletions in case 1 are underlined. The VH gene sequences have been deposited to the EMBL database with following accession numbers: patient 1, AJ235649-AJ235661; patient 2,AJ235662; patient 3, AJ235663; patient 4, AJ235664-AJ23566; patient 5,AJ235671-AJ235678.
Deduced amino acid sequences of VH regions of PCNSL. Comparisons were made with the most homologous germ line VH genes. Amino acid numbering is according to Kabat et al.46 Dashes represent identity with the representative germ line sequence. Uppercase, replacement mutation; lowercase, silent mutation.
Deduced amino acid sequences of VH regions of PCNSL. Comparisons were made with the most homologous germ line VH genes. Amino acid numbering is according to Kabat et al.46 Dashes represent identity with the representative germ line sequence. Uppercase, replacement mutation; lowercase, silent mutation.
Distributions of Mutations in Tumor-Derived VH Sequences
| Patient . | Region . | Expected . | Observed . | P4-150 . | ||
|---|---|---|---|---|---|---|
| R . | S . | R . | S . | |||
| 1 | FR | 42 | 15 | 29 | 23 | <.01 |
| CDR | 13 | 3 | 16 | 5 | .07 | |
| 2 | FR | 33 | 11 | 19 | 15 | <.01 |
| CDR | 9 | 2 | 15 | 6 | .02 | |
| 3 | FR | 35 | 13 | 16 | 16 | <.01 |
| CDR | 11 | 2 | 5 | 4 | .12 | |
| 4 | FR | 31 | 11 | 23 | 14 | .02 |
| CDR | 9 | 2 | 10 | 5 | .14 | |
| 5 | FR | 26 | 9 | 14 | 16 | <.01 |
| CDR | 8 | 2 | 10 | 5 | .11 | |
| Patient . | Region . | Expected . | Observed . | P4-150 . | ||
|---|---|---|---|---|---|---|
| R . | S . | R . | S . | |||
| 1 | FR | 42 | 15 | 29 | 23 | <.01 |
| CDR | 13 | 3 | 16 | 5 | .07 | |
| 2 | FR | 33 | 11 | 19 | 15 | <.01 |
| CDR | 9 | 2 | 15 | 6 | .02 | |
| 3 | FR | 35 | 13 | 16 | 16 | <.01 |
| CDR | 11 | 2 | 5 | 4 | .12 | |
| 4 | FR | 31 | 11 | 23 | 14 | .02 |
| CDR | 9 | 2 | 10 | 5 | .14 | |
| 5 | FR | 26 | 9 | 14 | 16 | <.01 |
| CDR | 8 | 2 | 10 | 5 | .11 | |
P is the probability of obtaining the number of R mutations by chance.
Intraclonal nucleotide sequence variation.
In 3 cases (1, 4, and 5), intraclonal nucleotide variation was observed in VH genes (Fig 3). This sequence heterogeneity is considered mostly as the result of ongoing somatic mutation rather than being introduced in PCR amplification, as the levels of variation (>1 in 700 bp) were much higher than the PCR error frequency (≈1 in 5,000 bp in our laboratory) and several identical base changes were seen in different clones. This is supported also by the fact that no heterogeneity was observed in cases 2 and 3 with similar numbers of clones sequenced. The level of nucleotide variation in the 3 cases differs and the variations appeared to be distributed randomly. Most nucleotide variations were single base substitutions, resulting in both silent and replacement mutations. However, deletion of in-frame triple nucleotides, ie, an entire codon, in 3 different positions in the CDR2 was found in a number of clones in patient 1. Single nucleotide deletion was also found in 2 clones in this case, 1 in the FR2 and 1 in the FR3, leading to aberrant sequences. No nucleotide insertions were observed.
DISCUSSION
PCNSL is a rare tumor with increasing incidence during the past decade, both in immunocompromised and in general populations.1PCNSL shows morphological and immunohistochemical features similar to those of malignant lymphomas arising in sites outside the CNS. The B-cell nature of most of these tumors has not only been demonstrated by immunohistochemistry, but by Southern blotting and PCR amplification of the short VH CDR3 sequences.19-21 In this small study of full-length VH gene sequences, VH gene usage appeared to be restricted as the V4-34 gene was used in 4 of 5 cases. The V4-34 gene is used in ≈5% of healthy adults27,28 and appears to be overexpressed in some autoimmune diseases and B-cell tumors. The V4-34 gene has been shown to be mandatory for encoding the IgM proteins of cold agglutinin disease.29,30 In this case, the red blood cell I/i antigens appear to bind to the FR1 of the Ig. This binding outside the CDRs is indication of a B-cell superantigen. Recently, Hsu and Levy15 reported that in DLCL there was a preferential usage of genes from the VH4 family, especially the V4-34 gene (11 of 17 cases, 65%), although a recent study31using the V4-34 gene product-specific antibody, 9G4, showed that only 30% of DLCL expressed the V4-34 gene; and the sequence analysis by Küppers et al17 and a study of 11 cases of DLCL in this laboratory did not show bias toward V4-34 gene (C. Ottensmeier, personal communication, September 1998). VH4 family genes, but not V4-34, also appear to be preferentially used in the B cells from cerebrospinal fluid in multiple sclerosis.32 33 If such bias were to be confirmed in PCNSL, it might suggest a role for superantigen drive in lymphoma development. It is interesting that certain infections, including EBV, cytomegalovirus, and mycoplasma pneumoniae, cause a selective increase in serum levels of V4-34 encoded Ig. Although the cerebral tumors described are EBV-genome negative, this intriguing association may indicate a potential influence of pathogens on B cells expressing V4-34 encoded Ig.
The VH genes of all 5 cases were very heavily mutated. In fact, the somatic mutation frequency in these cases (mean ± standard deviation [SD]% = 18.4 ± 3.7%) is higher than in all of the other B-cell malignancies studied so far (summarized by Klein et al10). In DLCL, which involves cells morphologically and phenotypically similar to PCNSL, a mean mutation frequency of 11% (range, 4% to 22%) was reported.17 Despite high levels of somatic mutation in PCNSL, there is good evidence for selection against R mutations in the FRs, suggesting that functional expression of the Ig receptor may be advantageous for tumor growth. Some shared nucleotide substitutions leading to silent as well as replacement mutations were seen among the V4-34 encoded sequences. The intrinsic mutational “hot spots” are likely to be responsible for these common mutations, as the same changes in the V4-34 gene have been observed in different studies,15,34 and biased nucleotide substitution is a general feature of both human35 and murine IgV genes.36
Intraclonal heterogeneity was observed in 3 cases, suggesting that the tumor cells retained the capacity to undergo further somatic hypermutation. In case 1, which was the most mutated, deletion of in-frame triple nucleotides occurred in 2 different positions in the CDR2; therefore the gene could still be expressed. However, in 2 clones, the reading-frame was disrupted by single nucleotide deletion and therefore no Ig could be produced. Possibly, these sequences were derived from the cells that contained deleterious mutations and were due to die, but had not yet been eliminated from the tumor population. Nucleotide deletion and insertion have also been reported in other B-cell tumors.37-39 However, insertional or deletional events are not unique to tumor cells because there is strong evidence that nucleotide insertion and deletion are frequently introduced in the somatic hypermutation process, and in-frame insertion and deletion have been found in normal B cells.40 41
The pattern of somatic mutation and intraclonal variation indicates that the cell of origin of PCNSL has been exposed to the mutational mechanism, both before and after neoplastic transformation. This pattern is characteristic of tumors of germinal center origin, such as FCL, and is also seen in Burkitt’s lymphoma, mucosa-associated lymphoid tissue (MALT) lymphoma and DLCL.9,10 Stimulation of somatic mutation is generally considered to require a GC environment where activated T cells and cytokines are present.7,8 Cross-linking of surface Ig also may increase the rate of somatic mutation, possibly reflecting a role for antigen.42
For PCNSL, there is no evidence for GC structure in the brain. However, PCNSL cells do express BCL-6 protein.43 PCNSLs are mainly located in the cerebral parenchyma and can infiltrate the pial or ependymal surfaces to spread along the edge of CSF pathways, with no particular anatomical location. One possibility is that the somatic mutation is occurring independently of GC structure. Support for some GC-independent mutational activity comes from mice deficient in lymphotoxin-α44 and Lyn kinase.45 Although these mice cannot form GCs, they are able to accumulate somatic mutations in V genes. However, levels are low and would not be expected to reach the very high levels found in the B cells that give rise to PCNSL.
An alternative possibility is that the B cells of PCNSL have undergone somatic mutation and neoplastic transformation in a draining cervical lymph node. These events could have occurred after antigenic stimulation, perhaps due to infection. However, the neoplastic cells have then to leave the lymph node and enter the cerebral parenchyma where they apparently remain. The puzzle is that cervical lymph nodes are not obviously involved at presentation. Availability of VH CDR3 sequence tags should now allow an approach to these questions by probing of blood and bone marrow of patients to track possible migration of tumor cells from the brain.
Supported by Tenovus and the Cancer Research Campaign, UK.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Delin Zhu, PhD, Molecular Immunology Group, Tenovus Research Laboratory, Southampton University Hospitals Trust, Tremona Rd, Southampton SO16 6YD, UK; e-mail: dz1@soton.ac.uk.

![Fig. 1. PCNSL histology. (A) Brain at the periphery of a PCNSL characteristically shows accumulation of neoplastic cells in the perivascular spaces (periodic acid Schiff; original magnification [OM] × 250). (B) Invasion of surrounding brain by neoplastic cells occurs from the perivascular space (periodic acid Schiff; OM × 500). (C) A sheet of neoplastic lymphoid cells characterizes this intraparenchymal large cell PCNSL. Moderate nuclear pleomorphism, a variable number of nucleoli per cell, and mitotic figures are evident (hematoxylin and eosin; OM × 500). (D) All tumors in this study were positive for CD20 (L-26 antibody, immunoperoxidase; OM × 500).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/5/10.1182_blood.v94.5.1738/5/m_blod41710001ay.jpeg?Expires=1769153571&Signature=j1Pbtb-KsrjK2otPVw~zl5xcBd21s9p9-w03GOApiR6W3Eu8MAxObhuNJ6JlPsVB9DQlqay5ZXFz4L2jipAHwasj7LVPNwe4Zi~wX8S9Z58HjPvBrilOw7Q~5FGHWzci5-tS-7dyup-4bnhip1LrbeaVGe8hhag279osvJefAZ9w4TStlVwAoL4gmnq2zIkK8VpQPfcMN6X~Xx0GKpnVbabgCimnplXcoGK2Zxj-ttmJgGSVESC2GVI2guMPO1miNULdOXqaE2L3Q4A4WBwi4f5I1CnDZMR1lr3yNj039~gyAEjwl~bNdhTzSO4-ajJGFNoKZBEv76zjJsiQO8Mrvw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 1. PCNSL histology. (A) Brain at the periphery of a PCNSL characteristically shows accumulation of neoplastic cells in the perivascular spaces (periodic acid Schiff; original magnification [OM] × 250). (B) Invasion of surrounding brain by neoplastic cells occurs from the perivascular space (periodic acid Schiff; OM × 500). (C) A sheet of neoplastic lymphoid cells characterizes this intraparenchymal large cell PCNSL. Moderate nuclear pleomorphism, a variable number of nucleoli per cell, and mitotic figures are evident (hematoxylin and eosin; OM × 500). (D) All tumors in this study were positive for CD20 (L-26 antibody, immunoperoxidase; OM × 500).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/5/10.1182_blood.v94.5.1738/5/m_blod41710001by.jpeg?Expires=1769153571&Signature=nw~JPjc-W4tzzh1vo3UThiS~UmA3QGUvBvIVXBhbCsGu-32P9SIW74Uhn2m4PoWKd2U-EAr4dYiDX7aczPk1bXZW945RiQNCcmX1MDRZuqGxwllHN69GwhzA6eDWhQh5e2MGNjfmCw8D29ySbd4QD1u-PtL0f1e5tX6Hmt6BmEE~6BfXFtOu2~zcz1Gjj4z-sfCjeME2ifdzDV6wCuEIuvHZwb~xy3qmUxCaJIO-2FGRwy6aI6zCfsaU~2JRZ08CsiK4xGYH3nNmAQvM9WNmdTU0jM1hxUHZmIJIIGslJ~wwwmlXbHa-IEjZ9bl5B8b6QWT6NxAlh-FYsAIF5vpuwg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 1. PCNSL histology. (A) Brain at the periphery of a PCNSL characteristically shows accumulation of neoplastic cells in the perivascular spaces (periodic acid Schiff; original magnification [OM] × 250). (B) Invasion of surrounding brain by neoplastic cells occurs from the perivascular space (periodic acid Schiff; OM × 500). (C) A sheet of neoplastic lymphoid cells characterizes this intraparenchymal large cell PCNSL. Moderate nuclear pleomorphism, a variable number of nucleoli per cell, and mitotic figures are evident (hematoxylin and eosin; OM × 500). (D) All tumors in this study were positive for CD20 (L-26 antibody, immunoperoxidase; OM × 500).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/5/10.1182_blood.v94.5.1738/5/m_blod41710001cy.jpeg?Expires=1769153571&Signature=SkBctCXzsRSPwg6SDGqro~ksRqEj2jCRTOh5aqeVnwkNsLeQlE30R-dKjEVfGDFkgs8j4m3r0m9lPXuESpFLTWVj3sCUXaHhmBUjPdKrf4dGbSjaFdd5GMdnM0a0iLDKPI31QUI1XlhHdpL2o7dpXkVMy1zq5CHR8qA~deT63tso8gKb-4Z7UAhp87NDh8qmbBu1uKTOWDea08MXAuCJKk9CndtWF1zxKvdF~Aj6xAudK9YAp8kfcQYF~qxPpGzgK6Yd8u6HV6A0W~lRtyDXh6atp03dULljT1Q1FcPX755YWZARr2A1ZQUe6EAerh3BoBw-EokIrV-4KXrIbIQ~qw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 1. PCNSL histology. (A) Brain at the periphery of a PCNSL characteristically shows accumulation of neoplastic cells in the perivascular spaces (periodic acid Schiff; original magnification [OM] × 250). (B) Invasion of surrounding brain by neoplastic cells occurs from the perivascular space (periodic acid Schiff; OM × 500). (C) A sheet of neoplastic lymphoid cells characterizes this intraparenchymal large cell PCNSL. Moderate nuclear pleomorphism, a variable number of nucleoli per cell, and mitotic figures are evident (hematoxylin and eosin; OM × 500). (D) All tumors in this study were positive for CD20 (L-26 antibody, immunoperoxidase; OM × 500).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/5/10.1182_blood.v94.5.1738/5/m_blod41710001dy.jpeg?Expires=1769153571&Signature=f~jtgdwCPwkSu630vWWm8-NEPeokfpi9TmastlVDiqbNws7EKqZgMVpOBBUrgq1r0-1bKhMhV1gQJoYdKoRKkRVlaPaXm4m2bbvsNIngkL8bYQktkvfmuedK0UTvP7u00Qm6~WdaDoFwiydnZwrOSiFUSexxmcwJXdbe5RNyVTOf6UhuvXgJ9lrA-RC94AVnsrFXitGWbnoL2xYZ83UG-Qf95Qjxn2tIGLSK-WRhVjcATnz4hF11acb0s~9h9VPpfcChv1uQi3IhxxdQYZoisSydi3V0r4VUadYrvc8xP3X2496dj1DrfwsDmQdwbKM7D89MGW0ljiphhP-EMbU5cg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal