Abstract
The E2A-HLF fusion gene, generated by t(17;19)(q22;p13) in acute lymphoblastic leukemia (ALL), encodes a chimeric transcription factor in which the trans-activating domains of E2A are fused to the DNA-binding and dimerization domains of hepatic leukemic factor (HLF). To investigate its biological role, we generated transgenic mice expressing E2A-HLF using Ig enhancer and promoter, which direct transgene expression in cells committed to the lymphoid lineage. The transgenic mice exhibited abnormal development in the thymus and spleen and were susceptible to infection. The thymus contained small numbers of thymocytes, and TUNEL staining showed that higher population of thymocytes were undergoing apoptosis. The spleen exhibited a marked reduction in splenic lymphocytes and the flow cytometric analyses and the in vitro colony formation assays showed that the B-cell maturation was blocked at a very early developmental stage. These findings indicated that the expression ofE2A-HLF induced T-cell apoptosis and B-cell maturation arrest in vivo and that the susceptibility of the transgenic mice to infection was due to immunodeficiency. Moreover, several transgenic mice developed acute leukemia, classified as T-ALL based on the surface marker analysis and DNA rearrangements, suggesting that an additional event is required for malignant transformation of lymphoid cells expressing E2A-HLF. Our findings provide insight into the biological function of E2A-HLF in lymphoid development and also its role in leukemogenesis.
CHROMOSOMAL TRANSLOCATIONS are recurrent features of human hematopoietic malignancies.1 Consistently observed chromosomal abnormalities in specific tumors indicate that the underlying genetic events are essentially implicated in the causation of the diseases. In acute leukemias, several characteristic chromosomal translocations have been noted, and studies on the chromosomal breakpoints have identified candidates of genes whose dysregulated or aberrant expression would be responsible for the tumor development.1
Transcription factors are frequent target of chromosomal translocations observed in acute lymphoblastic leukemia (ALL).2 Among them, E2A gene, encoding a basic helix-loop-helix (bHLH) transcription factor on chromosome 19, has been known to be involved in two chromosomal translocations, t(1;19)(q23;p13)3,4 and t(17;19)(p22;q13)5,6 that are observed in human B-lineage leukemias. As a result of t(1;19)(q23;p13), the C-terminal region ofE2A gene, including the bHLH DNA-binding and dimerization domains, is replaced with the DNA-binding domain of PBX1homeobox gene on chromosome 1.3,4 On the other hand, following the t(17;19)(p22;q13), the same region of the E2Agene is fused to the DNA-binding and dimerization domains of hepatic leukemic factor (HLF) gene belonging to the basic region/leucine zipper (bZIP) family on chromosome 17.5 6These events create novel fusion gene products, E2A-PBX1 andE2A-HLF, respectively, and the expression of these chimeric transcription factors with altered structural and functional features would play a substantial role in the leukemogenic process(es).
One of the E2A-involving chimeric molecules, E2A-HLF,was cloned in t(17;19)(p22;q13)-bearing childhood ALL with pro-B immuophenotype.5,6 Structurally, E2A-HLF is composed of the trans-activation domains of E2A and the DNA-binding and dimerization domains of HLF.5,6E2A was originally identified as an Ig κ chain enhancer-binding protein7 and has been shown to be expressed in many different types of cells.8 On the other hand, HLF is normally expressed in the liver, kidney, and lung, but not in lymphoid cells.5,6 Although both E2A-HLFand HLF have been shown to bind to the same consensus sequence, 5′-GTTACGTAAT-3′, a core dyad-symmetric motif characteristic of bZIP family DNA-binding proteins,9 studies have shown thatE2A-HLF and HLF possess different transcriptional and transforming properties.10E2A-HLF has an ability to transactivate a reporter gene in various types of cells, such as HepG2 hepatocellular carcinoma cells, NIH 3T3 fibroblasts, and 293 fetal kidney cells, whereas transactivating ability of wild-typeHLF was detected only in 293 cells but was silent in HepG2 and NIH 3T3 cells.10 In addition, expression of E2A-HLFin NIH 3T3 cells induced anchorage-independent cell growth in soft agar and rendered these cells tumorigenic in nude mice, while overexpression of HLF has not been reported to be oncogenic.10 A subsequent study showed that E2A-HLF has an anti-apoptotic effect in hematopoietic cell lines.11 Introduction of a dominant-negative form of E2A-HLF, which lacks thetrans-activation domains of E2A and contains a defective HLF basic domain, in E2A-HLF–expressing human leukemic cells rapidly induced apoptosis.11 In addition, the expression of E2A-HLF in mouse factor-dependent Ba/F3 cells blocked factor-depleted– and radiation–induced apoptosis.11 These findings suggest that E2A-HLFwould activate genes that are normally quiescent in lymphoid development and that the aberrant gene expression would contribute to the development of ALL.12
Although these studies have provided us with insights in the biological properties of E2A-HLF, the in vivo role of E2A-HLF in lymphoid development and leukemogenesis has not been fully understood. To address this issue, we generated and analyzed mice expressingE2A-HLF in cells committed to the lymphoid lineage.
MATERIALS AND METHODS
Construction of the transgene and generation of the transgenic mice.
The E2A-HLF cDNA5 was subcloned into an expression vector which contains Ig enhancer and promoter as regulatory elements. A fragment encompassing the Ig enhancer/promoter, the E2A-HLFcDNA, and the SV40 early splicing and polyadenylation signals was microinjected into pronuclei of eggs from C57BL/6XDBA/2 F2 mice as described earlier.13 The transgenic mice were identified by hybridizing tail DNA with the injected fragment13 and the transgenic progeny were generated by successive mating of the founder mice or transgenic descendants with C57BL/6XDBA/2 F1 mice. The mice were kept under a conventional maintaining condition or in a separated clean isolator.
Pathological examination.
After gross examination, all the tissues were fixed in 10% phosphate-buffered saline (PBS)-buffered formaldehyde, embedded in paraffin, and stained with hematoxylin-eosin (HE). Smears of peripheral blood cells were stained with Wright-Giemsa (WG).
Protein extraction, immunoprecipitation, and Western blotting.
Tissues were homogenized in RIPA lysis buffer (150 mmol/L NaCl, 50 mmol/L Tris-Cl (pH 7.4), 1% Triton X-100, 0.05% sodium dodecyl sulfate, 1% sodium deoxycholate) with 50 U/mL aprotinin. For detectingE2A-HLF protein, 1 mg of protein aliquots were incubated with 1:1,000 diluted anti-E2A antibody5 and immunoprecipitated proteins were probed with 1:1,000 diluted anti-HLF antibody.5 The positive signals were visualized using the ProtoBlot Western AP system (Promega, Madison, WI).
Immunofluorescent staining.
Transgenic spleens were fixed in 10% PBS-buffered formalin and embedded in paraffin. Specimens were mounted on saline-coated slides, dewaxed in xylin, rehydrated through a graded series of alcohols, and treated with 20 μg/mL proteinase K in 10 mmol/L Tris-Cl (pH 7.4) at 37°C for 15 minutes. After washing two times with PBS, specimens were stained with 1:100 diluted fluorescein isothiocyanate (FITC)-conjugated anti-B220 monoclonal antibody (MoAb) (Pharmingen, Osaka, Japan) and 1:50 diluted anti-HLF polyclonal antibody5 coupled with 1:50 diluted Texas-red–conjugated anti-rabbit antibody (Jackson ImmunoResearch Laboratories Inc, West Grove, PA). After washing three times with PBS, specimens were observed under a confocal microscopic system, MRC1024 (BioRad, Hercules, CA).
Detection of apoptotic cells with TdT-mediated dUTP nick end labeling (TUNEL).
Transgenic and nontransgenic thymuses were fixed in 10% PBS-buffered formalin and embedded in paraffin. Specimens mounted on saline-coated slides were dewaxed as described above. After washing two times with PBS, specimens were subjected to TUNEL reaction using in situ cell death detection kit (Boehringer, Mannheim, Germany) according to the manufacturer’s instructions and were observed under a confocal microscopic system, MRC1024 (BioRad).
Flow cytometric analysis.
Cells were stained with FITC- or phycoerythrin-conjugated commercial MoAbs including anti–Thy-1.2, anti-B220, anti-CD4, anti-CD8, and anti-CD43 (Pharmingen) according to the manufacturer’s instructions. The stained cells were washed three times with PBS and analyzed on a FACScan (Becton Dickinson, Sunnyvale, CA).
Colony assays.
Bone marrow cells of transgenic mice and nontransgenic littermates were subjected to the in vitro colony formation assays as described elsewhere.14
RNA extraction and reverse transcription-polymerase chain reaction (RT-PCR).
RESULTS
Expression of the E2A-HLF transgene product in T and B lymphocytes in the transgenic mice.
We planned to generate transgenic mice expressing E2A-HLF in hematopoietic cells committed to the lymphoid lineage. With this aim, Ig enhancer and promoter were chosen as regulatory elements. The schematic model of the transgene is shown in Fig1A. Microinjection resulted in generation of three founder mice containing approximately 10 copies of the transgene in a head-to-tail manner (data not shown) and two of these three founders (named 1-17 and 2-17) transmitted the transgene stably to their progeny.
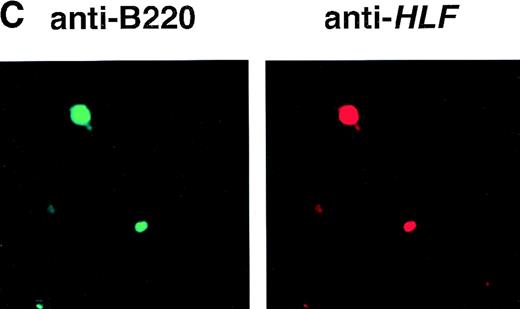
(A) Schematic model of the injection fragment for generating E2A-HLF transgenic mice. The Ig enhancer/promoter, the E2A-HLF cDNA, and the SV40 early splicing and poly(A) signals are shown as white, black, and shaded boxes, respectively. The positions of primers used for RT-PCR (P1-P4) are also indicated. The bar shows 1 kb. (B) Western blot analysis for the expression of theE2A-HLF transgene product in the thymus and spleen of a control mouse (C) and a transgenic mouse from two independent lines (1-17 and 2-17). The positions of E2A-HLF protein and Ig are indicated by arrows, and the positions of the molecular marker are shown on the left. (C) Immunofluorescent staining of the transgenic spleen with anti-B220 and anti-HLF antibodies. The B220-expressing and HLF-expressing cells are visualized by green and red signals, respectively.
(A) Schematic model of the injection fragment for generating E2A-HLF transgenic mice. The Ig enhancer/promoter, the E2A-HLF cDNA, and the SV40 early splicing and poly(A) signals are shown as white, black, and shaded boxes, respectively. The positions of primers used for RT-PCR (P1-P4) are also indicated. The bar shows 1 kb. (B) Western blot analysis for the expression of theE2A-HLF transgene product in the thymus and spleen of a control mouse (C) and a transgenic mouse from two independent lines (1-17 and 2-17). The positions of E2A-HLF protein and Ig are indicated by arrows, and the positions of the molecular marker are shown on the left. (C) Immunofluorescent staining of the transgenic spleen with anti-B220 and anti-HLF antibodies. The B220-expressing and HLF-expressing cells are visualized by green and red signals, respectively.
To examine the expression of the transgene product in the lymphoid tissues, proteins extracted from the thymus and the spleen of a transgenic mouse and a nontransgenic littermate were immunoprecipitated with anti-E2A antibody and immunoprecipitated proteins were blotted with anti-HLF antibody. As shown in Fig 1B, the expression of E2A-HLF protein was detected in the thymus but was undetectable in the spleen in both lines, raising a question of whether B lymphocytes expressed the E2A-HLF transgene product. To address this issue, we performed immunofluorescent staining of the transgenic spleen using FITC-conjugated anti-B220 antibody and anti-HLF antibody coupled with Texas-red–conjugated anti-rabbit antibody. The results showed that cells expressing B220 were positive for HLF (Fig1C). BecauseHLF has not been shown to be expressed in the hematopoietic cells,5HLF+ lymphocytes are considered to express E2A-HLF protein. These results showed that both T and B lymphocytes expressed the E2A-HLF transgene product in the transgenic mice.
E2A-HLF transgenic mice were susceptible to infection.
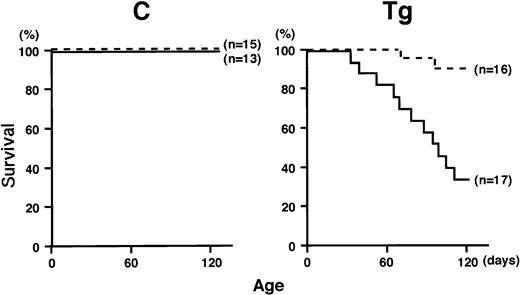
The transgenic mice were initially kept under a conventional maintaining condition. Under this condition, although the transgenic mice grew normally until weaning (4 weeks of age), they gradually exhibited emaciation and almost all the transgenic mice died within several months after birth, in contrast to the fact that no nontransgenic littermates died within the same period. Autopsies of the dead animals showed abscesses, strongly suggesting that the transgenic mice were susceptible to infection. To address this issue, newly weaned transgenic and nontransgenic littermates (4 weeks of age) were divided into two groups and mice in one group were kept under the same conventional condition, whereas mice in the other group were maintained in a separated clean isolator. As a result of about 3 months of observation, we found that the transgenic mice that grew under the conventional condition also showed a high mortality rate (5 of 17 transgenic mice were alive more than 4 months of age), whereas the transgenic mice that grew in a clean isolator exhibited much better surviving rate (14 of 16 transgenic mice are surviving over 4 months of age) (Fig 2, right). During this period, no nontransgenic mice died under either condition (Fig 2, left). To investigate the cause of the death of the transgenic mice, tissues of the dead or moribund mice were pathologically examined. In about half of the mice examined, macroscopic abscesses were found in several major organs such as the liver, spleen, and intestine. Microabscesses were detected in all the cases and were accompanied by tissue necrosis and infiltration of granulocytes. Staining of stamp specimens of the abscesses showed proliferation of bacteria. These results indicated that the transgenic mice were susceptible to infection and that the major cause of early death of the transgenic mice was bacterial infection.
Survival curves of transgenic mice (Tg, right) and nontransgenic controls (C, left) that are kept under a conventional condition (solid lines) or in a clean isolator (dotted lines). The life span is presented by the Kaplan-Meier method.
Survival curves of transgenic mice (Tg, right) and nontransgenic controls (C, left) that are kept under a conventional condition (solid lines) or in a clean isolator (dotted lines). The life span is presented by the Kaplan-Meier method.
Decrease in thymocytes and marked reduction in splenic lymphocytes inE2A-HLF transgenic mice.
To examine whether the expression of E2A-HLF affected the development of the lymphoid tissues, transgenic mice and nontransgenic littermates at 4 weeks of age were autopsied and macroscopically examined. Consistent and remarkable changes were detected in the thymus and in the spleen. The thymuses of the transgenic mice were apparently smaller than those of the nontransgenic littermates. The spleens of the transgenic mice were consistently larger than those of the nontransgenic littermates. The representative macroscopic appearances of a transgenic mouse and a nontransgenic littermate are shown in Fig3.
Macroscopic appearances of a transgenic mouse (Tg) and a nontransgenic control (C) (4 weeks of age). The thymuses and spleens are indicated by (▴) and (▵), respectively. The transgenic mouse exhibits regressed thymus and enlarged spleen.
Macroscopic appearances of a transgenic mouse (Tg) and a nontransgenic control (C) (4 weeks of age). The thymuses and spleens are indicated by (▴) and (▵), respectively. The transgenic mouse exhibits regressed thymus and enlarged spleen.
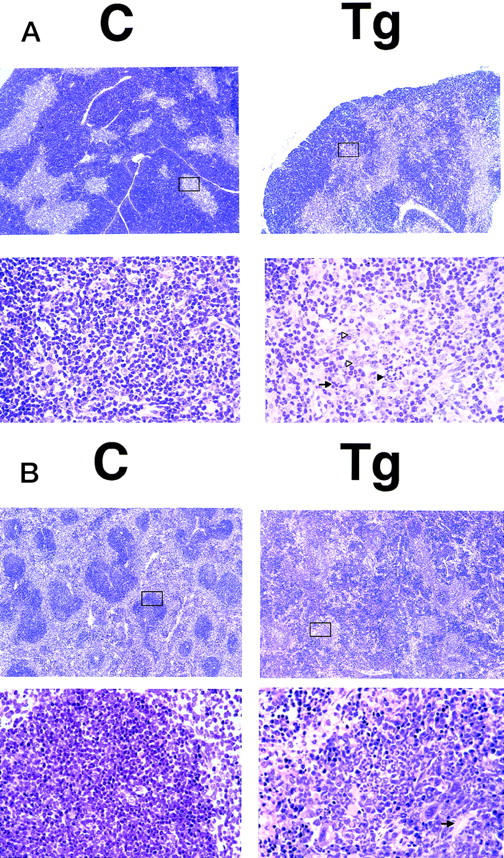
To investigate the histological changes underlying the phenotypic abnormalities, the thymuses and spleens of transgenic mice and nontransgenic littermates at 4 weeks of age were subjected to microscopic examination. Significant changes were observed in the medulla of the thymus and in the white pulps of the spleen. In comparison with the nontransgenic thymus, the transgenic thymus showed enlargement of the medulla, and obscurity and irregularity of the medulla/cortex junction (Fig 4A, top panels). The medulla of the transgenic thymus contained small numbers of lymphoid cells and consisted mainly of epithelial cells and interdigitating cells. Infiltration of neutrophils, multinucleated giant cells, and tingeble-body macrophages were frequently seen (Fig 4A, bottom panels). As for the spleen, the fundamental structure of the spleen (integrity of white and red pulps) was disorganized in the transgenic mice in comparison with the nontransgenic littermates (Fig 4B, top panels). The periarteriolar lymphoid sheaths in the transgenic spleen were irregularly shaped where lymphoid cells were markedly depleted and neutrophils and megakaryocytes infiltrated (Fig 4B, bottom panels). These findings showed that the thymic regression was due to decrease in thymocyte number and that the splenic enlargement was associated with marked depletion of splenic lymphocytes and diffuse proliferation of other hematopoietic cells.
HE-stained sections of the thymus (A) and spleen (B) of a transgenic mouse (Tg) and a nontransgenic control (C) (4 weeks of age). The boxed areas in the top panels are magnified in the bottom panels. (A) Thymus. In the lower magnification (top panels), transgenic thymus shows enlargement of the medulla and obscurity and irregularity of the medulla/cortex junction. In the higher magnification (bottom panels), the medulla of the transgenic thymus contains fewer thymocytes and consists of epithelial and interdigitating cells. Infiltrated neutrophils and multi-nucleated giant cells are indicated by black and white triangles, respectively, and the tingeble-body macrophages are indicated by an arrow. (B) Spleen. In the lower magnification (top panels), transgenic spleen shows marked disorganization in the fundamental structures. In the higher magnification (bottom panels), in a white pulp of the transgenic spleen, lymphocytes are markedly reduced and proliferation of neutrophils and megakaryocytes is observed. The central splenic artery, which is located in the middle of the white pulp, is indicated by an arrow.
HE-stained sections of the thymus (A) and spleen (B) of a transgenic mouse (Tg) and a nontransgenic control (C) (4 weeks of age). The boxed areas in the top panels are magnified in the bottom panels. (A) Thymus. In the lower magnification (top panels), transgenic thymus shows enlargement of the medulla and obscurity and irregularity of the medulla/cortex junction. In the higher magnification (bottom panels), the medulla of the transgenic thymus contains fewer thymocytes and consists of epithelial and interdigitating cells. Infiltrated neutrophils and multi-nucleated giant cells are indicated by black and white triangles, respectively, and the tingeble-body macrophages are indicated by an arrow. (B) Spleen. In the lower magnification (top panels), transgenic spleen shows marked disorganization in the fundamental structures. In the higher magnification (bottom panels), in a white pulp of the transgenic spleen, lymphocytes are markedly reduced and proliferation of neutrophils and megakaryocytes is observed. The central splenic artery, which is located in the middle of the white pulp, is indicated by an arrow.
Increased thymocyte apoptosis in E2A-HLF transgenic mice.
The decrease in thymocyte number is suggestive of either reduction of thymocyte production or alteration in thymocyte survival. The pathological findings that the thymocyte decrease was associated with macrophage infiltration is in agreement with the latter possibility. To confirm this, the thymuses of transgenic mice and nontransgenic littermates were subjected to TUNEL staining, which detects apoptotic cells by in situ labeling using TdT and dUTP.16 As shown in Fig 5 (see page 2785), the number of positively stained cells was significantly increased in the transgenic thymus in comparison with the nontransgenic thymus, showing that the decrease in thymocyte number was caused by increased apoptosis. To investigate whether the thymocyte apoptosis was due to a differentiation block at a specific developmental stage, the transgenic thymocytes were subjected to flow cytometric analysis using anti-CD4 and anti-CD8 antibodies. Repeated experiments showed that the percentage of each CD4/CD8 subset (CD4−/CD8−, CD4+/CD8+, CD4+/CD8−, and CD4−/CD8+) was not significantly altered between the transgenic and nontransgenic thymuses (data not shown). These results indicated that the expression of E2A-HLFinduced thymocyte apoptosis with no obvious interference in thymocyte differentiation.
TUNEL staining of the thymus of a transgenic mouse (Tg) and a nontransgenic control (C) (4 weeks of age). Apoptotic thymocytes are detected as yellow signals and background cells are seen as green signals. The transgenic thymus exhibits increased number of thymocytes undergoing apoptosis.
TUNEL staining of the thymus of a transgenic mouse (Tg) and a nontransgenic control (C) (4 weeks of age). Apoptotic thymocytes are detected as yellow signals and background cells are seen as green signals. The transgenic thymus exhibits increased number of thymocytes undergoing apoptosis.
Maturation block of B-cell progenitors in E2A-HLF transgenic mice.
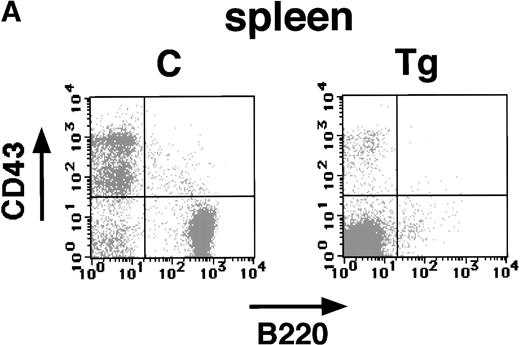
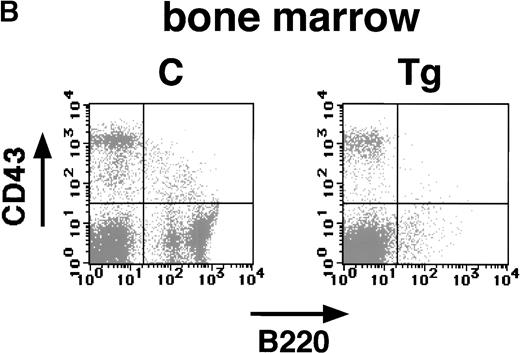
The pathological analysis of the transgenic spleen showed a marked decrease in splenic lymphocytes (Fig 4B). These findings strongly suggest that the expression of E2A-HLF impaired B-cell development as well as induced T-cell apoptosis. To address this possibility and to identify the stages in which the B-cell development was impaired, splenic and bone marrow cells of the transgenic mice and nontransgenic littermates were subjected to flow cytometric analysis with anti-B220 and anti-CD43 antibodies. The B220+CD43+ population contains mainly early B-cell progenitors, whereas B220+ CD43− population consists of pre-B and mature B lymphocytes.17 As shown in Fig6A and B, significant changes were observed in the B220+ CD43+ as well as the B220+ CD43− population both in the spleen and in the bone marrow. The nontransgenic spleen and bone marrow contained approximately 6% to 10% of B220+ CD43+ cells, whereas only 2% to 3% cells were positive for both B220 and CD43 in the transgenic spleen and bone marrow (Fig 6A and B). More remarkable is the reduction in B220+ CD43− late progenitor population. In the transgenic spleen and bone marrow, this population was reduced to 5% to 8%, as compared with 30% to 40% in the nontransgenic spleen and bone marrow (Fig 6A and B). In addition, no B cells expressing high levels of B220 were detected in the transgenic spleen or bone marrow (Fig 6A and B). These results showed that both early and late B-cell progenitors were severely reduced in the transgenic mice.
Flow cytometric analysis of the spleen (A) and bone marrow (B) of a transgenic mouse (Tg) and a nontransgenic control (C) with B220 and anti-CD43 antibodies (4 weeks of age). The transgenic spleen and bone marrow show marked reduction of both B220+ CD43+ and B220+CD43− populations.
Flow cytometric analysis of the spleen (A) and bone marrow (B) of a transgenic mouse (Tg) and a nontransgenic control (C) with B220 and anti-CD43 antibodies (4 weeks of age). The transgenic spleen and bone marrow show marked reduction of both B220+ CD43+ and B220+CD43− populations.
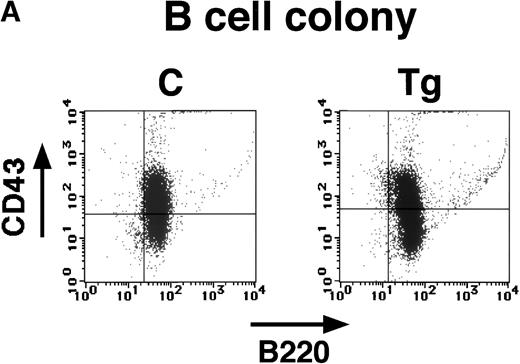
We then investigated whether hematopoietic progenitor cells having a potential to differentiate into B220+ cells might exist in the transgenic bone marrow. The bone marrow cells from transgenic mice and nontransgenic littermates were cultured with interleukin-7 (IL-7) and stem cell factor (SCF), which can recapitulate B-cell differentiation in vitro.14 Interestingly, although the colony numbers varied among experiments, we found that the transgenic bone marrow could develop B-cell colonies as well as the nontransgenic bone marrow (Table 1). To confirm that the colonies generated with IL-7 and SCF were committed to the B-cell lineage, cells from the colonies were subjected to flow cytometric analysis with anti-B220 and anti-CD43 antibodies. As shown in Fig7A, both colonies were positive for B220 and showed the same broad pattern for CD43, demonstrating that they both consisted of early and late B-cell progenitors. To further confirm that the B-cell colonies developed from transgenic bone marrow expressed E2A-HLF transgene product, mRNA extracted from the colonies were subjected to RT-PCR analysis. By use of the primers encompassing the splicing signal (P1-P4, see Fig 1), the mRNA-derived PCR product (160 bp) can be distinguished from any contaminating genome-derived PCR product (230 bp), because 70 bp should be removed by the splicing. As shown in Fig 7B, we detected the 160-bp band in addition to the 230-bp band in the PCR products generated from the transgenic colonies, indicating that the B-cell colonies developed from transgenic bone marrow expressed transgene-derived mRNA. The overall results indicated that although hematopoietic progenitor cells having a potential to differentiate into the B-cell progenitors did exist in the transgenic bone marrow, their commitment to the B-cell lineage was blocked at a very early developmental stage.
In Vitro B-Cell Colony Formation by IL-7 + SCF
| . | +FCS . | −FCS . |
|---|---|---|
| C-1 | 10.5 ± 3.3 | 10.5 ± 4.4 |
| C-2 | 3.8 ± 1.0 | 3.5 ± 1.7 |
| Tg-1 | 25.5 ± 5.5 | 16.8 ± 3.0 |
| Tg-2 | 7.0 ± 1.4 | 5.5 ± 0.6 |
| . | +FCS . | −FCS . |
|---|---|---|
| C-1 | 10.5 ± 3.3 | 10.5 ± 4.4 |
| C-2 | 3.8 ± 1.0 | 3.5 ± 1.7 |
| Tg-1 | 25.5 ± 5.5 | 16.8 ± 3.0 |
| Tg-2 | 7.0 ± 1.4 | 5.5 ± 0.6 |
Colonies were generated with IL-7 + SCF with or without fetal calf serum (FCS).
Abbreviations: C, control; Tg, transgenic.
(A) Flow cytometric analysis of the B-cell colonies generated from bone marrow of a transgenic mouse (Tg) and a nontransgenic control (C) with anti-B220 and anti-CD43 antibodies (4 weeks of age). Colonies established from transgenic mice and nontransgenic controls are similarly positive for B220 and show the same broad pattern for CD43. (B) Expression of transgene-derived mRNA in the transgenic B-cell colonies. The RT-PCR products of the transgenic colonies (Tg) and nontransgenic colonies (C) were electrophoresed in an agarose gel, stained with ethidium bromide, and photographed. In the top panel (E2A-HLF), products of genomic and mRNA amplification are indicated by arrows and the position of the DNA size markers are shown on the left. In the bottom panel (K-ras), K-ras RT-PCR products are shown as an internal control.
(A) Flow cytometric analysis of the B-cell colonies generated from bone marrow of a transgenic mouse (Tg) and a nontransgenic control (C) with anti-B220 and anti-CD43 antibodies (4 weeks of age). Colonies established from transgenic mice and nontransgenic controls are similarly positive for B220 and show the same broad pattern for CD43. (B) Expression of transgene-derived mRNA in the transgenic B-cell colonies. The RT-PCR products of the transgenic colonies (Tg) and nontransgenic colonies (C) were electrophoresed in an agarose gel, stained with ethidium bromide, and photographed. In the top panel (E2A-HLF), products of genomic and mRNA amplification are indicated by arrows and the position of the DNA size markers are shown on the left. In the bottom panel (K-ras), K-ras RT-PCR products are shown as an internal control.
Acute lymphoblastic leukemia in E2A-HLF transgenic mice.
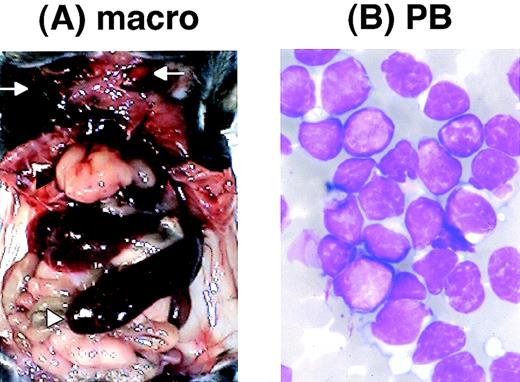
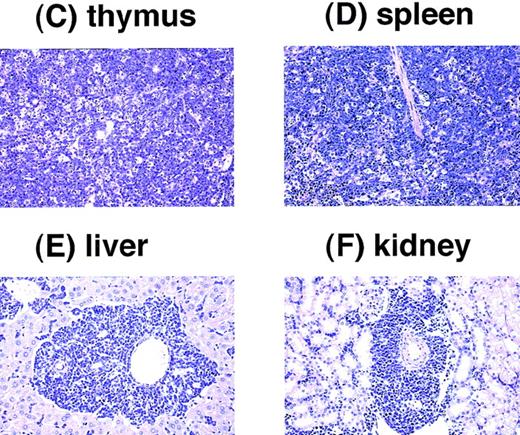
We found that several transgenic mice developed acute leukemia. Among 26 transgenic mice that survived more than 4 months, 5 have developed acute leukemia. All the leukemic mice exhibited massive thymic enlargement, splenomegaly, and lymph node swelling (Fig 8A [see page 2785]). The peripheral blood smears of these mice showed massive proliferation of blast cells with no granules, having the appearance of lymphoblasts (Fig 8B). Pathological analyses showed infiltration of leukemic cells into almost all tissues examined, especially in the thymus, spleen, liver, and kidney (Fig 8C through F). The characteristics of the leukemic mice are summarized in Table2. Surface marker analysis of the leukemic cells showed that all the leukemic cells were positive for Thy1.2 but negative for B220, indicating that the leukemias were of a T-cell phenotype. Staining of the leukemic cells with anti-CD4 and anti-CD8 antibodies showed that one tumor was double positive (CD4+CD8+) but three cases were single positive for CD4 (CD4+ CD8−) and the other one case was single positive for CD8 (CD4− CD8+). The clonality of the leukemic cells was shown by Southern blot analysis using T-cell receptor (TCR)-γ as a probe. These results indicated that the leukemias developed in the transgenic mice were clonal T-cell tumors.
Pathological analysis of acute leukemia developed inE2A-HLF transgenic mice. (A) Macroscopic appearance of a leukemic mouse. The enlarged thymus and spleen are indicated by black and white triangles, respectively, and subcutaneous lymph node swellings are indicated by arrows. (B) WG-stained peripheral blood smear of a leukemic mouse. Massive proliferation of lymphoblasts is apparent. (C through F) HE-stained thymus (C), spleen (D), liver (E), and kidney (F) of a leukemic mouse. The fundamental structures of the thymus and the spleen are destructed due to massive proliferation of leukemic cells (C and D, respectively) and infiltration of leukemic cells is detected around the blood vessels in the liver and the kidney (E and F, respectively).
Pathological analysis of acute leukemia developed inE2A-HLF transgenic mice. (A) Macroscopic appearance of a leukemic mouse. The enlarged thymus and spleen are indicated by black and white triangles, respectively, and subcutaneous lymph node swellings are indicated by arrows. (B) WG-stained peripheral blood smear of a leukemic mouse. Massive proliferation of lymphoblasts is apparent. (C through F) HE-stained thymus (C), spleen (D), liver (E), and kidney (F) of a leukemic mouse. The fundamental structures of the thymus and the spleen are destructed due to massive proliferation of leukemic cells (C and D, respectively) and infiltration of leukemic cells is detected around the blood vessels in the liver and the kidney (E and F, respectively).
Characteristics of E2A-HLF Leukemic Mice
| Mouse No. (sex) . | Age at Diagnosis (mo) . | Macroscopic Tumor Sites . | Hematological Examination . | Surface Marker Analysis . | DNA Status (TCR) . | ||
|---|---|---|---|---|---|---|---|
| WBC (×103/μL) . | Hb (g/dL) . | Plt (×103/μL) . | |||||
| Thy1.2 (+) | |||||||
| 1-17-9 | 5.5 | Thy, Spl, LN | 43.8 | 4.5 | 6.2 | CD4 (−) CD8 (+) | G/R |
| (M) | (>90% lymphoblasts) | B220 (−) | |||||
| Thy1.2 (+) | |||||||
| 2-17-2 | 6.0 | Thy, Spl, LN | 20.5 | 8.0 | 10.5 | CD4 (+) CD8 (+) | G/R |
| (F) | (>90% lymphoblasts) | B220 (−) | |||||
| Thy1.2 (+) | |||||||
| 2-17-8-2 | 4.2 | Thy, Spl, LN | 19.8 | 7.7 | 22.2 | CD4 (+) CD8 (−) | G/R |
| (F) | (>90% lymphoblasts) | B220 (−) | |||||
| Thy1.2 (+) | |||||||
| 2-17-8-4 | 5.2 | Thy, Spl, LN | 15.6 | 8.5 | 23.3 | CD4 (+) CD8 (−) | G/R |
| (F) | (>90% lymphoblasts) | B220 (−) | |||||
| Thy1.2 (+) | |||||||
| 2-17-10-8 | 3.1 | Thy, Spl, LN | 52.3 | 8.1 | 30.1 | CD4 (+) CD8 (−) | G/R |
| (M) | (>90% lymphoblasts) | B220 (−) | |||||
| Mouse No. (sex) . | Age at Diagnosis (mo) . | Macroscopic Tumor Sites . | Hematological Examination . | Surface Marker Analysis . | DNA Status (TCR) . | ||
|---|---|---|---|---|---|---|---|
| WBC (×103/μL) . | Hb (g/dL) . | Plt (×103/μL) . | |||||
| Thy1.2 (+) | |||||||
| 1-17-9 | 5.5 | Thy, Spl, LN | 43.8 | 4.5 | 6.2 | CD4 (−) CD8 (+) | G/R |
| (M) | (>90% lymphoblasts) | B220 (−) | |||||
| Thy1.2 (+) | |||||||
| 2-17-2 | 6.0 | Thy, Spl, LN | 20.5 | 8.0 | 10.5 | CD4 (+) CD8 (+) | G/R |
| (F) | (>90% lymphoblasts) | B220 (−) | |||||
| Thy1.2 (+) | |||||||
| 2-17-8-2 | 4.2 | Thy, Spl, LN | 19.8 | 7.7 | 22.2 | CD4 (+) CD8 (−) | G/R |
| (F) | (>90% lymphoblasts) | B220 (−) | |||||
| Thy1.2 (+) | |||||||
| 2-17-8-4 | 5.2 | Thy, Spl, LN | 15.6 | 8.5 | 23.3 | CD4 (+) CD8 (−) | G/R |
| (F) | (>90% lymphoblasts) | B220 (−) | |||||
| Thy1.2 (+) | |||||||
| 2-17-10-8 | 3.1 | Thy, Spl, LN | 52.3 | 8.1 | 30.1 | CD4 (+) CD8 (−) | G/R |
| (M) | (>90% lymphoblasts) | B220 (−) | |||||
Abbreviations: M, male; F, female; Thy, thymus; Spl, spleen; LN, lymph node; WBC, white blood cell count; G, germline; R, rearranged.
DISCUSSION
Generation of transgenic mice has successfully been used for investigating gene functions in vivo.18 Transgenic mice have provided significant insights into the biological properties of the chimeric gene product, especially for genes that are activated by chromosomal translocations, such as Bcr/Abl andPML/RARα.13,19-22 Thus, we applied this technique to investigate the biological function ofE2A-HLF. The use of Ig enhancer/promoter allowed us to express the E2A-HLF transgene product in both T and B lymphocytes (Fig1B and C), as shown in previous reports.23-25 Failure in detecting the E2A-HLF protein in the transgenic spleen by immunoprecipitation/Western blot (Fig 1B) was probably due to the reduction in B lymphocytes (Figs 4B and 6A).
Histological analysis showed remarkable changes in the thymus and in the spleen (Fig 4A and B). The overall decrease in thymocytes and marked reduction in splenic lymphocytes indicated that the expression of E2A-HLF impaired normal lymphocyte development in both T and B lineages. These results enabled us to explain the cause of the high mortality rate observed in the transgenic mice. Reduction in the lymphocyte number would render the transgenic mice immunodeficient and highly susceptible to fatal infections. The finding that transgenic mice kept in a clean isolator survived much longer than those maintained under a conventional condition supports this idea. The reason why the phenotypic changes (emaciation and death) were not observed before weaning could be explained by the notion that the maternal Ig would be transmitted through the milk and rescue the immunodeficiency of the transgenic mice, as suggested in a previous report.26
The TUNEL staining showed that the decrease in thymocyte number was caused by increased apoptosis (Fig 5). In addition, the flow cytometric analysis demonstrated that the marked reduction in splenic lymphocytes was caused by the maturation block of B lymphocytes at a very early developmental stage (Fig 6A and B). Thus, the expression ofE2A-HLF in lymphoid cells induced T-cell apoptosis and B-cell maturation arrest in vivo. Although the precise mechanism is not clear, it could be supposed that E2A-HLF induced expression of genes regulating cell survival and cell death and led to impairment of normal lymphoid development. Thus, we compared the expression levels of several candidate genes, including bcl-2,27bcl-x,28 and Fas,29 in the transgenic and nontransgenic thymuses. Because we did not detect any differences in their expression levels (data not shown), the abnormal lymphocyte development would be caused by other mechanism(s) than we examined. Interestingly, although B-cell maturation was blocked at a very early stage in vivo, transgenic bone marrow cells restored the ability of developing B-cell progenitors in vitro (Fig 7 and Table 1), suggesting that some in vivo factor(s) might contribute to the abnormal lymphoid development in cooperation with E2A-HLF.
Increased T-cell apoptosis observed in E2A-HLF transgenic mice seems to be inconsistent with the results of experiments using in vitro cell systems, which demonstrated an anti-apoptotic function ofE2A-HLF.11,30 One possible explanation is the difference in the cell lineages. We observed that UOC-B1 cells, which was established from human pro-B leukemia expressing E2A-HLF,underwent apoptosis by the induction of a dominant-negative form ofE2A-HLF.11 In addition, two murine IL-3–dependent pro-B cell lines, Ba/F3 and FL5.12, became resistant to apoptosis caused by IL-3 depletion when E2A-HLF was expressed in these cells.11,30 However, E2A-HLF failed to reverse apoptosis in cytokine-derived 32D cells, a murine IL-3–dependent myeloid cell line (our unpublished results). These results suggest that the anti-apoptotic function of E2A-HLF might be confined to B-lineage lymphocytes. Maturation arrest of B lymphocytes, on the other hand, may agree with our recent result obtained using an in vitro experimental system that CD20, a well-known cell-surface marker for B-cell maturation, is induced by the expression of a dominant-negative form of E2A-HLF in UOC-B1 cells.31
It is to be noted that several transgenic mice developed acute leukemia. The leukemic mice exhibited thymic enlargement, splenomegaly, and lymph node swelling (Fig 8A). The leukemic cells, having the appearance of lymphoblasts (Fig 8B), were highly malignant, showing massive infiltration into almost all the tissues examined (Fig 8C through F). The leukemic cells were positive for Thy1.2 and carried rearrangements in TCR loci (Table 2), indicating that they were clonal T-cell tumors. However, the expression patterns of CD4 and CD8 in each leukemia were heterogeneous; one tumor was double-positive for CD4 and CD8, three tumors were single-positive for CD4, and the other one case was single-positive for CD8. Thus, E2A-HLF rendered an oncogenic activity to T cells that are in transition from double-positive to single-positive stage of development. These results indicated that E2A-HLF possesses an oncogenic potential in addition to inducing T-cell apoptosis and B-cell maturation block. The finding that 5 of 26 mice that survived more than 4 months developed ALL may suggest that an additional genetic event might be required for disease development. This idea is in agreement with the findings thatE2A-HLF exhibited an oncogenic ability in NIH 3T3 cells10 and showed an anti-apoptotic activity in Ba/F3 cells,11 since the transfected cells are immortalized and would carry a genetic abnormality(ies) for immortalization. Therefore, it is likely that E2A-HLF contributes to leukemogenesis in cooperation with another or more genetic event(s).
Interestingly, the phenotypic aspects observed in the E2A-HLFtransgenic mice (T-cell apoptosis, B-cell maturation block, and T-cell leukemia) closely resemble those reported in the transgenic mice expressing another E2A-involving chimeric protein,E2A-PBX1, under the control of the same Ig promoter/enhancer.32 If aberrant gene transcription induced by E2A-HLF or E2A-PBX1 is responsible for the phenotypes, the phenotypic coincidence between both types of transgenic mice seems to be anomalous, because the DNA-binding activities ofE2A-HLF and E2A-PBX1 are dependent on HLF- andPBX1-derived regions, respectively, and their experimentally defined binding motifs are completely different (5′-GTTACGTAAT-3′ forHLF and 5′-ATCAATCAA-3′ for PBX1).9,33 One possible explanation for this is that a gene(s) that is a common target of both E2A-HLF and E2A-PBX1 exists, and its aberrant expression in lymphoid cells impaired normal lymphoid development and led to phenotypic abnormalities. Another possibility, which seems to be more likely, is that the phenotypes commonly observed in both transgenic mice were caused by a dominant-negative inhibition ofE2A activity. This idea could be supported by the finding that mice lacking E2A also exhibited very similar phenotypes.E2A-deficient mice failed to generate B220+ cells because of maturation arrest of B cells at an early developmental stage.34,35 In addition, they contained a low number of thymocytes and eventually developed T-cell lymphomas.36Thus, it would be possible to consider that the biological properties of E2A-HLF and E2A-PBX1 are caused, at least in part, by interfering E2A function.
The results of functional studies of E2A-HLF andE2A-PBX1 using cell lines support this idea. TheE2A-derived regions of E2A-HLF and E2A-PBX1have been shown to be required for the transactivating and transforming abilities in both chimeric proteins10,37-39 and also for the anti-apoptotic function of E2A-HLF in IL-3–dependent murine lymphocytes.30 In contrast, the bZIP domain ofHLF and the homeodomain of PBX1, which mediate site-specific DNA-binding capacities of E2A-HLF andE2A-PBX1, respectively, have been shown to be unnecessary for the transforming ability of E2A-PBX138,39 or for anti-apoptotic function of E2A-HLF.30Tal-1(also called as SCL), which is a transcription factor belonging the bHLH family and is involved in human T-ALL, is shown to directly bind to E2A40,41 and is considered to contribute to leukemogenesis by interfering normal E2A function through forming a heterodimer with E2A.42 However, it is not the case of E2A-HLF and E2A-PBX1, because the dimerization domain of E2A is deleted in both proteins.3-6 Thus, although it is likely that the abnormal lymphocyte development observed in the transgenic mice would be induced by a dominant-negative effect on E2A function, the underlying mechanisms have remained to be established. One possibility to be investigated is that E2A-HLF and E2A-PBX1 might competitively bind to a specific interaction partner of the activation domain of E2A that is expressed in lymphoid cells and is required for E2A activity, through which they inhibitE2A function and induce an E2A-deficient phenotype.
There remains a question concerning the difference in the immunophenotypes of the leukemias between humans and mice. In humans,E2A-PBX1 is detected in pre-B cell leukemias3,4 andE2A-HLF is associated with pro-B type leukemias,5,6whereas the leukemias developed in the transgenic mice have so far been diagnosed as T-ALL32 (and this report). Although the reason is not clear, one possibility is that Ig enhancer/promoter allowed transgene expression in both in T and B lymphocytes, and T cells might be more susceptible to the oncogenic properties of the chimeric proteins. The generation of knock-in mice expressing E2A-HLFand E2A-PBX1 under the control of native E2A promoter might clarify the problem concerning the phenotypic disparity between humans and mice.
ACKNOWLEDGMENT
We thank Dr T.W. Mak for providing the mouse TCR-γ probe. We thank Yoshikazu Oh-hira for preparing pathological specimens, and we also thank Haruki Kume for photographs.
Supported in part by Grants-in-Aids from the Ministry of Education, Science and Culture of Japan.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Hisamaru Hirai, MD, Third Department of Internal Medicine, Faculty of Medicine, University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113, Japan; e-mail: hhirai-tky@umin.ac.jp.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal