The human immunodeficiency virus type 1 (HIV-1) Tat gene, a potent transactivator of viral and cellular genes, has been proposed as a key agent in the pathogenesis of acquired immune deficiency syndrome related disorders, including nonHodgkin’s lymphoma. In cultured cells, the HIV-1 Tat protein can induce the expression of the cytokines interleukin-6 (IL-6) and IL-10, which are known to induce proliferation and differentiation of lymphoid cells. Such alterations in cytokine expression, together with a secondary genetic event, are thought to ultimately lead to oncogenic transformation. To address the influence of Tat on lymphoid development in the context of the whole organism, we produced several transgenic mouse lines that express the Tat gene under the control of an actin promoter. We show here that this promoter directs expression to a variety of sites, including spleen, bone marrow, and lymph nodes. Approximately 25% to 30% of the Tat-transgenic population developed enlarged spleens within 1 year after birth. On histological examination, a significant number of spleens from Tat-transgenic mice exhibited malignant lymphoma of B-cell origin. IgG heavy chain rearrangement confirmed the clonal B-cell nature of these lymphoproliferations. In contrast, T-cell receptor genes exhibited a germline (unrearranged) structure. Reverse transcription polymerase chain reaction analysis of transgenic spleens revealed that mRNA encoding cytokines IL-6 and IL-10 was upregulated, suggesting a possible mechanism for the B-cell expansion in vivo.
HUMAN IMMUNODEFICIENCY virus type 1 (HIV-1) infection causes several clinical and immunological abnormalities, including lymphadenopathy, activation of polyclonal B cells, Kaposi’s sarcoma, and nonHodgkin’s lymphoma.1,2Approximately 5% to 10% of acquired immune deficiency syndrome (AIDS) patients develop such lymphomas, which are almost invariably derived from B cells.3 Although the pathogenesis of AIDS-related lymphoma is poorly understood, experiments in cell culture models have shown that the HIV-1 Tat gene can cause changes in cytokine levels, which, in turn, stimulate the growth of lymphoid cells.4
The Tat gene encodes a transregulatory protein with important functions in the expression of HIV-1 genes.5 Tat is required for efficient viral gene expression6 and functions by enhancing transcription elongation through interactions with the cis-acting transactivation response element (TAR).7,8 An interaction between Tat and cyclin T, a novel C-type cyclin, enhances the affinity and specificity of the Tat-TAR interaction.8 Tat can also interact with a region of the HIV-1 promoter that binds AP-1/NF-κB.9 Cotransfection studies have shown that the Tat gene product can activate a number of cellular genes including interleukin-6 (IL-6), which is known to stimulate lymphoid development.10-12
Transgenic approaches have been used to address the function of Tat. Transgenic mice carrying both the BK virus early region and the HIV-1 Tat gene develop a variety of neoplasms, including B-cell lymphoma,13 although interpretation of these findings is complicated by the coexpression of BK T-antigen together with Tat. Expression of Tat under the control of the HIV-1 long terminal repeat (LTR) causes skin lesions that resemble Kaposi’s sarcoma.14 Lymphoid hyperplasia results from the expression of Tat directed by the cellular proteolipoprotein promoter.15 Intriguingly, targeted inactivation of the IL-6 gene reduces susceptibility to B-lineage neoplasms (plasmacytomas),16 consistent with the view that IL-6 may function in the Tat pathway or parallel with Tat to modulate lymphoid development.
Findings that Tat is sufficient to cause at least some of the features of AIDS, and that it may do so by affecting cytokine gene expression raise the possibility that expression of Tat in lymphoid cells may be a precipitating event in the development of nonHodgkin’s lymphoma. In an effort to develop an animal model that will provide a direct approach to this issue, we created transgenic mice in which HIV-1 Tat is expressed under the control of a chicken β-actin promoter. We show that Tat is expressed in lymphoid cells of such β-actin–Tat mice. We show that β-actin–Tat mice exhibit B-cell hyperplasia and clonal rearrangement of Ig heavy chain genes, consistent with B-cell lymphoma. Finally, we show that IL-6 and IL-10 mRNA levels are elevated in spleens of β-actin–Tat transgenic mice, suggesting that a Tat effect on cytokine gene expression may contribute to lymphomagenesis.
MATERIALS AND METHODS
Generation of β-actin/Tat-transgenic mice.
A 0.34-kb SalI-SalI chicken β-actin promoter fragment from the plasmid PUC β-actin17 was first subcloned in pSP73 (Promega, Madison, WI) vector. The vector was previously modified with two NotI sites by inserting linkers at the PvuII and EcoRV sites. A 300-bp BglII-SalI fragment of Tat cDNA from pSVTat18 was subcloned into theSmaI site of the pSP73 β-actin plasmid. The 3.8-kb pSVTat plasmid was a kind gift from Dr B.M. Peterlin (UCSF, San Francisco, CA). A 2.5-kb fragment containing SV40 intron and poly A addition site was excised from pKSV10 vector (Pharmacia, Piscataway, NJ) and inserted downstream of the Tat cDNA to make the final plasmid. To generate transgenic mice, a 3.14-kb NotI-NotI fragment containing the full transcriptional unit was purified by CsCl density gradient and microinjected into fertilized eggs from superovulated B6CBA F1 females (Jackson Laboratories, Bar Harbor, ME) previously mated to C57BL/6 males (Jackson Laboratories) as described.19 Zygotes were implanted into B6CBA F1 foster females. Founder transgenic animals were crossed with C57BL/6 mice to generate stable transgenic lines. Additional breedings were performed in each line to generate mice homozygous for the Tat transgene.
Screening of transgenic mice.
The β-actin–Tat transgene was detected in the founders and their offspring by Southern blot.20 Ten micrograms of mouse genomic tail DNA was digested overnight at 37°C with KpnI (which cuts once in the transgene construct), electrophoresed in a 0.7% agarose gel, and transferred to a Hybond-N nylon membrane (Amersham, Arlington Heights, IL) overnight. Hybridization was performed in a solution containing 6X SSC (1X SSC: 150 mmol/L NaCl, 15 mmol/L Na-Citrate), 5× Denhardt’s solution (1 mg/mL polyvinylpyrrolidone, 1 mg/mL bovine serum albumin and 1 mg/mL Ficoll), 0.5% sodium dodecyl sulfate (SDS), 100 μg/mL salmon sperm DNA and32P-labeled Tat cDNA at 68°C overnight. The filters were washed once in 2× SSC/0.1% SDS at room temperature for 15 minutes, once in 0.1× SSC and 0.1% SDS at 68°C for 30 minutes, and exposed to Kodak X-OMAT films (Eastman-Kodak, Rochester, NY).
Isolation of subsets of B cells and T cells.
Spleens from normal and transgenic mice were harvested for isolation of B cells, monocytes, CD4 and CD8 T cells by flow cytometry. One million cells were labeled with either fluorescein isothiocyanate (FITC)-conjugated antibody to CD4 (Pharmingen, San Diego, CA) or phycoerythrin-conjugated antibody to CD8 (Pharmingen) followed by labeling with biotinylated secondary antibody anti-CD25 (Pharmingen) and phycoerythrin-FITC conjugated streptavidin (R & D Systems, Minneapolis, MN).
Northern analysis of Tat mRNA.
Total RNA was extracted from different tissues by the acid guanidinium/phenol-chloroform method.21 Twenty micrograms of total RNA of each sample was electrophoresed on a 1% agarose-formaldehyde gel and blotted onto a Hybond-N nylon membrane (Amersham). Hybridization was performed with Quikhyb rapid hybridization solution (Stratagene, La Jolla, CA) containing 100 μg/mL of salmon sperm DNA and 106 cpm/mL of32P-labeled 340-bp Tat cDNA fragment at 65°C for 2 hours. Membranes were washed twice for 15 minutes at room temperature with 2× SSC/0.1% SDS and once for 30 minutes at 60°C with 0.1× SSC/0.1% SDS. Signals were detected with Kodak X-OMAT film. The mouse GAPDH and β-actin cDNA were used as probes to control the amount of RNA present in the gel.
Histological analysis.
Transgenic and control animals were anesthetized and sacrificed and spleens were perfused with phosphate buffered saline. Spleens were later fixed in formalin or B5 fixative and embedded in paraffin. Sections (5 μm) were stained with hematoxylin and eosin.
Immunocytochemistry.
A mouse monoclonal anti-Tat antibody was purchased from ABL (American Bio-technologies Inc, Cambridge, MA). The processing of the tissue section and staining with the anti-Tat antibody were performed with a kit obtained from Zymed Laboratories (San Francisco, CA) according to the supplier’s instructions. Antigen retrieval was performed with methods previously described.22
Ig gene heavy chain rearrangement study.
Spleen DNA was extracted from control and transgenic mice.21 The IgJ heavy chain probe and T-cell receptor (TCR) β probe were obtained from the plasmids p2-123and p86T5,24 which were kind gifts from Dr Huang Fan (University of California, Irvine, CA). Genomic DNA was digested with EcoRI for JH and with HpaI for analysis of TCR-β. The hybridization and washing conditions for these two probes were identical to those described in Screening of Transgenic Mice.
Cytokine mRNA analysis by reverse transcription-polymerase chain reaction (RT-PCR).
A range of dilutions of total RNA (1 to 5 μg) from splenocytes of normal and transgenic mice was reverse transcribed with the cDNA cycle kit (Invitrogen, Carlsbad, CA). An equal amount of cDNA product from the reaction mixes was amplified with forward and reverse primers for IL-6, IL-10, and GAPDH mRNAs. PCR primers used for the different cDNAs were as follows: IL-6 forward, 5′ TTC CAT CCA GTT GCC TTC TTG G-3′; IL-6 reverse, 5′ CTT CAT GTA CTC CAG GTA G-3′; IL-10 forward, 5′ GGA CAA CAT ACT GCT AAC CGA CTC 3′; IL-10 reverse, 5′ AAA ATC ACT CTT CAC CTG CTC CAC TT3′(24); GAPDH forward, GAA TCT ACT GGC GTC TTC ACC 3′; and GAPDH reverse, 5′ GTC ATG AGC CCT TCC ACG ATG C 3′. PCR programs were 2 minutes at 94°C followed by 1 minute at 94°C, 1.5 minutes at 50°C (IL-6), 55°C (IL-10), and 56°C (GAPDH), and 1 minute at 72°C for 21 to 30 cycles with a final extension of the PCR products for 10 minutes at 72°C.
RESULTS
Creation of HIV Tat transgenic mouse lines.
As a first approach to Tat function in a mouse model, we sought to express Tat at moderate to high levels in a variety of tissue types, including the components of the lymphoid system. We used the chicken β-actin promoter, which seemed likely to have the requisite expression properties.17 This promoter was fused with a full-length Tat cDNA sequence and a SV40 3′ UTR and polyadenylation sequence (Fig 1). The transgene construct was used to create three transgenic founder mice, which were bred into a C57Bl/6 background to generate stable lines. Southern blot analysis of DNA of each of these lines revealed multiple copies of the Tat transgene integrated at a single site and oriented in a head to tail tandem array (data not shown). The lines were designated 21, 24, and 29.
Schematic diagram showing the structure of the β-actin–Tat transgene. The Tat cDNA was placed under the transcriptional control of the chicken β-actin promoter. Transcriptional splicing and termination functions were provided by an SV40-derived 3′ UTR and polyadenylation signal. (▩), Chicken β-actin promoter; (▩), Tat cDNA.
Schematic diagram showing the structure of the β-actin–Tat transgene. The Tat cDNA was placed under the transcriptional control of the chicken β-actin promoter. Transcriptional splicing and termination functions were provided by an SV40-derived 3′ UTR and polyadenylation signal. (▩), Chicken β-actin promoter; (▩), Tat cDNA.
The Tat transgene is expressed in lymphoid tissues.
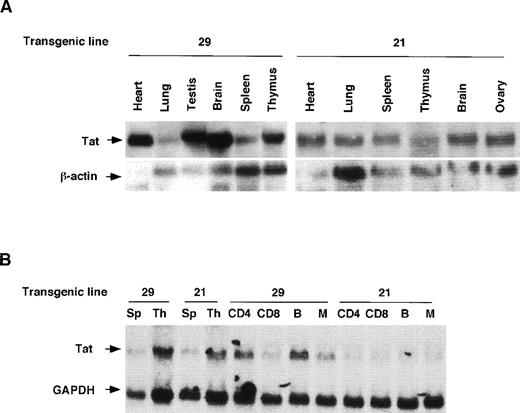
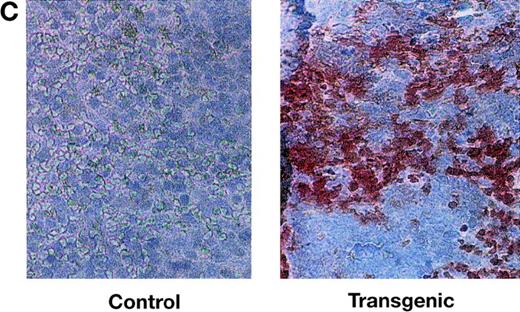
We examined Tat expression in lines 21 and 29 by Northern blot analysis. Tat transcripts of the expected size were evident in several tissues, including spleen and thymus (Fig2A). Immunostaining of transgenic spleen tissue with an anti-Tat antibody showed widespread expression of the Tat protein (Fig 2C). To identify more specifically the lymphoid cell types in which the Tat transgene was expressed, we fractionated lymphoid cells into T, B, and monocyte compartments. Northern analysis of RNA derived from sorted cells showed that T cells, B cells, and monocytes contained Tat transcripts in readily detectable amounts (Fig2B).
Expression of HIV Tat mRNA and protein in tissues of transgenic mice. (A) Northern blot analysis Tat mRNA in two independent transgenic lines, 21 and 29. Whole-cell RNA was extracted from spleen, thymus, and other tissues. The RNA samples (20 μg per lane) were size-separated, blotted onto a membrane, and incubated with radiolabeled Tat cDNA probe and, subsequently, with a β-actin cDNA probe. The blot was washed and exposed to x-ray film. (B) Expression of Tat mRNA in lymphoid cells. Lymphocytes were sorted into T-cell and B-cell components, from which total RNA was prepared, size separated, and blotted. A mouse GAPDH cDNA probe was used as an internal control. Sp, spleen; Th, thymus; CD4, CD4-positive T cell; CD8, CD8-positive T cell; B, B cell; M, monocyte. (C) Immunodetection of Tat in histologic sections of transgenic spleens. Spleens of Tat transgenic animals and littermate controls were processed for histology, sectioned, and the sections incubated with an anti-Tat monoclonal antibody. The specificity of this antibody for Tat was documented previously.33 The secondary antibody was a horseradish peroxidase-streptavidin conjugate (×25).
Expression of HIV Tat mRNA and protein in tissues of transgenic mice. (A) Northern blot analysis Tat mRNA in two independent transgenic lines, 21 and 29. Whole-cell RNA was extracted from spleen, thymus, and other tissues. The RNA samples (20 μg per lane) were size-separated, blotted onto a membrane, and incubated with radiolabeled Tat cDNA probe and, subsequently, with a β-actin cDNA probe. The blot was washed and exposed to x-ray film. (B) Expression of Tat mRNA in lymphoid cells. Lymphocytes were sorted into T-cell and B-cell components, from which total RNA was prepared, size separated, and blotted. A mouse GAPDH cDNA probe was used as an internal control. Sp, spleen; Th, thymus; CD4, CD4-positive T cell; CD8, CD8-positive T cell; B, B cell; M, monocyte. (C) Immunodetection of Tat in histologic sections of transgenic spleens. Spleens of Tat transgenic animals and littermate controls were processed for histology, sectioned, and the sections incubated with an anti-Tat monoclonal antibody. The specificity of this antibody for Tat was documented previously.33 The secondary antibody was a horseradish peroxidase-streptavidin conjugate (×25).
The Tat transgene causes lymphoproliferative anomalies, including B-cell lymphoma.
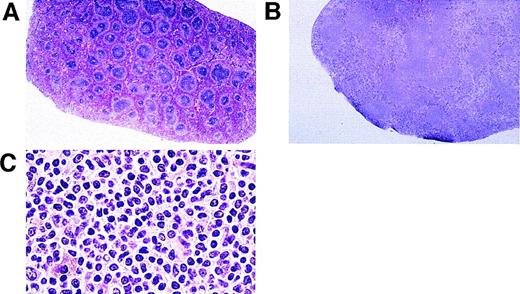
We examined the brain, heart, liver, lung, spleen, and testis of Tat transgenic and control mice. All tissues appeared normal except the spleen. Tat transgenic mice (20 of 80 mice, representing three independent lines) developed enlarged spleens between 12 and 15 months of age (Table 1). These spleens weighed an average of 2.5 times the weight of a normal spleen. Control mice, which were either littermates of Tat transgenics or age-matched controls identical to the transgenics in strain background, did not have enlarged spleens.19 Histological examination of spleens of HIV–Tat-transgenic mice showed several histopathologic differences relative to controls. There was an increase in the size of white-pulp nodules as well as fusion of adjacent lymphoid nodules in a substantial percentage of mice (Fig3A and B). Higher magnification of the nodules (Fig 3C) showed malignant follicular center cell lymphomas composed predominantly of irregularly shaped small lymphoid cells with a few larger follicular center cells. This contrasted with the spleen histology reported by Vellutini et al15 in transgenic mice bearing the Tat transgene under the control of proteolipoprotein promoter. In these mice, the red pulp showed extramedullary hematopoiesis with varying degrees of hyperplasia of erythroid, myeloid, and megakaryocytic precursors.
Spleen Weight in Tat Transgenic and Control Mice
| Line . | No. of Mice Analyzed . | Mean Age (mo) . | Age Range (mo) . | No. of Mice With Enlarged Spleen . | Mean Spleen Weight (g ± SD) . |
|---|---|---|---|---|---|
| 29 | 50 | 13 | 12-15 | 14 | 0.45 ± 0.13 |
| 21 | 20 | 13 | 12-15 | 4 | 0.42 ± 0.10 |
| 24 | 10 | 13 | 12-15 | 2 | 0.46 ± 0.14 |
| Control | 19 | 13 | 12-15 | 0 | 0.16 ± 0.05 |
| Line . | No. of Mice Analyzed . | Mean Age (mo) . | Age Range (mo) . | No. of Mice With Enlarged Spleen . | Mean Spleen Weight (g ± SD) . |
|---|---|---|---|---|---|
| 29 | 50 | 13 | 12-15 | 14 | 0.45 ± 0.13 |
| 21 | 20 | 13 | 12-15 | 4 | 0.42 ± 0.10 |
| 24 | 10 | 13 | 12-15 | 2 | 0.46 ± 0.14 |
| Control | 19 | 13 | 12-15 | 0 | 0.16 ± 0.05 |
Histological analysis of spleens of Tat-transgenic and control mice. Tat-transgenic mice (lines 21 and 29) and littermate control mice were killed at 12 to 15 months of age. Spleens were removed and processed for histology. Shown are the representative hematoxylin/eosin stains of (A) a control spleen (10×), (B) a Tat-transgenic spleen (10×), (C) a Tat-transgenic spleen at high magnification (25×).
Histological analysis of spleens of Tat-transgenic and control mice. Tat-transgenic mice (lines 21 and 29) and littermate control mice were killed at 12 to 15 months of age. Spleens were removed and processed for histology. Shown are the representative hematoxylin/eosin stains of (A) a control spleen (10×), (B) a Tat-transgenic spleen (10×), (C) a Tat-transgenic spleen at high magnification (25×).
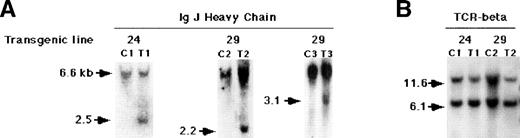
To confirm the involvement of B cells and to determine whether the B-cell expansion was clonal, we tested for the rearrangement of Ig heavy chain genes in the affected spleens of β-actin–Tat transgenic mice. Southern blot analysis with a JH heavy-chain probe showed that such rearrangements did occur in a significant fraction of Tat transgenic mice. Several examples are shown in Fig4A. The data are summarized in Table2. In addition to the germ line DNA fragment of 6.6 kb, fragments of sizes ranging from 2.2 to 3.1 kb were evident, indicative of a heavy chain rearrangement (Fig 4A). The TCR-β gene was not rearranged, ruling out a substantial clonal expansion of T cells (Fig 4B). JH rearrangements were detected in 8 of 40 total animals representing three independent transgenic lines. No such rearrangements were observed in control animals (Table 2). Because B-cell expansion was observed in three independent transgenic lines, it could not have been a consequence of an insertional mutagenesis event. Thus, our data show that overexpression of the HIV-Tat gene is sufficient to cause B-cell lymphoma.
Southern blot showing Ig heavy-chain rearrangement in splenic DNA of Tat-transgenic mice. DNA was extracted from spleens of two independent lines of Tat-transgenic mice, 24 and 29. The DNA was digested with EcoRI for IgJ heavy-chain analysis orHpaI for TCR-β analysis, blotted, and hybridized with an (A) IgJ heavy-chain probe or (B) TCR-β probe. C, control mouse; T, Tat-transgenic mouse. C1-C3 and T1-T3 refer to individual control and transgenic mice. Transgenic lines from which the mice were derived are indicated above the blots. The sizes of the bands in kilobase are indicated on the left. The 2.2-kb, 2.5-kb, and 3.1-kb bands in the Tat-transgenic samples indicate a JH rearrangement; there was no apparent rearrangement of the TCR-β gene.
Southern blot showing Ig heavy-chain rearrangement in splenic DNA of Tat-transgenic mice. DNA was extracted from spleens of two independent lines of Tat-transgenic mice, 24 and 29. The DNA was digested with EcoRI for IgJ heavy-chain analysis orHpaI for TCR-β analysis, blotted, and hybridized with an (A) IgJ heavy-chain probe or (B) TCR-β probe. C, control mouse; T, Tat-transgenic mouse. C1-C3 and T1-T3 refer to individual control and transgenic mice. Transgenic lines from which the mice were derived are indicated above the blots. The sizes of the bands in kilobase are indicated on the left. The 2.2-kb, 2.5-kb, and 3.1-kb bands in the Tat-transgenic samples indicate a JH rearrangement; there was no apparent rearrangement of the TCR-β gene.
Incidence of Heavy-Chain Rearrangement in Tat-Transgenic Mice
| . | Controls . | Transgenic Lines (total) . |
|---|---|---|
| N (number) | 39 | 40 |
| Age (mo) | 14 | 14 |
| Age (mo) (median range) | 9 | 9 |
| JHrearrangement* | 0 | 8 (20%)† P < .005 |
| . | Controls . | Transgenic Lines (total) . |
|---|---|---|
| N (number) | 39 | 40 |
| Age (mo) | 14 | 14 |
| Age (mo) (median range) | 9 | 9 |
| JHrearrangement* | 0 | 8 (20%)† P < .005 |
JH rearrangement was assessed by Southern blotting as described in Fig 2.
The heavy-chain rearrangement was observed in all three transgenic lines.
Levels of IL-6 and IL-10 transcripts are elevated in spleens of Tat-transgenic mice.
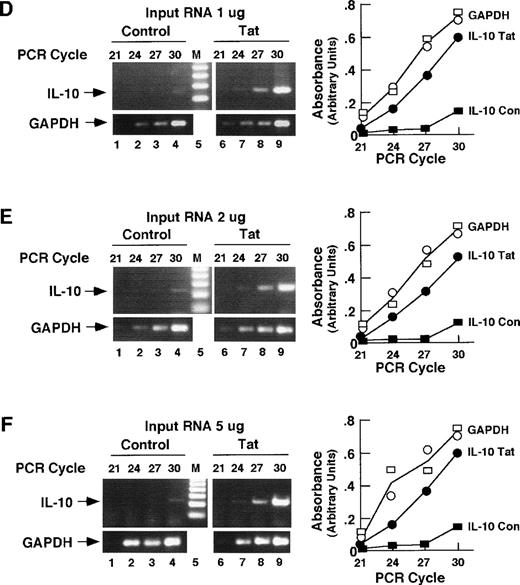
A number of studies have shown that Tat regulates the IL-6 and IL-10 genes.11,25,26 Transcripts of these genes are elevated in lymphoid tissues of HIV-1–infected patients,16,27 and cotransfection assays have shown that Tat can transactivate these genes in cultured lymphoid cells.10 As an initial test of the hypothesis that the Tat effect on B cells in Tat-transgenic mice is mediated through IL-6, IL-10, or both, we performed a semiquantitative RT-PCR analysis on RNA from transgenic spleens. Primers specific for IL-6 or IL-10 were used against reverse-transcribed RNA from transgenic and control spleens. Both the amount of input RNA and the number of PCR cycles were varied to identify conditions under which the RT-PCR signal was quantitatively related to the amount of input RNA (Fig5). PCR amplification of GAPDH sequences provided an internal control. As can be seen in Figs 5A and B, the amounts of GAPDH PCR products were similar in RNA samples from control versus Tat-transgenic mice. The amount of IL-6 PCR product was substantially elevated in samples derived from Tat-transgenic mice compared with controls. This elevation was evident at several different PCR cycles and at three different levels of input RNA: 1, 2, and 5 μg. Analysis of IL-10 mRNA yielded similar results. Levels of IL-10 PCR product were substantially higher in samples from Tat-transgenic mice compared with nontransgenic controls. Comparison of the relative amounts of PCR products in Tat transgenic versus control samples showed increases of approximately threefold for IL-6 and fourfold for IL-10. We conclude that IL-6 and IL-10 mRNA levels are substantially higher in splenic tissues of Tat-transgenic mice compared with littermate or age-matched controls, which is consistent with the view that the HIV-Tat gene may affect B cells through the cytokines IL-6 and IL-10.
RT-PCR detection of IL-6 and IL-10 mRNA in spleens of Tat-transgenic mice. Whole-cell RNA from the spleens of age-matched control and β-actin–Tat-transgenic mice was reverse transcribed and subjected to PCR using primers specific for IL-6 mRNA (A through C), IL-10 mRNA (D through F), and GAPDH mRNA (internal control; A through F). RT-PCR was performed with 1 μg of input RNA (A and D), 2 μg (B and E), and 5 μg (C and F). PCR products were size-separated and visualized by staining with ethidium bromide. Stained gels were photographed and the PCR products quantitated by scanning with a laser densitometer. M, 100-bp ladder. The areas of the IL-6 (A through C), IL-10 (D through F), and GAPDH (control) peaks are plotted against PCR cycle number. IL-6 Tat, measurement of IL-6 mRNA in Tat-transgenic mice. IL-6 Con, measurement of IL-6 mRNA in control mice. IL-10 Tat, measurement of IL-10 mRNA in Tat-transgenic mice. IL-10 Con, measurement of IL-10 mRNA in control mice. Open circles, GAPDH in Tat-transgenic mice; Open squares, GAPDH in control mice; The analyses of IL-6, IL-10, and GAPDH were performed at the same time on the same splenic RNA samples. The analyses shown in (A) and (B) were performed on Tat-transgenic line 29. Parallel experiments conducted on line 21 yielded similar results.
RT-PCR detection of IL-6 and IL-10 mRNA in spleens of Tat-transgenic mice. Whole-cell RNA from the spleens of age-matched control and β-actin–Tat-transgenic mice was reverse transcribed and subjected to PCR using primers specific for IL-6 mRNA (A through C), IL-10 mRNA (D through F), and GAPDH mRNA (internal control; A through F). RT-PCR was performed with 1 μg of input RNA (A and D), 2 μg (B and E), and 5 μg (C and F). PCR products were size-separated and visualized by staining with ethidium bromide. Stained gels were photographed and the PCR products quantitated by scanning with a laser densitometer. M, 100-bp ladder. The areas of the IL-6 (A through C), IL-10 (D through F), and GAPDH (control) peaks are plotted against PCR cycle number. IL-6 Tat, measurement of IL-6 mRNA in Tat-transgenic mice. IL-6 Con, measurement of IL-6 mRNA in control mice. IL-10 Tat, measurement of IL-10 mRNA in Tat-transgenic mice. IL-10 Con, measurement of IL-10 mRNA in control mice. Open circles, GAPDH in Tat-transgenic mice; Open squares, GAPDH in control mice; The analyses of IL-6, IL-10, and GAPDH were performed at the same time on the same splenic RNA samples. The analyses shown in (A) and (B) were performed on Tat-transgenic line 29. Parallel experiments conducted on line 21 yielded similar results.
DISCUSSION
We provide evidence that expression of the HIV-Tat gene in lymphoid tissues is associated with the development of B-cell lymphoma in transgenic mice. With a chicken β-actin promoter to drive a Tat transgene, we show that Tat mRNA is expressed in B cells, T cells, and monocytes. We document in these β-actin–Tat-transgenic mice several morphologic and molecular features characteristic of follicular center cell-derived B-cell lymphoma. These mice had grossly enlarged spleens, which on histologic analysis exhibited an increase in the size and number of white-pulp nodules. These nodules contained monomorphic populations of small lymphoid cells primarily with irregular nuclei. Southern blot analysis performed on DNA from transgenic spleens revealed rearranged Ig heavy chain genes in approximately 20% of transgenic mice, but did not show evidence of rearrangements in TCR gene. Therefore, our data show a clonal expansion of B cells and not T cells. Such rearrangements were not observed in age-matched control mice and were, thus, specifically associated with the Tat transgene.
Splenomegaly and other features of lymphoma appeared in β-actin–Tat-transgenic mice between 12 and 15 months after birth. Because control mice did not develop lymphomas within this same time frame, we can conclude that the Tat transgene plays a causative role in lymphomagenesis. The Tat-transgenic mice are in a C57Bl/6 background, which has a moderate incidence of lymphoma in older animals (39% of animals aged 30 months or more for both males and females).28 Thus, C57Bl/6 may provide a permissive genetic background for the development of Tat-induced lymphomas.
Transgenic approaches have implicated Tat in the development of lymphoproliferative anomalies. Corallini et al13reported B-cell lymphomas in transgenic mice expressing both the HIV-1–Tat gene and the BKV T-antigen under the control of a BKV promoter. However, because the BKV T-antigen gene was coexpressed with Tat, it was not possible to determine whether Tat was sufficient to cause B-cell lymphoma. The use of the cellular lipoprotein promoter to drive Tat expression produced mice with lymphoid hyperplasia.15 In addition to these effects on lymphoid tissues, expression of Tat under the control of the HIV LTR resulted in skin lesions that resembled Kaposi’s sarcoma.14
Our data extend these observations by showing that Tat is sufficient to cause B-cell lymphoma when expressed under the control of the chicken β-actin promoter. It remains unclear why B-cell lymphomas were not observed when Tat expression was driven by the cellular lipoprotein promoter or the HIV-1 LTR. The relative strength of these promoters in lymphoid tissues is one likely possibility, although direct quantitative comparisons are lacking. Wei et al8 have shown that cyclin T has a central role in the Tat:TAR interaction and that overexpression of human cyclin T in NIH 3T3 and CHO cells significantly enhances Tat activity in vivo. Species differences in the activity of cyclin T may, thus, explain the inability of HIV to replicate in murine cells. It is unclear whether cyclin T plays any part in the lymphoid phenotypes exhibited by Tat-transgenic mice. However, it is attractive to speculate that the introduction of a human cyclin T into the β-actin–Tat-transgenic mice might enhance the frequency of lymphomas and other Tat-induced phenotypes.
Our demonstration by semiquantitative RT-PCR that mRNAs encoding the lymphokines IL-6 and IL-10 are upregulated in spleens of β-actin–Tat-transgenic mice is consistent with the results of cotransfection experiments showing that Tat can activate both promoters.11,12 These findings, both in transgenic mice and in cultured cells, together with findings that IL-6 and IL-10 can stimulate the proliferation of B-lineage cells,4 suggest that lymphomagenesis in Tat-transgenic mice may be a direct result of Tat-mediated enhancement of IL-6 and IL-10 transcription. Under this model, elevated levels of IL-6 and IL-10 would lead first to enhanced proliferation of B cells and, ultimately, after a secondary genetic change, to neoplastic growth. The availability of mice bearing null mutations in IL-616 29 or IL-10 will allow a direct test of the roles of IL-6 and IL-10 in lymphomagenesis in Tat-transgenic mice.
To what extent do the Tat-transgenic mice provide a model of AIDS-related lymphoma? Because hematopoiesis is sufficiently different in mice and in humans, making histologic comparisons of types of lymphomas may be problematic.30,31The most common types of B-cell lymphoma in patients with AIDS are either aggressive Burkitt’s or immunoblastic cell types that arise from B cells. This contrasts with the B-cell lymphoma of the Tat-transgenic mice, which was composed predominantly of irregularly shaped small lymphoid cells arising from follicular center cells. On the other hand, the molecular cascade leading to lymphomagenesis may have common features: the roles of IL-6 and IL-10 as modulators of B-cell proliferation and differentiation are likely to be conserved between mice and humans.26,27,32 Moreover, IL-6 and IL-10 are elevated in lymphoid cells of patients with AIDS-related lymphoma16 27 similar to our findings in the Tat-transgenic mice. There is reason to believe, therefore, that the effect of Tat on B cells operates by a similar mechanism in Tat-transgenic mice and humans. Our Tat transgenic mice may thus serve as a model with which to dissect the molecular pathways of Tat function. In the long term, these mice may also provide a means to investigate therapeutic approaches to HIV infection based on interference with Tat activity.
ACKNOWLEDGMENT
We thank Anna Perez for pronuclear injections and the late C. Nugyen Huu for initiating the project.
Supported by grants from the National Institute of Health (NIH) to R.M. (HD 22416) and D.R.H. (PO1 NS 26991) and a University of Southern California/Norris translational research grant to R.M.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Robert Maxson, PhD, Department of Biochemistry and Molecular Biology, and USC/Norris Hospital and Research Institute, 1441 Eastlake Ave, Los Angeles, CA 90089-9176; e-mail: maxson@zygote.hsc.usc.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal