Abstract
TT virus (TTV) is a newly discovered transfusion-transmissible DNA virus, which may cause posttransfusion hepatitis. The virus was detected in 12% of Japanese blood donors. The aim of the study is to investigate the prevalence and clinical influence of TTV in bone marrow transplant (BMT) recipients. Sera from 25 BMT recipients obtained 6 to 12 weeks after the transplant were examined for TTV-DNA by the seminested polymerase chain reaction. Serial samples were additionally analyzed in patients with TTV-DNA. Fifteen of 25 recipients (60%) were positive for TTV-DNA after transplant, whereas it was detected in only two of 20 BMT donors (10%). In patients positive for TTV-DNA before BMT, the amount of TTV-DNA decreased to an undetectable level during the myelosuppressed period after BMT. We also found that there was a novel group of TTV, G3, classified by the nucleotide sequences. The median peak alanine aminotransferase (ALT) levels were 135.0 IU/L and 116.5 IU/L (normal range, 4 to 36 IU/L) in TTV-positive and TTV-negative recipients, respectively. In one of the seven TTV-positive patients who developed hepatic injury (ALT > 150 IU/L), a serial change in the serum TTV titer showed a good correlation with the ALT level. We concluded that (1) the prevalence of TTV is high in BMT recipients, (2) TTV might be replicated mainly in hematopoietic cells, (3) transfusion-transmitted TTV may cause persistent infection, (4) a novel genetic group of TTV, G3, was discovered, and (5) TTV does not seem to frequently cause hepatic injury, although one patient was strongly suggested to have TTV-induced hepatitis.
HEPATIC DYSFUNCTION IS one of the frequent complications after bone marrow transplantation (BMT). Some of them are caused by drug toxicity or tumor infiltration. Hepatic venoocclusive disease (VOD), which is characterized by painful hepatomegaly, ascites, and jaundice, is considered to be a toxicity of preparative regimen before BMT. Graft-versus-host disease (GVHD) also causes severe hepatic injury, which is difficult to treat. We, however, sometimes meet with hepatic dysfunction of unknown etiology. Posttransfusion hepatitis, which has been reduced by screening for hepatitis B virus (HBV) and hepatitis C virus (HCV), is still responsible for a part of hepatic problems. Hepatitis G virus (HGV), also called GB virus C, is a recently discovered transfusion-transmissible flavivirus and had been expected to be a responsible agent for posttransfusion hepatitis.1 However, accumulating data showed that this virus rarely causes hepatitis.2 Studies in bone marrow transplant recipients also failed to show the relationship between HGV infection and liver injury, although immunosuppression associated with BMT might increase the risk of HGV infection after transfusion-related exposure.3-6 Nishizawa et al7 cloned a novel DNA virus from serum of a patient with posttransfusion hepatitis. The virus was designated TT virus (TTV) after the patient from whom it was derived. The name also stands for a “transfusion-transmitted virus”. TTV-DNA was detected in sera from three of five patients with posttransfusion non-A to G hepatitis, and the increase in serum TTV-DNA coincided with the elevation of serum alanine aminotransferase (ALT).7 Now, it is highlighted as a candidate for the causative agent for such hepatitis. TTV is an unenveloped single-stranded DNA virus of at lease 3,700 bases and resembles parvovirus in some features.8 However, little is known about the clinical characteristics of the virus except for the 12% prevalence in Japanese blood donors.8 Whether TTV is replicated in the liver is also unknown. In this study, we retrospectively investigated the prevalence and clinical impact of TTV in 25 BMT recipients at our institute.
MATERIALS AND METHODS
Patients.
Frozen sera from 25 BMT recipients transplanted between June 1995 and January 1998 were retrospectively analyzed for TTV-DNA. There were 20 men and five women with a median age of 35.0 years (range, 18 to 48 years). Nine patients with chronic myelocytic leukemia, six with acute lymphoblastic leukemia, six with acute myeloblastic leukemia, three with myelodysplastic syndrome, and one with severe aplastic anemia were included. Eighteen patients had a history of transfusion before BMT. Seventeen patients were transplanted from HLA-identical siblings, three from one-locus–mismatched related donors, and five from HLA-matched unrelated donors. Preparative regimens among the cases were various including 18 irradiating regimens (mainly, 120 mg/kg cyclophosphamide and 12 Gy total body irradiation) and seven nonirradiating regimens (mainly, 16 mg/kg busulfan and 120 mg/kg cyclophosphamide). Prophylaxis for GVHD was performed with cyclosporin A and short-course methotrexate. Fluconazole, tosufloxacin, sulfamethoxazole-trimethoprim, and aciclovir were administered prophylactically.
First, sera obtained between 6 and 12 weeks after BMT were subjected to the analyses. Next, sera just before BMT, during the myelosuppressed period after BMT, and more than 6 months after BMT were additionally examined, if available, for patients with TTV-DNA. Sera from bone marrow donors were also examined.
Detection of TTV-DNA by seminested polymerase chain reaction.
DNA was extracted from sera by a modified method originally described by Okamoto et al.9 Serum (50 μL) was mixed with 150 μL of 1.3x lysis buffer (13.3 mmol/L Tris-HCl pH 8.0, 6.7 mmol/L EDTA, 0.67% sodium dodecyl sulfate, and 133 μg/mL proteinase K) and incubated at 70°C for 3 hours. After thorough mixing by Vortex with 200 μL of phenol-chloroform, the aliquots were centrifuged at 15,000 rpm for 10 minutes. The supernatants were collected and incubated with 20 μL of 3 mol/L NaOAc, 500 μL of ethanol, and 7.5 μg of carrier tRNA (final, 10 μg/mL) in dry ice for 5 minutes. After a centrifugation at 15,000 rpm for 15 minutes at 4°C, the supernatants were discarded. The pellet was washed with 70% ethanol, dried up, and dissolved with 20 μL of TE (10 mmol/L Tris-HCl pH 8.0, 1 mmol/L EDTA). A half portion of the extracts was subjected to seminested polymerase chain reaction (PCR) for TTV-DNA. The first-round PCR was performed with NG059 primer (5′-ACA GAC AGA GGA GAA GGC AAC ATG-3′) and NG063 primer (5′-CTG GCA TTT TAC CAT TTC CAA AGT T-3′) for 35 cycles (94°C, 30 seconds; 60°C, 45 seconds; 72°C, 45 seconds).8 The second-round PCR was performed with NG061 primer (5′-GGC AAC ATG YTR TGG ATA GAC TGG-3′ [Y = T or C; R = A or G]) and NG063 primer for 25 cycles at the same condition for the amplification of a 271-bp product.8 One third of the products was electrophoresed on 2% agarose gel and stained with ethidium bromide. For the positive samples, amplified products were cloned into pBluescript plasmid, and the nucleotide sequence of several clones from each PCR product was determined by the dideoxy chain termination method. Amplified products were also digested with an endonuclease, ScaI, or EcoRI, electrophoresed on 3% agarose gel, and stained with ethidium bromide. The digestion patterns were helpful to confirm the genetic classification. TTV titer was estimated by PCR using the serially 10-fold diluted template DNA. The titer was described as the highest dilution giving a positive result.
Screening for HBV, HCV, and HGV.
HBV and HCV were screened by the conventional serological methods. Detection of HGV-RNA was performed by the method described previously.10
Statistical analyses.
Results are described as median values with the ranges of distribution. Statistical comparison between groups was performed using the Fisher’s exact test or the Mann-Whitney U test.
RESULTS
Prevalence of TTV.
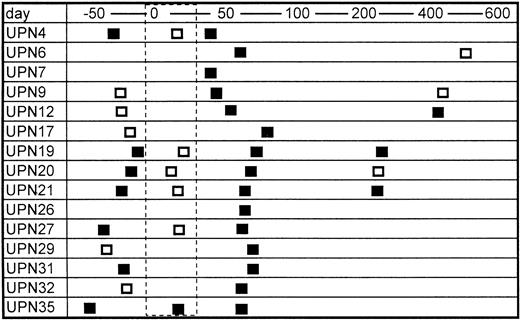
Sera from only two of 20 bone marrow donors were positive for TTV-DNA (10%). The prevalence is almost equivalent with the data in normal blood donors in a previous report (12%).8 In contrast, 15 of 25 patients were positive for TTV-DNA after BMT (60%) (Table 1). The mean numbers of blood transfusion donors, including concentrated red blood cells and platelet rich plasma, were 43.4 and 27.0 in the TTV-positive and TTV-negative patients, respectively, the difference of which was not statistically significant. None of the recipients enrolled were positive for HBV and HCV, but six patients were positive for HGV-RNA. The prevalence of HGV-RNA was 33% and 10% in TTV-positive and TTV-negative patients, respectively (not significant). Serial samples from TTV-positive BMT recipients were further analyzed, if available. TTV was detected in seven of 12 (67%) available samples obtained before BMT (Fig 1). It was also detected in three of six (50%) samples obtained several months (280 to 568 days) after BMT (Fig 1). In contrast, only one of six patients (17%) with positive TTV before BMT was shown to have TTV during the myelosuppressed period after BMT (Fig 1). This finding suggested that the amount of TTV might decrease to an undetectable level in the myelosuppressed period after BMT.
Patient Characteristics and Results of Virus Surveillance
| UPN . | Age/Sex . | Disease . | BTF . | TTV . | Donor TTV . | HBV . | HCV . | HGV . |
|---|---|---|---|---|---|---|---|---|
| 1 | 20/M | ALL | (+)/(+) | (−) | (−) | (−) | (−) | (−) |
| 4 | 24/M | AML | (+)/(+) | (+) | (−) | (−) | (−) | (+) |
| 6 | 47/F | CML | (−)/(+) | (+) | (−) | (−) | (−) | (−) |
| 7 | 24/M | AML | (+)/(+) | (+) | (−) | (−) | (−) | (+) |
| 9 | 36/M | AML | (+)/(+) | (+) | (−) | (−) | (−) | (−) |
| 10 | 35/M | CML | (−)/(+) | (−) | (−) | (−) | (−) | (−) |
| 12 | 23/M | SAA | (+)/(+) | (+) | (−) | (−) | (−) | (−) |
| 15 | 41/M | CML | (−)/(+) | (−) | (−) | (−) | (−) | (−) |
| 17 | 27/M | MDS | (+)/(+) | (+) | (+) | (−) | (−) | (+) |
| 19 | 27/M | AML | (+)/(+) | (+) | (−) | (−) | (−) | (−) |
| 20 | 20/M | CML | (−)/(+) | (+) | (−) | (−) | (−) | (−) |
| 21 | 35/M | CML | (−)/(+) | (+) | (+) | (−) | (−) | (−) |
| 22 | 18/F | ALL | (+)/(+) | (−) | (−) | (−) | (−) | (−) |
| 26 | 18/M | ALL | (+)/(+) | (+) | NT | (−) | (−) | (−) |
| 27 | 26/M | ALL | (+)/(+) | (+) | (−) | (−) | (−) | (−) |
| 28 | 25/M | ALL | (+)/(+) | (−) | (−) | (−) | (−) | (+) |
| 29 | 40/F | MDS | (+)/(+) | (+) | (−) | (−) | (−) | (−) |
| 31 | 48/M | MDS | (+)/(+) | (+) | (−) | (−) | (−) | (+) |
| 32 | 28/M | CML | (+)/(+) | (+) | (−) | (−) | (−) | (−) |
| 33 | 46/F | ALL | (+)/(+) | (−) | NT | (−) | (−) | (−) |
| 34 | 45/F | CML | (−)/(+) | (−) | NT | (−) | (−) | (−) |
| 35 | 47/M | CML | (+)/(+) | (+) | (−) | (−) | (−) | (+) |
| 36 | 37/M | AML | (+)/(+) | (−) | NT | (−) | (−) | (−) |
| 38 | 39/M | AML | (+)/(+) | (−) | NT | (−) | (−) | (−) |
| 39 | 35/M | CML | (−)/(+) | (−) | (−) | (−) | (−) | (−) |
| UPN . | Age/Sex . | Disease . | BTF . | TTV . | Donor TTV . | HBV . | HCV . | HGV . |
|---|---|---|---|---|---|---|---|---|
| 1 | 20/M | ALL | (+)/(+) | (−) | (−) | (−) | (−) | (−) |
| 4 | 24/M | AML | (+)/(+) | (+) | (−) | (−) | (−) | (+) |
| 6 | 47/F | CML | (−)/(+) | (+) | (−) | (−) | (−) | (−) |
| 7 | 24/M | AML | (+)/(+) | (+) | (−) | (−) | (−) | (+) |
| 9 | 36/M | AML | (+)/(+) | (+) | (−) | (−) | (−) | (−) |
| 10 | 35/M | CML | (−)/(+) | (−) | (−) | (−) | (−) | (−) |
| 12 | 23/M | SAA | (+)/(+) | (+) | (−) | (−) | (−) | (−) |
| 15 | 41/M | CML | (−)/(+) | (−) | (−) | (−) | (−) | (−) |
| 17 | 27/M | MDS | (+)/(+) | (+) | (+) | (−) | (−) | (+) |
| 19 | 27/M | AML | (+)/(+) | (+) | (−) | (−) | (−) | (−) |
| 20 | 20/M | CML | (−)/(+) | (+) | (−) | (−) | (−) | (−) |
| 21 | 35/M | CML | (−)/(+) | (+) | (+) | (−) | (−) | (−) |
| 22 | 18/F | ALL | (+)/(+) | (−) | (−) | (−) | (−) | (−) |
| 26 | 18/M | ALL | (+)/(+) | (+) | NT | (−) | (−) | (−) |
| 27 | 26/M | ALL | (+)/(+) | (+) | (−) | (−) | (−) | (−) |
| 28 | 25/M | ALL | (+)/(+) | (−) | (−) | (−) | (−) | (+) |
| 29 | 40/F | MDS | (+)/(+) | (+) | (−) | (−) | (−) | (−) |
| 31 | 48/M | MDS | (+)/(+) | (+) | (−) | (−) | (−) | (+) |
| 32 | 28/M | CML | (+)/(+) | (+) | (−) | (−) | (−) | (−) |
| 33 | 46/F | ALL | (+)/(+) | (−) | NT | (−) | (−) | (−) |
| 34 | 45/F | CML | (−)/(+) | (−) | NT | (−) | (−) | (−) |
| 35 | 47/M | CML | (+)/(+) | (+) | (−) | (−) | (−) | (+) |
| 36 | 37/M | AML | (+)/(+) | (−) | NT | (−) | (−) | (−) |
| 38 | 39/M | AML | (+)/(+) | (−) | NT | (−) | (−) | (−) |
| 39 | 35/M | CML | (−)/(+) | (−) | (−) | (−) | (−) | (−) |
The histories of transfusion before and after BMT are shown at the left and right side of the oblique line in the column of BTF, respectively.
Abbreviations: ALL, acute lymphoblastic leukemia; AML, acute myeloblastic leukemia; CML, chronic myelocytic leukemia; SAA, severe aplastic anemia; MDS, myelodysplastic syndrome; BTF, blood transfusion; NT, not tested.
Serial changes in the results of PCR for TTV-DNA. Open and closed boxes indicate TTV-DNA negative and positive samples, respectively. The area surrounded by the dashed line represents the myelosuppressed period after transplant.
Serial changes in the results of PCR for TTV-DNA. Open and closed boxes indicate TTV-DNA negative and positive samples, respectively. The area surrounded by the dashed line represents the myelosuppressed period after transplant.
Nucleotide sequence of the amplified TTV-DNA.
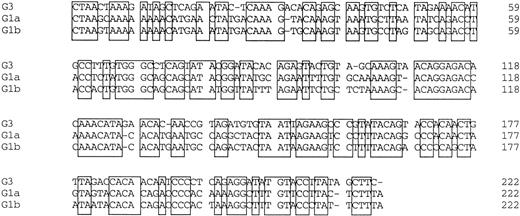
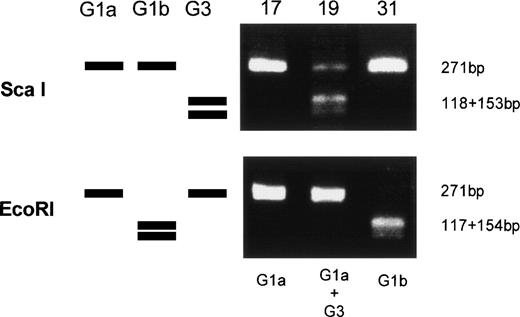
Amplified products were cloned into the pBluescript plasmid, and nucleotide sequences of several clones per each product were determined by the dideoxy chain termination method. Okamoto et al8classified the nucleotide sequences of TTV into four genetic subgroups; G1a, G1b, G2a, and G2b. All of the sequences in our series belonged to subgroups of G1a and G1b, except for 3, designated G3-group, which belonged to none of the known four subgroups. The sequences of G3-group differed by 0% to 3.6% in sequence to each other and by 36% to 42% to G1a (Fig 2). It was also found that UPN19, UPN35, and the donor of UPN21 simultaneously had two distinct subgroups of TTV in their sera. To confirm these findings, amplified products were digested with an endonuclease, ScaI, or EcoRI. The G1b-subgroup and the G3-group have an internal digestion site for EcoRI and ScaI, respectively, whereas the G1a-subgroup has neither of the digestion sites. Thus, the genotype can be distinguished by the pattern of digestion (Fig3). From the nucleotide sequence and the digestion patterns, TTV in each individual was classified as shown in Table 2.
Comparison of nucleotide sequences in genetic groups of TTV. Nucleotide sequences of 222 bp spanning 1959-2180 are shown. Primer sequences at both ends are excluded. Conserved nucleotides between the groups are boxed.
Comparison of nucleotide sequences in genetic groups of TTV. Nucleotide sequences of 222 bp spanning 1959-2180 are shown. Primer sequences at both ends are excluded. Conserved nucleotides between the groups are boxed.
Genetic classification of TTV by digesting the amplified products with endonucleases. Three lanes on the left side schematically represents digestion patterns of the amplified products of G1a, G1b, and G3 subtypes. Three lanes on the right side are photographs of the gel stained with ethidium bromide on which amplified products from UPN 17, 19, and 31 were electrophoresed after digestion with an endonuclease, ScaI, or EcoRI.
Genetic classification of TTV by digesting the amplified products with endonucleases. Three lanes on the left side schematically represents digestion patterns of the amplified products of G1a, G1b, and G3 subtypes. Three lanes on the right side are photographs of the gel stained with ethidium bromide on which amplified products from UPN 17, 19, and 31 were electrophoresed after digestion with an endonuclease, ScaI, or EcoRI.
Classification of TTV by the Nucleotide Sequence
| UPN . | Donor . | Before BMT . | After BMT . |
|---|---|---|---|
| 4 | (−) | G1a | G1a |
| 6 | (−) | NT | G1a |
| 7 | (−) | NT | G1a |
| 9 | (−) | (−) | G1a |
| 12 | (−) | (−) | G1a |
| 17 | G1a | (−) | G1a |
| 19 | (−) | G3 | G1a + G3 |
| 20 | (−) | G1a | G1a |
| 21 | G1a + G1b | G1a | G1b |
| 26 | NT | NT | G1a |
| 27 | (−) | G1a | G1a |
| 29 | (−) | (−) | G1b |
| 31 | (−) | G1a | G1b |
| 32 | (−) | (−) | G1b |
| 35 | (−) | G1a | G1a + G3 |
| UPN . | Donor . | Before BMT . | After BMT . |
|---|---|---|---|
| 4 | (−) | G1a | G1a |
| 6 | (−) | NT | G1a |
| 7 | (−) | NT | G1a |
| 9 | (−) | (−) | G1a |
| 12 | (−) | (−) | G1a |
| 17 | G1a | (−) | G1a |
| 19 | (−) | G3 | G1a + G3 |
| 20 | (−) | G1a | G1a |
| 21 | G1a + G1b | G1a | G1b |
| 26 | NT | NT | G1a |
| 27 | (−) | G1a | G1a |
| 29 | (−) | (−) | G1b |
| 31 | (−) | G1a | G1b |
| 32 | (−) | (−) | G1b |
| 35 | (−) | G1a | G1a + G3 |
Abbreviation: NT, not tested.
Relationship between TTV infection and liver dysfunction.
First, we investigated the relationship between pretransplant TTV infection and liver dysfunction in the early period (0 to 5 weeks) after BMT. The diagnosis of VOD was established according to McDonald’s criteria.11 Patients who satisfied at lease two of the following three criteria were diagnosed with VOD: jaundice (total bilirubin > 2.0 mg/dL), hepatomegaly and upper abdominal pain, ascites and/or body weight gain (at least 5% gain from the baseline body weight).11 As shown in Table 3, the incidence of VOD was almost equivalent in TTV-positive and TTV-negative patients. The median values of peak total bilirubin levels and peak ALT (normal range, 4 to 36 IU/L) levels during the period were 1.0 mg/dL and 117 IU/L in the TTV-positive patients and 1.4 mg/dL and 103 IU/L in the TTV-negative patients, respectively. The differences were not statistically significant. Thus, there seemed to be no relationship between the regimen-related toxicity of the liver and pretransplant TTV infection.
Association Between TTV Infection and Liver Diseases
| TTV Before BMT . | VOD . | Peak T.Bil (day 0 to 41) . | Peak ALT (day 0 to 41) . |
|---|---|---|---|
| Positive (n = 7) | 1 | 1.0 (0.7-2.4) | 117 (64-1331) |
| Negative (n = 5) | 1 | 1.4 (0.9-6.4) | 103 (31-314) |
| TTV Before BMT . | VOD . | Peak T.Bil (day 0 to 41) . | Peak ALT (day 0 to 41) . |
|---|---|---|---|
| Positive (n = 7) | 1 | 1.0 (0.7-2.4) | 117 (64-1331) |
| Negative (n = 5) | 1 | 1.4 (0.9-6.4) | 103 (31-314) |
| TTV After BMT . | GVHD (Grade II-IV) . | Peak ALT (day 42 to 83) . |
|---|---|---|
| Positive (n = 15) | 3 | 135.0 (42-685) |
| Negative (n = 10) | 3 | 116.5 (33-241) |
| TTV After BMT . | GVHD (Grade II-IV) . | Peak ALT (day 42 to 83) . |
|---|---|---|
| Positive (n = 15) | 3 | 135.0 (42-685) |
| Negative (n = 10) | 3 | 116.5 (33-241) |
The peak levels of T.Bil and ALT are described in median value and the range of distribution.
Abbreviations: VOD, hepatic venoocclusive disease; T.Bil, total bilirubin; ALT, alanine aminotransferase.
Next, we analyzed the relationship between posttransplant TTV infection and liver dysfunction between 6 and 12 weeks after BMT. The median peak serum ALT levels were 135.0 IU/L (range, 42 to 685 IU/L) and 116.5 IU/L (range, 33 to 241 IU/L) in TTV-positive and TTV-negative patients, respectively, the difference of which was not statistically significant (Table 3). The incidence of grade II-IV GVHD was not different between the groups. The serum ALT exceeded 150 IU/L in seven of the 15 TTV-positive recipients and in three of the 10 TTV-negative recipients (not significant). Considering the clinical course, graft-versus-host reaction was considered to be responsible for the liver injury in most of the patients, although some of them did not fulfill the criteria of the diagnosis of GVHD. However, laboratory data suggested that the bile duct damage in UPN21 was moderate compared with the hepatocyte injury, which is atypical in hepatic GVHD. This patient had received liver biopsy and the pathological findings of the specimen showed that there was severe hepatocyte injury despite mild bile duct damage, which also conflicted with the diagnosis of GVHD. Drug adverse reaction was denied from the clinical course. We considered the hepatic injury of the patient a result of TTV-associated hepatitis and conducted further analyses.
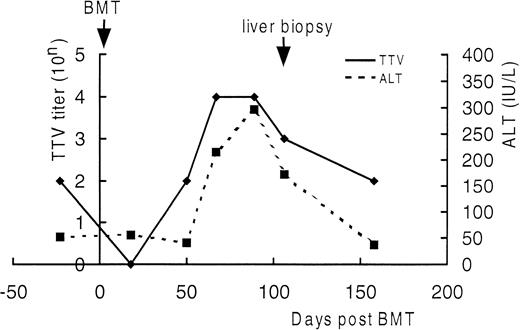
Serial semiquantitation of TTV-DNA in sera of UPN21.
DNA extracted from sera of UPN21 obtained before BMT and 18, 50, 67, 89, 106, and 158 days after BMT were serially 10-fold diluted and subjected to PCR. TTV titers were estimated according to the highest dilution giving a positive result. As shown in Fig4, the serum TTV titer closely correlated with the serum ALT level. The TTV titer decreased just after BMT and increased in the ensuing period as the bone marrow recovered along with the elevation of the serum ALT level. After day 150, both returned to the level before BMT without treatment. The trough blood concentration of cyclosporine A was maintained between 150 and 250 ng/mL throughout the period. These findings strengthened the diagnosis of TTV-associated hepatitis.
Detection of TTV-DNA in the liver specimen from UPN21.
DNA was extracted from the formalin-fixed liver specimens from UPN21 and subjected to PCR for TTV-DNA. The liver sample from UPN1, who had suffered from chronic GVHD of the liver, was used as a negative control. TTV-DNA was detected in the liver specimen from UPN21, but not in the sample from UPN1. We performed in situ hybridization of the TTV-DNA using the specimen, because PCR might detect TTV-DNA from contaminated blood. However, we could not detect TTV-DNA by in situ hybridization, probably due to its low sensitivity by use of formalin-fixed tissue.
DISCUSSION
In the present study, we investigated the relationship between TTV and the clinical course in BMT recipients. TTV-DNA was detected in 60% of the BMT recipients. The prevalence is far higher than that (12%) in normal blood donors in the previous report,8 as well as that (10%) in the bone marrow donors in our series (P < .001). The TTV-negative recipients had received transfusion from 27.0 donors on average, suggesting that the infused TTV did not always cause infection. Seven of the 12 TTV-positive patients were positive for TTV before BMT and two (UPN20 and UPN21) of seven patients had no history of transfusion before BMT. The bone marrow donor of UPN21, who is a sister of UPN21, was also positive for TTV without a history of transfusion. Therefore, there are routes of transmission other than blood transfusion. Nucleotide sequencing showed that UPN21 had TTV of G1a-subgroup before BMT and the donor of UPN21 had two types of TTV, G1a and G1b. The sequences of G1a-subgroup TTV from the two individuals were completely identical, suggesting that the TTV was acquired vertically or environmentally (household contact, oral intake of infected foods, and so on). The infection route of G1b-TTV in the donor was unknown, because her sexual activity was normal, she was not a drug abuser, and she did not have acupuncture or tattoos. TTV seemed to be transmissible other than vertically or by transfusion. UPN21 had received administration of interferon-α (IFN-α) as a treatment for chronic myelocytic leukemia for 2 years before BMT, first at 3 million units three times weekly and then at 6 million units daily. This implies that IFN-α did not eliminate TTV in the patient. The effect of IFN-α on eliminating TTV needs further investigation in a large scale study.
We also found that TTV decreased to an undetectable level in the myelosuppressed period after BMT in five of the six patients who were positive for TTV before BMT. The same nucleotide sequences of TTV present before BMT were not detected after BMT in two (UPN 21 and UPN 31) of the seven patients who were TTV-positive before BMT (Table 2). These findings suggested a possibility that TTV is replicated mainly in the hematopoietic cells. Three of the six patients analyzed remained positive for TTV more than 6 months after BMT, when the immunosuppressants were discontinued. Two of the three had acquired TTV after BMT, because the sequence of TTV after BMT was not detected before BMT, suggesting that transfusion-transmitted TTV may cause persistent infection.
Most of the nucleotide sequences of the amplified products belonged to G1a or G1b-subgroups according to the classification by Okamoto et al,8 although three sequences, designated G3-group, belonged to none of the known subgroups. They had 96.4% to 100% homology to each other and 58% to 62% homology to the G1a sequence. There were no published sequences with a high homology to the G3 sequence. The relationship between these subtypes and the characteristics of the virus should be evaluated in further studies. We have found persons who have two distinct strains simultaneously. In such cases, direct sequencing of the amplified products may result in detecting only one of the strains or in reading incorrect sequence. Thus, subcloning of the amplified products into a vector and sequencing several clones is necessary for the correct genetic classification. Digesting the amplified products with an endonuclease, ScaI, or EcoRI and distinguishing the digestion pattern is also helpful to confirm the genetic subtype. By this method, we can estimate the proportion of the coexisting clones. Nucleotide sequencing, however, cannot be avoided because there may be a mutation in the internal digestion site for these endonucleases.
In the BMT recipients, TTV infection neither before nor after BMT significantly affected the clinical course after BMT because the median peak ALT levels were almost equivalent in TTV-positive and TTV-negative BMT recipients. The incidence of VOD or GVHD was not influenced by the TTV infection. Most of the hepatic injury (ALT > 150 IU/L) in TTV-positive recipients was considered to be due to graft-versus-host reaction. One patient, however, developed hepatic injury 55 days after BMT and the serum ALT level showed significant correlation with the serum TTV titer. The pathological findings of the liver specimen agreed with the diagnosis of viral hepatitis. TTV was detected in the specimen by PCR, but not by in situ hybridization, probably due to its low sensitivity by use of formalin-fixed tissue. The sequence of TTV at the time of hepatitis was different from that before BMT, suggesting that the hepatitis was induced by the infused TTV from the bone marrow donor or the blood donors. Because the nucleotide sequence of G1b in UPN 21 after BMT was identical to that of the BMT donor of UPN 21, it was likely that the TTV was derived from the BMT donor. The hepatitis spontaneously regressed in several weeks, whereas TTV-DNA had remained positive thereafter at low titers for more than 6 months. Cyclosporin A did not affect the course of hepatitis. While it has been suggested that HBV carriers have a risk of fulminant hepatitis after BMT,12 HCV and HGV are relatively safe in BMT.13 14 Although TTV does not seem to frequently cause hepatitis in BMT recipients, a larger study is required to determine the safety of the virus in BMT because we cannot deny the possibility that TTV might be synergistically implicated in the hepatic injury of the patients with GVHD. In addition, why TTV causes hepatitis in some patients and does not in others remains to be resolved. There were some patients whose TTV titer was higher than UPN 21, but who did not develop liver injury. To determine whether TTV is replicated in the liver is important to show that TTV is really a “hepatitis virus” because our findings suggested that TTV might be replicated mainly in the hematopoietic cells.
We concluded from these findings that (1) the prevalence of TTV is extremely higher in BMT recipients than in the normal population, (2) TTV decreased to an undetectable level during the myelosuppressed period after BMT, suggesting a possibility that TTV might be replicated mainly in hematopoietic cells, (3) transfusion-transmitted TTV may cause persistent infection, (4) a novel genetic group, G3, was discovered by the nucleotide sequencing, and (5) there was one patient who was strongly suspected to have TTV-induced hepatitis.
ACKNOWLEDGMENT
We thank Dr Makoto Mayumi for his instructive advise and for providing us the primers and control samples for PCR.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Hisamaru Hirai, MD, Department of Cell Therapy and Transplantation Medicine, Faculty of Medicine, University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113, Japan; e-mail:hhirai-tky@umin.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal