Dendritic cells (DCs) are professional antigen-presenting cells in the immune system and can be generated in vitro from hematopoietic progenitor cells in the bone marrow, CD34+ cord blood cells, precursor cells in the peripheral blood, and blood monocytes by culturing with granulocyte-macrophage colony-stimulating factor (GM-CSF), interleukin-4, and tumor necrosis factor-. We have performed serial analysis of gene expression (SAGE) in DCs derived from human blood monocytes. A total of 58,540 tag sequences from a DC complementary DNA (cDNA) library represented more than 17,000 different genes, and these data were compared with SAGE analysis of tags from monocytes (Mo) and GM-CSF–induced macrophages (M◊). Many of the genes that were differentially expressed in DCs were identified as genes encoding proteins related to cell structure and cell motility. Interestingly, the highly expressed genes in DCs encode chemokines such as TARC, MDC, and MCP-4, which preferentially chemoattract Th2-type lymphocytes. Although DCs have been considered to be very heterogeneous, the identification of specific genes expressed in human Mo-derived DCs should provide candidate genes to define subsets of, the function of, and the maturation stage of DCs and possibly also to diagnose diseases in which DCs play a significant role, such as autoimmune diseases and neoplasms. This study represents the first extensive gene expression analysis in any type of DCs.
DENDRITIC CELLS (DCs) PLAY a pivotal role in the immune system by processing and presenting antigens to CD4+ naive T cells.1 DCs have also been reported to be involved in directly inducing cytotoxic T cell (CTL), Ig production by B cells, and T-cell tolerance. DCs in lymphoid and nonlymphoid organs vary in their surface markers and functions and, therefore, have different names (eg, the Langerhans cell in the skin; the interdigitating DC in the lymph nodes; the interstitial DC in heart, lung, kidney, and intestine; and the thymic DC in the thymus). DCs are thought to belong to a lineage distinct from monocytes (Mo)/macrophages (Mφ). However, it has been reported that Mφ and DCs share a common progenitor.2 Human DCs have been generated from CD34+ precursor cells isolated from cord blood3 and bone marrow4 in the presence of granulocyte-macrophage colony-stimulating factor (GM-CSF) and tumor necrosis factor-α (TNF-α). Moreover, blood monocytes cultured with GM-CSF and interleukin-4 (IL-4) differentiate into nonadherent CD1a+CD14low/− cells with morphologic and functional characteristics of DCs.5,6 DCs become mature DCs expressing CD83 by stimulating with TNF-α, CD40 ligand, lipopolysaccharide (LPS),7 or monocyte-conditioned medium.8 However, the process of differentiation from monocytes to DCs has not been systematically explored. In this study, we have applied the recently developed serial analysis of gene expression (SAGE) method to allow quantitative analysis of an extremely large number of transcripts in human DCs. The SAGE method has provided a powerful means for the quantitative cataloging and comparison of expressed genes in the cells or tissues from various physiological, developmental, and pathological states.9-12 This may provide not only useful information to define the development and function of DCs, but also a greater understanding of the differentiation pathway and of the immunological relationship between monocytes/macrophages and DCs by comparison between SAGE analysis of tags from DCs and from Mo or GM-CSF–induced Mφ. Furthermore, many sets of genes, including the novel genes identified to be selectively expressed in DCs, should provide further understanding of the biological function of DCs in the host defense system.
MATERIALS AND METHODS
Preparation of cells.
Peripheral blood mononuclear cells (PBMCs) were isolated from venous blood drawn from normal healthy volunteers at the Tokyo Metropolitan Red Cross Blood Center (Tokyo, Japan). Briefly, PBMCs were isolated by centrifugation on a Ficoll-Metrizoate density gradient (d = 1.077 g/mL; Lymphoprep; Nycomed, Oslo, Norway) and suspended in RPMI 1640 medium containing 7.5% heat-inactivated fetal calf serum (FCS; GIBCO/Life Technologies, Tokyo, Japan), 100 μg/mL streptomycin, and 100 U/mL penicillin. The FCS contained 3 pg of LPS per milliliter as assessed by a Limulus amebocyte lysate. PBMCs were incubated with anti-CD14 monoclonal antibody (MoAb) coated with microbeads, and Mo were isolated by passing the PBMCs through a magnetic cell separation system (MACS; Miltenyi Biotec, Bergisch Gladbach, Germany) with column type VR. These cell suspensions were then aliquotted into plastic tissue culture plates and incubated for 30 minutes at 37°C, 5% CO2 to obtain the highly purified cells. More than 99% of the cells were judged to be Mo by morphology, by positive staining for CD14 (LeuM3; Becton Dickinson, San Jose, CA) in a flow cytometric analysis, and by nonspecific esterase staining.
Phenotyping with MoAbs.
The expression of leukocyte cell surface markers and cytoplasmic antigens was assessed. The cells were sequentially incubated with optimal concentrations of biotinylated anti-CD86 (2331; PharMingen, San Diego, CA) and anti-CD1a MoAbs (HI 149; PharMingen), followed by fluorescein isothiocyanate (FITC)-labeled streptavidin and by phycoerythrin (PE)-conjugated goat antimouse IgG, respectively, or directly stained with FITC-labeled anti–HLA-DR (4C3; PharMingen), mouse anti-CD80 (MAB104; Coulter, Fullerton, CA), CD83 (HB15a; Coulter), and PE-labeled anti-CD14 (M5E; PharMingen), CD33 (B8.12.2; Immunotech, Marseille Cedex, France) MoAbs. To block nonspecific FcR-mediated binding of MoAbs, cells were incubated for 60 minutes at 4°C with normal goat serum (Cedarlane Laboratories Ltd, Hornby, Ontario, Canada) before staining. For all experiments, irrelevant control MoAbs of the same IgG isotype and second-step controls were included. Stained cells were fixed with 1% paraformaldehyde (Sigma, St Louis, MO). For all labeling experiments, analysis was performed using the EPICS ELITE (Coulter Electronics, Hialeah, FL).
SAGE protocol.
mRNAs of monocytes, macrophages, and monocyte-derived DCs were purified from a mixture of total RNA of 8 donors. Monocytes were incubated with IL-4 (100 U/mL; Ono Pharmaceutical Co, Ltd, Osaka, Japan) or GM-CSF (500 U/mL; Kirin Brewery Co, Ltd, Tokyo, Japan) in RPMI1640 containing 7.5% FCS in 5% CO2 at 37°C for 7 days. Total RNA from these cells was isolated by direct lysis in RNAzol B (TEL-TEST, Inc, Friendswood, TX). Poly(A)+ RNA were isolated using the FastTrac mRNA purification kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. SAGE was performed as described.9-12 SAGE libraries were generated using 2.5 μg poly(A)+. RNA was converted to complementary DNA (cDNA) with a BRL synthesis kit (GIBCO BRL) following the manufacturer’s protocol, with the inclusion of primer biotin-5′-T18-3′. The cDNA was cleaved with the restriction enzyme Nla III, and the 3′-terminal cDNA fragments were bound to streptavidin-coated magnetic beads (Dynal, Oslo, Norway). After ligation to oligonucleotides containing recognition sites for BsmF1, the linkered cDNAs were released from the beads by digestion with BsmF1. The released tags were ligated to one another, concatemerized, and cloned into the Sph I site of pZero 1.0 (Invitrogen). Colonies were screened with polymerase chain reaction (PCR) using M13 forward and M13 reverse primers. PCR products containing inserts of greater than 400 bp were sequenced with the TaqFS Dyeterminater kit and analyzed using a 377 ABI automated sequencer (Perkin-Elmer, Branchburg, NJ). All electropherograms were reanalyzed by visual inspection to check for ambiguous base and to correct misreads.
SAGE was performed on mRNA from human Mo, GM-CSF–induced Mφ, and Mo-derived DCs. Sequence files were analyzed with the SAGE software,10 CGAP SAGEdatabase (http://www.ncbi.nlm.nih.gov/SAGE/), the NCBI’s sequence search tool (Advanced BLAST search, http://www.ncbi.nlm.nih.gov/BLAST/), and DNAsis software (Takara, Shiga, Japan). After the elimination of linker sequences and the repeated ditags, a total of 173,563 tags, representing 57,560, 57,463, and 58,540 from human Mo, GM-CSF–induced Mφ, and monocyte-derived DC, respectively, were analyzed.
Reverse transcriptase-PCR (RT-PCR).
Total RNAs (200 ng) were prepared using RNAzol B. The RNA was reverse-transcribed in 50 μL of 10 mmol/L Tris-HCl (pH 8.3), 6.5 mmol/L MgCl2, 50 mmol/L KCl, 10 mmol/L dithiothreitol, 1 mmol/L of each dNTP, 2 μmol/L random hexamer, and 2.4 U/μL of Moloney murine leukemia virus reverse transcriptase for 1 hour at 42°C. cDNA, corresponding to 40 ng of total RNA, was boiled for 3 minutes and quenched on ice before amplification by PCR. The conditions for PCR were as follows: in a 50 μL reaction, 15 μmol/L of each primer; 125 μmol/L each of dGTP, dATP, dCTP, and dTTP (Toyobo, Osaka, Japan); 50 mmol/L KCl; 10 mmol/L Tris-HCl, pH 8.3; 1.5 mmol/L MgCl2; and AmplyTaq (Perkin-Elmer). Primers used were as follows: TARC: sense, 5′-ATGGCCCCACTGAAGATGCT-3′, and antisense, 5′-TGAACACCAACGGTGGAGGT-3′; MCP-4: sense, 5′-ATGACAGCAGCTTTCAACCCC-3′, and antisense, 5′-CTCCAAACCAGCAACAAGTCAAT-3′; Thymosin β 10: sense, 5′-TGGCAGACAAACCAGACATGG-3′, and antisense, 5′-ATTTGGCAGTCCGATTGGG-3′; phosphofructokinase: sense, 5′-TTTCAAGATGCGGTTCGACT-3′, and antisense, 5′-AATCCACCGATGATCAGCAG-3′; hepatocyte growth factor inhibitor type 2: sense, 5′-ATCCACGACTTCTGCCTGGT-3′, and antisense, 5′-CGGCAGCCTCCATAGATGAA-3′; metalloproteinase: sense, 5′-TTTTGCCCGTGGAGCTCAT-3′, and antisense, 5′-TTCCCACGGTAGTGACAGCA-3′; tristetraprolin: sense, 5′-CCCTGATGAATATGCCAGC-3′, and antisense, 5′-GGTTCATTGCCTCCCTTAAA-3′; cathepsin C: sense, 5′-TTTCTCAGCTCCCTGCAGCA-3′, and antisense, 5′-CATGCACCCACCCAGTCATT-3′; and CD14: sense, 5′-CGGTCTCAACCTAGAGCCGTTT-3′, and antisense, 5′-TGGGCAATGCTCAGTACCTTG-3′. Reaction mixtures were incubated in a Perkin-Elmer DNA Thermal Cycler for 25 to 30 cycles (denaturation for 60 seconds at 94°C, annealing for 60 seconds at 58°C, and extension for 120 seconds at 72°C).
Statistical analysis.
Statistical significance between samples was calculated as described previously.13
RESULTS
Surface phenotype of normal human blood Mo-derived DCs.
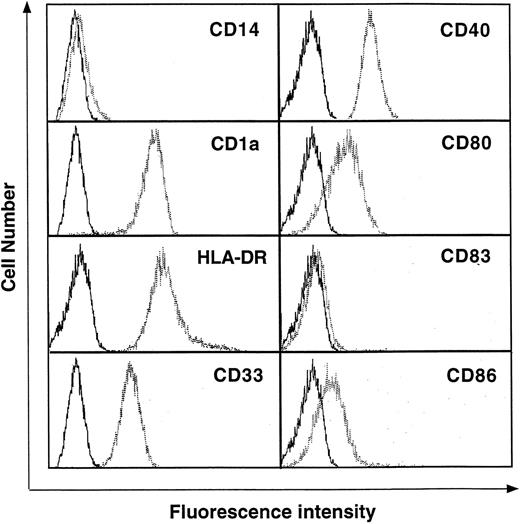
To identify genes specifically expressed in DCs, SAGE libraries were generated from highly purified resting human Mo-derived DCs differentiated by GM-CSF + IL-4 + TNF-α. Peripheral blood CD14+ Mo were cultured with GM-CSF + IL-4 + TNF-α for 5 days. Under these culture conditions, the cells differentiated into nonadherent CD14−, CD1a+, CD80low/−, CD86low/−, HLA-DR+, CD33+, and CD83−cells with the dendritic morphology of immature DCs (Fig 1).
Surface phenotype of normal human blood Mo-derived DCs. DCs were cultured in RPMI-1640 medium plus 7.5% FCS in the presence of GM-CSF (500 U/mL), IL-4 (100 U/mL), and TNF- (50 U/mL) for 5 days. After culture, the cells were washed and then stained with various antibodies as described in Materials and Methods. The data are shown as histograms depicting the number of cells exhibiting various fluorescence intensities. The dotted lines represent staining with specific antibodies and the solid lines represent the isotype-matched control. Results are representative of 3 independent experiments.
Surface phenotype of normal human blood Mo-derived DCs. DCs were cultured in RPMI-1640 medium plus 7.5% FCS in the presence of GM-CSF (500 U/mL), IL-4 (100 U/mL), and TNF- (50 U/mL) for 5 days. After culture, the cells were washed and then stained with various antibodies as described in Materials and Methods. The data are shown as histograms depicting the number of cells exhibiting various fluorescence intensities. The dotted lines represent staining with specific antibodies and the solid lines represent the isotype-matched control. Results are representative of 3 independent experiments.
SAGE tag abundance expression in Mo-derived Dcs.
A total of 58,540 tags sequences from the DC library allowed identification of more than 17,000 different genes, with more than 5,000 genes appearing more than two times. Next, the expressed genes were searched for through the GenBank database to identify individual genes. Table 1 shows the top 50 transcripts in Mo-derived DCs. The most expressed genes in human DCs were identified as HLA DR invariant chain (expression frequency, 1.67%) and ferritin L-chain (1.44%) and HLA DR α chain (1.17%). Overall, the genes expressed abundantly in the DC library consist of products associated with major histocompatibility complex (MHC) class I and class II, protein synthesis, and the cytoskeleton. These data agree with previous data showing high expression of MHC class I and class II genes in DCs, and expression of genes encoding cytoskeleton-associated molecules may be necessary for forming dendrites needed to interact with T cells and B cells.
Transcript Profiles in Human Monocyte-Derived DCs
| Abundance (%) . | Tag Sequence . | GenBank Match (accession no.) . |
|---|---|---|
| 1.66 | GTTCACATTA | HLA DR invariant chain (P33) (X00497) |
| 1.45 | CCCTGGGTTC | Ferritin L-chain (D14523) |
| 1.17 | GGGCATCTCT | HLA-DR α chain (V00523) |
| 0.85 | TTGGTGAAGG | Thymosin β4 (M17733) |
| 0.79 | CCTGTAATCC | Multiple match |
| 0.76 | TGCCTGCACC | Cystatin C (X05607) |
| 0.75 | CAAGCATCCC | No match |
| 0.75 | TTGGTCCTCT | Ribosomal protein L41 (AF02844) |
| 0.68 | TTGGGGTTTC | Ferritin H-chain (AF026844) |
| 0.57 | GTGAAACCCC | Multiple match |
| 0.56 | CTAAGACTTC | No match |
| 0.54 | GTGACCACGG | No match |
| 0.52 | CCACTGCACT | Multiple match |
| 0.48 | GGGGAAATCG | Thymosin β10 (M92381) |
| 0.45 | GTTGTGGTTA | β-2 microglobulin (AB021288) |
| 0.44 | TGTGTTGAGA | Elongation factor α1 subunit (X16871) |
| 0.42 | CCCATCGTCC | Cytochrom coxII (X55654) |
| 0.42 | TTCATACACC | No match |
| 0.35 | GTGAAACCCT | Multiple match |
| 0.33 | TCCAAATCGA | Vimentin (X56134) |
| 0.33 | TGGGTGAGCC | Cathepsin B (M14221) |
| 0.32 | TTGGCCAGGC | Multiple match |
| 0.32 | CACAAACGGT | Ribosomal protein S27 (U57847) |
| 0.30 | AGCCCTACAA | No match |
| 0.29 | GGCTGGGGGC | Profilin (J03191) |
| 0.29 | CACCTAATTG | No match |
| 0.26 | CTGACCTGTG | MHC class I (M54883) |
| 0.26 | ACTTTTTCAA | No match |
| 0.26 | TGGTGTTGAG | Ribosomal protein 18S (X69150) |
| 0.25 | GGAGGTGGGG | Granulin (M75161) |
| 0.25 | TGTACCTGTA | α Tubulin (K00558) |
| 0.24 | TGATTTCACT | No match |
| 0.22 | TGCACGTTTT | Ribosomal protein L32 (X03342) |
| 0.22 | TCACCGGTCA | Gelsolin (X04412) |
| 0.22 | CCTCAGGATA | No match |
| 0.22 | ATGGCTGGTA | LLR ep3 (X17206) |
| 0.21 | GGCACAAAGG | TARC (D43767) |
| 0.21 | GCCTGCTGGG | Phospholipid hydroperoxidase glutathione peroxidase (X71973) |
| 0.21 | AGAAGTGTCC | Lysosomal acid lipase (Z31690) |
| 0.21 | TTCCCTTCTT | MHC class II β chain (X03028) |
| 0.21 | CCAGAACAGA | Ribosomal protein L30 (X79238) |
| 0.21 | CCTAGCTGGA | Peptidyl-prolyl-cis transisomerase A (AA011066) |
| 0.21 | TAGGTTGTCT | Translationally controlled tumor protein (X16064) |
| 0.20 | GTGCTGAATG | Myosin alkali light chain (M22920) |
| 0.19 | AGGCTACGGA | 23-kD highly basic protein (X56932) |
| 0.19 | ACGCAGGGAG | Heat shock protein 90 (D87666) |
| 0.19 | GCTTTATTTG | β Actin (X00351) |
| 0.19 | AAAACATTCT | No match |
| 0.18 | CCCCCTGGAT | Growth factor inducible 2A9 gene (M14306) |
| 0.18 | TGAAAACTAC | MHC class II α chain (X00457) |
| Abundance (%) . | Tag Sequence . | GenBank Match (accession no.) . |
|---|---|---|
| 1.66 | GTTCACATTA | HLA DR invariant chain (P33) (X00497) |
| 1.45 | CCCTGGGTTC | Ferritin L-chain (D14523) |
| 1.17 | GGGCATCTCT | HLA-DR α chain (V00523) |
| 0.85 | TTGGTGAAGG | Thymosin β4 (M17733) |
| 0.79 | CCTGTAATCC | Multiple match |
| 0.76 | TGCCTGCACC | Cystatin C (X05607) |
| 0.75 | CAAGCATCCC | No match |
| 0.75 | TTGGTCCTCT | Ribosomal protein L41 (AF02844) |
| 0.68 | TTGGGGTTTC | Ferritin H-chain (AF026844) |
| 0.57 | GTGAAACCCC | Multiple match |
| 0.56 | CTAAGACTTC | No match |
| 0.54 | GTGACCACGG | No match |
| 0.52 | CCACTGCACT | Multiple match |
| 0.48 | GGGGAAATCG | Thymosin β10 (M92381) |
| 0.45 | GTTGTGGTTA | β-2 microglobulin (AB021288) |
| 0.44 | TGTGTTGAGA | Elongation factor α1 subunit (X16871) |
| 0.42 | CCCATCGTCC | Cytochrom coxII (X55654) |
| 0.42 | TTCATACACC | No match |
| 0.35 | GTGAAACCCT | Multiple match |
| 0.33 | TCCAAATCGA | Vimentin (X56134) |
| 0.33 | TGGGTGAGCC | Cathepsin B (M14221) |
| 0.32 | TTGGCCAGGC | Multiple match |
| 0.32 | CACAAACGGT | Ribosomal protein S27 (U57847) |
| 0.30 | AGCCCTACAA | No match |
| 0.29 | GGCTGGGGGC | Profilin (J03191) |
| 0.29 | CACCTAATTG | No match |
| 0.26 | CTGACCTGTG | MHC class I (M54883) |
| 0.26 | ACTTTTTCAA | No match |
| 0.26 | TGGTGTTGAG | Ribosomal protein 18S (X69150) |
| 0.25 | GGAGGTGGGG | Granulin (M75161) |
| 0.25 | TGTACCTGTA | α Tubulin (K00558) |
| 0.24 | TGATTTCACT | No match |
| 0.22 | TGCACGTTTT | Ribosomal protein L32 (X03342) |
| 0.22 | TCACCGGTCA | Gelsolin (X04412) |
| 0.22 | CCTCAGGATA | No match |
| 0.22 | ATGGCTGGTA | LLR ep3 (X17206) |
| 0.21 | GGCACAAAGG | TARC (D43767) |
| 0.21 | GCCTGCTGGG | Phospholipid hydroperoxidase glutathione peroxidase (X71973) |
| 0.21 | AGAAGTGTCC | Lysosomal acid lipase (Z31690) |
| 0.21 | TTCCCTTCTT | MHC class II β chain (X03028) |
| 0.21 | CCAGAACAGA | Ribosomal protein L30 (X79238) |
| 0.21 | CCTAGCTGGA | Peptidyl-prolyl-cis transisomerase A (AA011066) |
| 0.21 | TAGGTTGTCT | Translationally controlled tumor protein (X16064) |
| 0.20 | GTGCTGAATG | Myosin alkali light chain (M22920) |
| 0.19 | AGGCTACGGA | 23-kD highly basic protein (X56932) |
| 0.19 | ACGCAGGGAG | Heat shock protein 90 (D87666) |
| 0.19 | GCTTTATTTG | β Actin (X00351) |
| 0.19 | AAAACATTCT | No match |
| 0.18 | CCCCCTGGAT | Growth factor inducible 2A9 gene (M14306) |
| 0.18 | TGAAAACTAC | MHC class II α chain (X00457) |
The top 50 transcripts expressed in DCs are listed. The tag sequence represents the 10-bp SAGE tag. Probable GenBank matches are listed. More information is available on the Internet athttp://www.prevent.m.u-tokyo.ac.jp/SAGE.html.
Comparison of expression patterns in Mo-derived DCs.
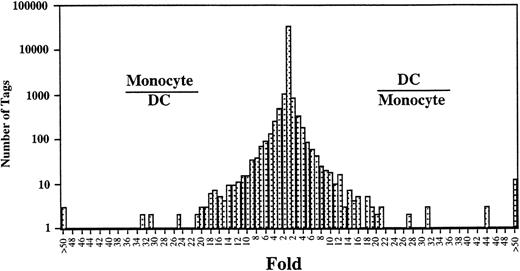
Comparison of the expressed genes between Mo14 and DCs showed that the expression levels of most of the transcripts (>20,000) in these cells were similar (Fig 2). However, the expression profiles also showed 313 transcripts in Mo that were different from those in DCs (P < .01). Expression levels of 181 of 313 genes were decreased in DCs as compared with those in Mo. Conversely, 132 transcripts were expressed at higher levels in the DCs than in Mo. Table 2 shows the top 50 that were increased transcripts in DCs compared with Mo. In addition, the gene expression compared between DCs and GM-Mφ14 shows a similar pattern (Table 2). The transcripts increased in DCs mainly consist of genes encoding proteins associated with cell structure such as gelsolin and vinculin, lipid metabolism such as lysosomal acid lipase and apolipoprotein C-1, and chemokines such as TARC, MDC, and MCP-4. The expression of gelsolin, lysosomal acid lipase, MDC, fatty acid binding protein homologue, acid phosphatase type 5, GA733-1, CD9, HSP27, etc, was increased in Mφ as well as in DCs. On the other hand, TARC, hepatocyte growth factor activator inhibitor type-2 (HAI-2), platelet-type phosphofructokinase, factor XIII,15CD23,16 cathepsin C, MCP-4, and metalloproteinase were highly specific for DCs (Fig 3).
Comparison of gene expression frequency in Mo and Mo-derived DCs. A semilogarithmic plot shows 105 tags that were decreased more than 10-fold in DCs compared with Mo, whereas 109 tags were increased more than 10-fold in DCs compared with Mo. The relative expression of each transcript was determined by dividing the number of tags observed in Mo or DCs, as indicated. To avoid division by 0, we used a tag value of 1 for any tag that was not detectable in 1 sample. These ratios are plotted on the abscissa. The number of genes displaying each ratio is plotted on the ordinate.
Comparison of gene expression frequency in Mo and Mo-derived DCs. A semilogarithmic plot shows 105 tags that were decreased more than 10-fold in DCs compared with Mo, whereas 109 tags were increased more than 10-fold in DCs compared with Mo. The relative expression of each transcript was determined by dividing the number of tags observed in Mo or DCs, as indicated. To avoid division by 0, we used a tag value of 1 for any tag that was not detectable in 1 sample. These ratios are plotted on the abscissa. The number of genes displaying each ratio is plotted on the ordinate.
Transcripts Increased in Human Monocyte-Derived DCs
| Fold . | Tag Sequence . | n* . | GenBank Match (accession no.) . | ||
|---|---|---|---|---|---|
| Mono . | GM . | DC . | |||
| 125 | TCACCGGTCA | 0 | 72 | 125 | Gelsolin (X04412) |
| 122 | GGCACAAAGG | 0 | 1 | 122 | TARC (D43767) |
| 61 | AGAAGTGTCC | 2 | 79 | 121 | Lysosomal acid lipase (Z31690) |
| 47 | AGCCACCGCA | 2 | 2 | 94 | No match |
| 41 | CGTGAGCCAC | 0 | 10 | 41 | No match |
| 40 | GCCTGCAGTC | 0 | 4 | 40 | Hepatocyte growth factor activator inhibitor type 2 (AB006534) |
| 36 | GCCCTGAAAG | 0 | 3 | 36 | No match |
| 35 | TTGGAACAAT | 0 | 0 | 35 | No match |
| 34 | CCTCTGGGCA | 0 | 0 | 34 | No match |
| 33 | AACGGGGCCC | 0 | 117 | 33 | MDC (U83171) |
| 33 | CCAAGACTTC | 0 | 3 | 33 | No match |
| 32 | CAGCTATTTC | 0 | 15 | 32 | Fatty acid binding protein homologue (M94856) |
| 31 | TTACTTCCCC | 0 | 5 | 31 | EST (AA927388) |
| 31 | CTTTCAGATG | 0 | 0 | 31 | Platelet-type phosphofructokinase (D25328) |
| 30 | TTGAGACCTC | 1 | 0 | 30 | Factor XIII subunit α (M14539) |
| 28 | CTGTTAGTGT | 2 | 20 | 55 | Malate dehydrogenase (U20352) |
| 25 | TGTCCCAGCC | 0 | 64 | 25 | Acid phosphatase type 5 (X14618) |
| 25 | TCTTGATTTA | 0 | 31 | 25 | α-2 macroglobulin (M11313) |
| 23 | CCCCCGTAAT | 0 | 3 | 23 | EST (AI203963) |
| 22 | ATGGAAGTCT | 0 | 6 | 22 | Inositol polyphosphate 5-phosphatase (X98429) |
| 21 | CTAAAAAAAA | 0 | 9 | 21 | No match |
| 21 | CAAGCATCCC | 21 | 166 | 430 | No match |
| 20 | TGTGAACAAC | 0 | 1 | 20 | EST (AI001049) |
| 20 | ACTCACCTTA | 0 | 0 | 20 | EST (AA742376) |
| 20 | GATACAGCCA | 0 | 0 | 20 | CD23 (M14766) |
| 20 | ACCCAGGGTA | 0 | 19 | 19 | EST (N30858) |
| 19 | CTGATCTCCA | 0 | 5 | 19 | No match |
| 19 | TTTGCTCTCC | 2 | 3 | 37 | Vinculin (M33308) |
| 18 | GATGACCCCC | 0 | 34 | 18 | No match |
| 18 | GCCTACCCGA | 0 | 15 | 18 | GA733-1 (X13425) |
| 18 | CCCTCGGTCC | 0 | 3 | 18 | No match |
| 18 | AACGAGGAAT | 0 | 2 | 18 | No match |
| 18 | CTATATTTTT | 0 | 1 | 18 | Cathepsin C (X87212) |
| 18 | AAGGGATGCT | 0 | 0 | 18 | MCP-4 (U46767) |
| 17 | AAGATTGGTG | 0 | 32 | 17 | CD9 (M38690) |
| 17 | CCCAAGCTAG | 1 | 9 | 17 | HSP27 (X54079) |
| 17 | CCGGGCGTGG | 0 | 5 | 17 | EST (R49492) |
| 17 | GGTGTGCTTG | 0 | 1 | 17 | EST (AF055021) |
| 17 | AGTGCAGGGC | 0 | 0 | 17 | No match |
| 17 | CCTCACTACC | 0 | 0 | 17 | EST (AI288509) |
| 17 | TGCAGAAGAA | 0 | 0 | 17 | Multiple match |
| 16 | TGGCGTACGG | 3 | 7 | 48 | No match |
| 16 | GCCAGCCCAG | 1 | 4 | 16 | Zinc finger protein (X97548) |
| 16 | GGTCCCCTAC | 0 | 4 | 16 | EST (AI343615) |
| 16 | TGTACCTGTA | 9 | 61 | 140 | No match |
| 16 | TGGCCCCAGG | 6 | 1,515 | 93 | Apolipoprotein C-1 (X00570) |
| 15 | TTTTCTGAAA | 0 | 6 | 15 | Thioredoxin (J04026) |
| 15 | CTCTGTAAGT | 0 | 0 | 15 | Metalloproteinase (L23808) |
| 15 | GGAGGCAGGA | 0 | 0 | 15 | No match |
| 15 | ATGAGCTGAC | 7 | 105 | 104 | Cystatin B (L03558) |
| Fold . | Tag Sequence . | n* . | GenBank Match (accession no.) . | ||
|---|---|---|---|---|---|
| Mono . | GM . | DC . | |||
| 125 | TCACCGGTCA | 0 | 72 | 125 | Gelsolin (X04412) |
| 122 | GGCACAAAGG | 0 | 1 | 122 | TARC (D43767) |
| 61 | AGAAGTGTCC | 2 | 79 | 121 | Lysosomal acid lipase (Z31690) |
| 47 | AGCCACCGCA | 2 | 2 | 94 | No match |
| 41 | CGTGAGCCAC | 0 | 10 | 41 | No match |
| 40 | GCCTGCAGTC | 0 | 4 | 40 | Hepatocyte growth factor activator inhibitor type 2 (AB006534) |
| 36 | GCCCTGAAAG | 0 | 3 | 36 | No match |
| 35 | TTGGAACAAT | 0 | 0 | 35 | No match |
| 34 | CCTCTGGGCA | 0 | 0 | 34 | No match |
| 33 | AACGGGGCCC | 0 | 117 | 33 | MDC (U83171) |
| 33 | CCAAGACTTC | 0 | 3 | 33 | No match |
| 32 | CAGCTATTTC | 0 | 15 | 32 | Fatty acid binding protein homologue (M94856) |
| 31 | TTACTTCCCC | 0 | 5 | 31 | EST (AA927388) |
| 31 | CTTTCAGATG | 0 | 0 | 31 | Platelet-type phosphofructokinase (D25328) |
| 30 | TTGAGACCTC | 1 | 0 | 30 | Factor XIII subunit α (M14539) |
| 28 | CTGTTAGTGT | 2 | 20 | 55 | Malate dehydrogenase (U20352) |
| 25 | TGTCCCAGCC | 0 | 64 | 25 | Acid phosphatase type 5 (X14618) |
| 25 | TCTTGATTTA | 0 | 31 | 25 | α-2 macroglobulin (M11313) |
| 23 | CCCCCGTAAT | 0 | 3 | 23 | EST (AI203963) |
| 22 | ATGGAAGTCT | 0 | 6 | 22 | Inositol polyphosphate 5-phosphatase (X98429) |
| 21 | CTAAAAAAAA | 0 | 9 | 21 | No match |
| 21 | CAAGCATCCC | 21 | 166 | 430 | No match |
| 20 | TGTGAACAAC | 0 | 1 | 20 | EST (AI001049) |
| 20 | ACTCACCTTA | 0 | 0 | 20 | EST (AA742376) |
| 20 | GATACAGCCA | 0 | 0 | 20 | CD23 (M14766) |
| 20 | ACCCAGGGTA | 0 | 19 | 19 | EST (N30858) |
| 19 | CTGATCTCCA | 0 | 5 | 19 | No match |
| 19 | TTTGCTCTCC | 2 | 3 | 37 | Vinculin (M33308) |
| 18 | GATGACCCCC | 0 | 34 | 18 | No match |
| 18 | GCCTACCCGA | 0 | 15 | 18 | GA733-1 (X13425) |
| 18 | CCCTCGGTCC | 0 | 3 | 18 | No match |
| 18 | AACGAGGAAT | 0 | 2 | 18 | No match |
| 18 | CTATATTTTT | 0 | 1 | 18 | Cathepsin C (X87212) |
| 18 | AAGGGATGCT | 0 | 0 | 18 | MCP-4 (U46767) |
| 17 | AAGATTGGTG | 0 | 32 | 17 | CD9 (M38690) |
| 17 | CCCAAGCTAG | 1 | 9 | 17 | HSP27 (X54079) |
| 17 | CCGGGCGTGG | 0 | 5 | 17 | EST (R49492) |
| 17 | GGTGTGCTTG | 0 | 1 | 17 | EST (AF055021) |
| 17 | AGTGCAGGGC | 0 | 0 | 17 | No match |
| 17 | CCTCACTACC | 0 | 0 | 17 | EST (AI288509) |
| 17 | TGCAGAAGAA | 0 | 0 | 17 | Multiple match |
| 16 | TGGCGTACGG | 3 | 7 | 48 | No match |
| 16 | GCCAGCCCAG | 1 | 4 | 16 | Zinc finger protein (X97548) |
| 16 | GGTCCCCTAC | 0 | 4 | 16 | EST (AI343615) |
| 16 | TGTACCTGTA | 9 | 61 | 140 | No match |
| 16 | TGGCCCCAGG | 6 | 1,515 | 93 | Apolipoprotein C-1 (X00570) |
| 15 | TTTTCTGAAA | 0 | 6 | 15 | Thioredoxin (J04026) |
| 15 | CTCTGTAAGT | 0 | 0 | 15 | Metalloproteinase (L23808) |
| 15 | GGAGGCAGGA | 0 | 0 | 15 | No match |
| 15 | ATGAGCTGAC | 7 | 105 | 104 | Cystatin B (L03558) |
The 50 transcripts displaying the biggest increase in expression in DCs are listed by fold induction. The tag sequence represents the 10-bp SAGE tag. The most probable GenBank matches are listed. n indicates the number of times the tag was identified. Fold changes in expression were calculated as described in Fig 2. More information is available on the Internet athttp://www.prevent.m.u-tokyo.ac.jp/SAGE.html.
Abbreviations: Mono, monocytes; GM, GM-CSF–induced macrophages.
In this table, the tag numbers are 57,560, 57,463, and 58,540 from Mo, GM-Mφ, and DCs, respectively.
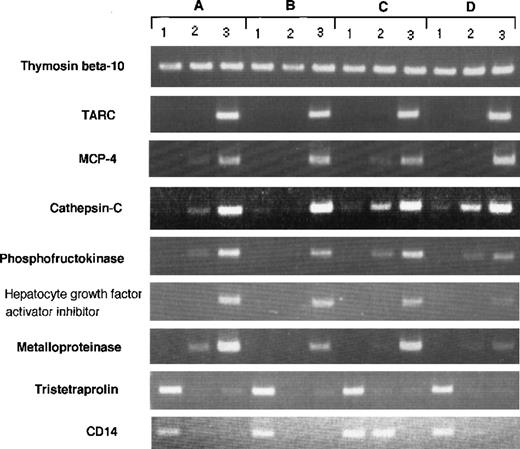
RT-PCR analysis of genes expressed differently in Mo, GM-CSF–induced M◊, and DCs. RT-PCR was performed on total RNA isolated from (1) human Mo, (2) GM-CSF–induced M◊, and (3) Mo-derived DCs as described in Materials and Methods. (A) through (D) indicate different donors.
RT-PCR analysis of genes expressed differently in Mo, GM-CSF–induced M◊, and DCs. RT-PCR was performed on total RNA isolated from (1) human Mo, (2) GM-CSF–induced M◊, and (3) Mo-derived DCs as described in Materials and Methods. (A) through (D) indicate different donors.
Table 3 shows the top 50 transcripts decreased in DCs. The most decreased mRNAs were identified complement proteins; ficolin and properdin; DNA-binding proteins; GOS3, GOS2, and c-fos; surface protein; and CD14. The decreased transcripts in DCs showed a similar tendency in GM-Mφ. Moreover, transcripts decreased more than 10-fold compared with GM-Mφ were MRP-14, macrophage inhibitory protein-1α (MIP-1α), CD14, α1 antitrypsin, HLA-B, and transletolase. The reduction of CD14 in DCs was also confirmed by a flow cytometric analysis (Fig 1).
Transcripts Decreased in Human Monocyte-Derived DCs
| Fold . | Tag Sequence . | n3-150 . | GenBank Match (accession no.) . | ||
|---|---|---|---|---|---|
| Mono . | GM . | DC . | |||
| 532 | GTGGCCACGG | 1,064 | 47 | 2 | MRP-14 (M21064) |
| 287 | TACCTGCAGA | 287 | 0 | 0 | MRP-8 (X06234) |
| 244 | CCCACAACCT | 244 | 3 | 0 | Ficolin-1 (S80990) |
| 105 | GCACCAAAGC | 105 | 19 | 0 | MIP-1α (D00044) |
| 97 | TGGAAGCACT | 97 | 0 | 0 | IL-8 (Y00787) |
| 84 | TGGTCCAGCG | 84 | 10 | 0 | CD14 (AA971438) |
| 77 | CTTGACATAC | 77 | 0 | 1 | CL100 mRNA for protein tyrosin phosphatase (X68277) |
| 66 | GGAAAAGTGG | 66 | 10 | 1 | α1 antitrypsin (X01683) |
| 65 | GGCCACGTAG | 65 | 2 | 0 | D component of complement (AI073930) |
| 65 | GCTGTTGCGC | 65 | 22 | 1 | Ribosomal protein S20 (AA969927) |
| 63 | TCTACACGTG | 63 | 0 | 0 | Properdin |
| 63 | CTGATGGCGA | 63 | 0 | 0 | No match |
| 55 | CTGTACTTGT | 55 | 0 | 0 | GOS3 (L49169) |
| 51 | TGGAAAGTGA | 51 | 0 | 0 | c-fos (V01512) |
| 48 | CTGACTTGTG | 48 | 10 | 1 | MHC class I HLA-B (M14539) |
| 46 | ACATTTCCAA | 46 | 0 | 0 | GOS2 (M69199) |
| 44 | TCACAGCTGT | 88 | 5 | 2 | B-cell translocation gene 1 (AI096550) |
| 44 | ATGGTGGGGG | 44 | 0 | 1 | Tristetraproline (M63625) |
| 44 | TTGGCAGCCC | 44 | 19 | 1 | Ribosomal protein L27a (H73560) |
| 34 | TGGGCAGCTG | 34 | 2 | 0 | Ribosomal protein S9 (AA833617) |
| 33 | TTGCGTGTGT | 33 | 20 | 0 | Protein phosphatase PAC-1 (AA922462) |
| 31 | CTCCATCCAG | 31 | 1 | 0 | G-CSF receptor (M59819) |
| 31 | AGTGCACGTG | 31 | 0 | 0 | Ribosomal protein L29 (R36372) |
| 31 | GGCCAGGACT | 31 | 0 | 0 | Formyl peptide receptor 1 (M60627) |
| 27 | CCCTGAGGCC | 27 | 0 | 0 | EST (AI148871) |
| 27 | GTCCCCCCAA | 27 | 2 | 1 | EST (AI217285) |
| 27 | GTGGGCCACG | 25 | 0 | 0 | No match |
| 25 | GCCGCCATCT | 70 | 20 | 3 | Protein disulfide isomerase (U79278) |
| 23 | CTGTTGGCAT | 112 | 16 | 5 | Ribosomal protein L21 (U14967) |
| 22 | ACCATTCTGC | 21 | 1 | 0 | Interferon-inducible mRNA (X02490) |
| 21 | ACTTTAATGA | 21 | 3 | 0 | C5a anaphylatoxin receptor (M62505) |
| 21 | AATTAAATTA | 21 | 11 | 0 | Multiple match |
| 21 | GCCGCCGTGC | 20 | 0 | 0 | No match |
| 20 | GGTAGCCCAC | 20 | 7 | 1 | EST (AI093040) |
| 20 | CTTTTTTCCC | 19 | 0 | 1 | CD48 (M59904) |
| 19 | CTTGGGATGT | 19 | 4 | 0 | EST (AA865970) |
| 19 | ATGGAGCGCA | 37 | 11 | 2 | TNFR2 (U52165) |
| 19 | TAGCCCCCTG | 18 | 0 | 0 | TNF (M10988) |
| 18 | TTGGAGCACT | 18 | 0 | 0 | EST (AA724726) |
| 18 | TTTCTGTATG | 18 | 0 | 0 | HISTONE H3.3 (AI033898) |
| 18 | ACGCTCTCGA | 18 | 2 | 1 | CD37 |
| 18 | ATGGCTTGGT | 18 | 6 | 0 | 40S ribosomal protein S2 (AA064604) |
| 17 | TAATGCTAAA | 17 | 0 | 0 | Helix-loop-helix basic phosphoprotein (L13463) |
| 16 | TGAAGTAACA | 48 | 0 | 3 | Translation factor SUI1 homolog (R15861) |
| 16 | GGGAAACAGG | 48 | 2 | 3 | EST (AA970435) |
| 16 | TGAGTCTGGC | 16 | 0 | 1 | EST (W63798) |
| 16 | GCGACAGCTC | 16 | 8 | 1 | Ribosomal protein L30 (M94314) |
| 16 | GCTGAACGCG | 79 | 18 | 5 | EST (AI024982) |
| 15 | TGTTTTCATA | 15 | 1 | 0 | No match |
| 15 | CTCACTTTTT | 15 | 0 | 0 | NF-IL6 β (M83667) |
| Fold . | Tag Sequence . | n3-150 . | GenBank Match (accession no.) . | ||
|---|---|---|---|---|---|
| Mono . | GM . | DC . | |||
| 532 | GTGGCCACGG | 1,064 | 47 | 2 | MRP-14 (M21064) |
| 287 | TACCTGCAGA | 287 | 0 | 0 | MRP-8 (X06234) |
| 244 | CCCACAACCT | 244 | 3 | 0 | Ficolin-1 (S80990) |
| 105 | GCACCAAAGC | 105 | 19 | 0 | MIP-1α (D00044) |
| 97 | TGGAAGCACT | 97 | 0 | 0 | IL-8 (Y00787) |
| 84 | TGGTCCAGCG | 84 | 10 | 0 | CD14 (AA971438) |
| 77 | CTTGACATAC | 77 | 0 | 1 | CL100 mRNA for protein tyrosin phosphatase (X68277) |
| 66 | GGAAAAGTGG | 66 | 10 | 1 | α1 antitrypsin (X01683) |
| 65 | GGCCACGTAG | 65 | 2 | 0 | D component of complement (AI073930) |
| 65 | GCTGTTGCGC | 65 | 22 | 1 | Ribosomal protein S20 (AA969927) |
| 63 | TCTACACGTG | 63 | 0 | 0 | Properdin |
| 63 | CTGATGGCGA | 63 | 0 | 0 | No match |
| 55 | CTGTACTTGT | 55 | 0 | 0 | GOS3 (L49169) |
| 51 | TGGAAAGTGA | 51 | 0 | 0 | c-fos (V01512) |
| 48 | CTGACTTGTG | 48 | 10 | 1 | MHC class I HLA-B (M14539) |
| 46 | ACATTTCCAA | 46 | 0 | 0 | GOS2 (M69199) |
| 44 | TCACAGCTGT | 88 | 5 | 2 | B-cell translocation gene 1 (AI096550) |
| 44 | ATGGTGGGGG | 44 | 0 | 1 | Tristetraproline (M63625) |
| 44 | TTGGCAGCCC | 44 | 19 | 1 | Ribosomal protein L27a (H73560) |
| 34 | TGGGCAGCTG | 34 | 2 | 0 | Ribosomal protein S9 (AA833617) |
| 33 | TTGCGTGTGT | 33 | 20 | 0 | Protein phosphatase PAC-1 (AA922462) |
| 31 | CTCCATCCAG | 31 | 1 | 0 | G-CSF receptor (M59819) |
| 31 | AGTGCACGTG | 31 | 0 | 0 | Ribosomal protein L29 (R36372) |
| 31 | GGCCAGGACT | 31 | 0 | 0 | Formyl peptide receptor 1 (M60627) |
| 27 | CCCTGAGGCC | 27 | 0 | 0 | EST (AI148871) |
| 27 | GTCCCCCCAA | 27 | 2 | 1 | EST (AI217285) |
| 27 | GTGGGCCACG | 25 | 0 | 0 | No match |
| 25 | GCCGCCATCT | 70 | 20 | 3 | Protein disulfide isomerase (U79278) |
| 23 | CTGTTGGCAT | 112 | 16 | 5 | Ribosomal protein L21 (U14967) |
| 22 | ACCATTCTGC | 21 | 1 | 0 | Interferon-inducible mRNA (X02490) |
| 21 | ACTTTAATGA | 21 | 3 | 0 | C5a anaphylatoxin receptor (M62505) |
| 21 | AATTAAATTA | 21 | 11 | 0 | Multiple match |
| 21 | GCCGCCGTGC | 20 | 0 | 0 | No match |
| 20 | GGTAGCCCAC | 20 | 7 | 1 | EST (AI093040) |
| 20 | CTTTTTTCCC | 19 | 0 | 1 | CD48 (M59904) |
| 19 | CTTGGGATGT | 19 | 4 | 0 | EST (AA865970) |
| 19 | ATGGAGCGCA | 37 | 11 | 2 | TNFR2 (U52165) |
| 19 | TAGCCCCCTG | 18 | 0 | 0 | TNF (M10988) |
| 18 | TTGGAGCACT | 18 | 0 | 0 | EST (AA724726) |
| 18 | TTTCTGTATG | 18 | 0 | 0 | HISTONE H3.3 (AI033898) |
| 18 | ACGCTCTCGA | 18 | 2 | 1 | CD37 |
| 18 | ATGGCTTGGT | 18 | 6 | 0 | 40S ribosomal protein S2 (AA064604) |
| 17 | TAATGCTAAA | 17 | 0 | 0 | Helix-loop-helix basic phosphoprotein (L13463) |
| 16 | TGAAGTAACA | 48 | 0 | 3 | Translation factor SUI1 homolog (R15861) |
| 16 | GGGAAACAGG | 48 | 2 | 3 | EST (AA970435) |
| 16 | TGAGTCTGGC | 16 | 0 | 1 | EST (W63798) |
| 16 | GCGACAGCTC | 16 | 8 | 1 | Ribosomal protein L30 (M94314) |
| 16 | GCTGAACGCG | 79 | 18 | 5 | EST (AI024982) |
| 15 | TGTTTTCATA | 15 | 1 | 0 | No match |
| 15 | CTCACTTTTT | 15 | 0 | 0 | NF-IL6 β (M83667) |
The 50 transcripts displaying the largest decrease in expression in DCs are listed by fold reduction. n indicates the number of times the tag was identified. Conditions are as described in Fig 2. More information is available on the Internet athttp://www.prevent.m.u-tokyo.ac.jp/SAGE.html.
In this table, the tag numbers are 57,560, 57,463, and 58,540 from Mo, GM-Mφ, and DCs, respectively.
RT-PCR of genes represented in the SAGE analysis.
Although our data represent the average gene expression on cells obtained from 8 donors, there could be differences in gene expression between individual donor-derived cells. To address this question, we arbitrarily selected 9 differently expressed genes and evaluated them in 4 donor-derived samples by RT-PCR (Fig 3). The expression of each transcript was compared with SAGE data. Thymosin β 10 was expressed in all cell types (Mo, 214; GM-Mφ, 246; and DCs, 273); TARC was highly expressed in DCs (Mo, 0; GM-Mφ, 1; and DCs, 122); MCP-4 was highly expressed in DCs (Mo, 0; GM-Mφ, 0; and DCs, 18); cathepsin-C was highly expressed in DCs (Mo, 0; GM-Mφ, 1; and DCs, 18); phosphofructokinase was highly expressed in DCs (Mo, 0; GM-Mφ, 0; and DCs, 31); HAI-2 was highly expressed in DCs (Mo, 0; GM-Mφ, 4; and DCs, 40); metalloproteinase was highly expressed in DCs (Mo, 0; GM-Mφ, 0; and DCs, 15); tristetraproline was highly expressed in Mo (Mo, 44; GM-Mφ, 0; and DCs, 1); and CD14 was highly expressed in Mo (Mo, 84; GM-Mφ, 10; and DCs, 0). These results confirm our SAGE data for Mo, GM-Mφ, and DCs and establish the general expression profile of the identified genes.
DISCUSSION
It is an important to clarify the origin and nature of DCs to exploit them as therapeutics and vaccines. The heterogeneity of DCs, which have various functions, may depend on differences in specific gene expression induced by growth factors, such as IL-4, TNF-α, transforming growth factor-β (TGF-β), and GM-CSF. Thus, to investigate the function of gene regulation in DCs, we performed SAGE in human blood Mo-derived DCs induced by GM-CSF, IL-4, and TNF-α.
A technology that yields identification of differently expressed genes can provide an important tool for the understanding of cell biology. Several methods, such as Northern blotting, RT-PCR, differential display, and subtraction, are useful for such studies. However, these technologies can analyze only a limited numbers of genes, and the quantitative analysis of the transcription of individual genes is difficult. SAGE allows both quantitative and simultaneous analysis of large numbers of transcripts. We investigated a total of 173,562 tags derived from 57,559, 57,463, and 58,540 tags from Mo, GM-Mφ, and DCs, respectively, which allowed for the identification of 36,605 different gene transcripts. Sequences from the DC library represented more than 17,000 different genes, with more than 5,000 genes appearing more than 2 times.
We observed many genes expressed in DCs that have not been identified before this analysis. One of these, HAI-2, which is highly specific for DCs and is a Kunitz-type serine protease inhibitor, has been found to be an inhibitor of hepatocyte growth factor activator (HGF) responsible for proteolytic activation of HGF.17,18 Another group has identified the same gene (kop, from pancreatic cancer), which may participate in tumor cell invasion and metastasis. Therefore, HAI-2 may be involved in the regulation of proteolytic activation of HGF in injured tissue or the migration of cells. Moreover, another differently expressed gene in DCs, a metalloproteinase that possesses elastolytic activity,19 is also thought to be a candidate molecule for the cause of diseases characterized by damage to the extracellular matrix or cell migration. It is predicted that alteration of expression of many cytoskeleton proteins is involved in the change of cell migration. In addition to the top 50 differently expressed genes in DCs, the expression of genes encoding proteins associated with cell structure such as thymosin β-4 (tag number; Mo, 223; and DCs, 486), β-actin (Mo, 53; and DCs, 108), α-actinin (Mo, 1; and DCs, 13), vimentin (Mo, 82; and DCs, 187), and profilin (Mo, 83; and DCs, 167) was also increased in DCs as compared with Mo. Taken together, all of these data suggest that differentiation from Mo into DCs is accompanied by a significant change in the expression of genes related to cell structure and motility.
Interestingly, increased expression of the genes encoding chemokines, such as TARC, MDC, and MCP-4, that selectively chemoattract CCR4- and CCR3-positive Th2-type lymphocytes,20,21 were observed in monocyte-derived DCs (Table 2 and Fig 3). In contrast, expression of the genes encoding chemokines that selectively chemoattract CCR5- and CXCR3-positive Th1-type lymphocytes, such as IP-10, Mig, and MIP-1α, was not observed in tags of the tested DC library (data not shown). These results suggests that DCs tested in this study may chemoattract Th2 cells selectively. Thus, the result was unexpected, because several groups have demonstrated that activated or mature DCs promote Th1 differentiation through IL-12 production.22,23 A recent study showed that immature DCs may promote Th2 responses.24Moreover, it has been reported that Th2 polarity in the resting mucosal immune system may be an inherent property of the resident DC population.25 Thus, we presume that Mo-derived DCs might be critical for polarization or amplification of Th2 cells and play an important role for Th2-dominated immune diseases such as asthma26,27 and atopic dermatitis.28 29
In conclusion, identification of the genes selectively expressed in human Mo-derived DCs should provide useful information for defining the development and function of DCs. Furthermore, many sets of genes, including the novel genes identified to be selectively expressed in DCs, should provide further understanding of the biological function of DCs in the host defense system and may also be useful for diagnosing or monitoring human diseases in which DCs may play a role.
ACKNOWLEDGMENT
The authors are very grateful to Drs V. Velculescu, L. Zhang, W. Zhou, B. Vogelstein, and K. Kinzler for their help in SAGE analysis and also to Dr H. Young and Dr C. Vestergaard for reviewing this manuscript.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Kouji Matsushima, MD, PhD, Department of Molecular Preventive Medicine, School of Medicine, University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan; e-mail:koujim@m.u-tokyo.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal