Abstract
Interleukin-10 (IL-10) helps maintain polarized T-helper cells in a T-helper lymphocyte 2 (Th2) phenotype. Part of this process involves the prevention of the development of Th1 cells, which are a primary source of interferon γ (IFNγ), a potent activator of monocytes and an inhibitor of Th2 proliferation. Because monocytes and macrophages are important mediators of Th1-type responses, such as delayed-type hypersensitivity, we sought to determine if IL-10 could directly mediate inhibition of IFNγ- and IFN-induced gene expression in these cells. Highly purified monocytes were incubated with IL-10 for 60 to 90 minutes before the addition of IFNγ or IFN. IL-10 preincubation resulted in the inhibition of gene expression for several IFN-induced genes, such as IP-10, ISG54, and intercellular adhesion molecule-1. The reduction in gene expression resulted from the ability of IL-10 to suppress IFN-induced assembly of signal transducer and activator of transcription (STAT) factors to specific promoter motifs on IFN- and IFNγ-inducible genes. This was accomplished by preventing the IFN-induced tyrosine phosphorylation of STAT1, a component of both IFN- and IFNγ-induced DNA binding complexes. Therefore, IL-10 can directly inhibit STAT-dependent early response gene expression induced by both IFN and IFNγ in monocytes by suppressing the tyrosine phosphorylation of STAT1. This may occur through the ability of IL-10 to induce expression of the gene, suppressor of cytokine signaling 3 (SOCS3).
INTERFERON α (IFNα) has been shown to be therapeutically effective in the treatment of certain types of leukemias and lymphomas1,2 and in some solid tumors such as melanoma.3 Although not used as frequently as IFNα, IFNγ has been shown to have some activity in renal cell carcinoma and is a major secretory product of T cells stimulated with interleukin-12 (IL-12).4 Obviously, not all tumors respond to IFNs, and the effects of IFNs on cells can be modulated by many different agents. A cytoplasmic protein is present in tumor cells that can block the formation of DNA-binding proteins that recognize the IFN-stimulated response element of IFN-induced genes.5 T-cell cytokines such as granulocyte-macrophage colony-stimulating factor (GM-CSF) can also effectively block the ability of IFNs to activate monocytes, cells known for their ability to mediate cytotoxic effects against tumor cells.6 Therefore, the effects that monocytes exert in any environment depend to a large extent on the type of T-cell cytokines present at the time they interact with other cells.
Immune responses mediated by monocytes and activated T cells can be divided into two types, based on the expression of groups of T-cell–derived cytokines.7 Although most clearly defined in murine models of parasitic diseases, such as Leishmania,8 the ability to classify certain human diseases into one polarized type or another is a useful tool to learn more regarding the immunologic basis of that particular disease. A cellular immune response dominated by a T-helper lymphocyte 1 (Th1) infiltration, in which IFNγ and IL-2 predominate, is observed in diseases such as rheumatoid arthritis9 or inflammatory bowel disease.10 This response generally evokes a delayed-type hypersensitivity reaction. Immune responses regulated by T-helper lymphocyte 2 (Th2) cells result in the secretion of IL-4 and IL-10. This response is important for humoral immunity and mast cell development seen in such diseases as hyperimmunoglobulin E secretion syndrome.
It has been appreciated that cytokines secreted by Th2 cells can dramatically modulate the responses of cells involved in a Th1 immune response. For example, IL-4 can modulate the expression of early response genes induced by IFNα and IFNγ.11 Likewise, IFNγ can affect the response of cells to IL-4.12 Whereas it is known that IL-10 can effectively block lipopolysaccharide (LPS) activation of monocytes,13 its effects on direct IFN activation of genes are poorly understood. IFNs and IL-10 bind to their cognate receptors and initiate a signal that results in the activation of Janus kinase (Jak) and signal transducer and activator of transcription (STAT) proteins, leading to transcription of early response genes.14 IL-10 activates Tyk2, Jak1, STAT1, and STAT3.15 IFNα activates Tyk2, Jak1, STAT1, STAT2, STAT3, and STAT5. IFNγ activates Jak1, Jak2, STAT1, and, in some cell lines, STAT5.16 Although there is some overlap among the proteins activated by these three cytokines, there is no clear explanation from this information as to why one IFN or one IL should inhibit the action of others. We hypothesized several years ago that GM-CSF could inhibit the expression of IFNγ-induced genes by activating a new STAT protein that bound to the same γ activation sequence (GAS) element as IFNγ-induced STAT1 and thus prevent the induction of the FcγRI gene.17 This protein was subsequently determined to be STAT5. Although it is well established that IL-10 can suppress LPS-induced responses, which are by and large driven by NFκB, its direct effect on the ability of IFNα and IFNγ to activate cells by an NFκB-independent mechanism is virtually unexplored. In this report, we describe the ability of IL-10 to inhibit the expression of specific sets of IFN response genes. We show that IL-10 can suppress IFN-induced expression of several genes by inhibiting STAT1 tyrosine phosphorylation. In addition, we have gone on to show that IL-10 markedly upregulates expression of the SOCS3 gene, which is known to suppress cytokine activation.
MATERIALS AND METHODS
Monocytes.
Human peripheral blood leukocytes from healthy donors were separated into a mononuclear cell fraction and then were subjected to countercurrent centrifugal elutriation. The fractions enriched for monocytes were cultured in Dulbecco’s modified Eagle medium (DMEM; GIBCO, Grand Island, NY) without serum for 45 to 60 minutes in 4-well tissue culture plates until adherent, washed, and then incubated with DMEM with 10% fetal bovine serum (Hyclone, Logan, UT) and gentamicin (50 μg/mL). Cells prepared in this manner are greater than 95% monocytes by morphology and staining for anti-CD14. Cells were incubated with recombinant human IL-10 (rhuIL-10; Schering-Plough, Madison, NJ) for 60 to 90 minutes before the incubation with IFNα (Roferon; Roche, Nutley, NJ) or IFNγ (Genentech, South San Francisco, CA) for the specified times.
RNAse protection assay.
Steady-state levels of mRNA induced by either IFNα or IFNγ were measured by RNAse protection assay, as previously described.18 For the assay, probes were constructed such that each lane of the gel contained several protected fragments. γ-Inducible gene-10 (IP-10), a chemokine with anti-angiogenesis properties, was linearized with Pvu II, which yielded a protected fragment of 360 bp.19 ISG54, a gene induced by IFNα (and in monocytes by IFNγ), was linearized withPvu II, and this yielded a protected fragment of 367 bp.20 Guanylate binding protein (GBP; an IFNα- and IFNγ-induced gene) was linearized by Asp718.21 For an internal control for the amount of RNA loaded onto the gel, glyceraldehyde-3-phosphate dehydrogenase (GAPDH; linearized with Sau3a) was used and had a protected fragment of 300 bp.22 All hybridizations were performed overnight at 56°C. The RNA was subjected to digestion with RNAse T1, followed by separation on a 6% polyacrylamide/urea sequencing gel, and analyzed by phosphorimager analysis and autoradiography.
Northern analysis.
Total RNA was isolated from cultured monocytes by the acid guanidinium thiocyanate-phenol-chloroform extraction method, as previously described.23 RNA precipitates were pelleted by microcentrifugation and redissolved in 100 μL of diethylpyrocarbonate (DEPC)-treated sterile water. Equivalent amounts of RNA (10 μg/lane) were size-fractionated by electrophoresis in 1% agarose gels containing 0.66 mol/L formaldehyde. The RNA was then blotted by overnight capillary transfer onto Nytran membranes (Schleicher & Schuell, Keene, NH) and cross-linked by exposure to UV light. The membranes were then prehybridized, hybridized, and washed according to standard procedures. The SOCS3 probe was a 681-bp Mlu I fragment of the full-length murine SOCS3 cDNA,24 kindly provided by Dr Douglas Hilton (Walter and Eliza Hall Institute, Parkville, Australia). Gel-purified insert DNA was radiolabeled by the random primer method of Feinberg and Vogelstein25 to a specific activity of 108 cpm/μg or greater.
Electrophoretic mobility assays (EMSA).
EMSA using the γ response region (GRR) of the FcγRI receptor were performed using either whole-cell extracts or nuclear extracts, as described previously.26,27 This element is similar to the GAS element found within many IFNγ-induced genes. This probe was also used for the study of IFNα-induced genes, in addition to the IFN-stimulated response element (ISRE) from the ISG15 gene, which is recognized by the ISGF3 complex and is specific for IFNα-induced genes.11
Immunoprecipitations.
After treatment with IL-10 and/or IFNs, monocytes were solubilized with 1% TX100-containing buffer, incubated on ice for 15 minutes, and then centrifuged for 13,000g for 12 minutes to generate a postnuclear extract as described.15 The postnuclear extracts were then incubated with anti-STAT1 antibodies. The immunoprecipitates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, transferred to a polyvinylidene difluoride membrane, and immunoblotted with antiphosphotyrosine. Alternately, some extracts were analyzed directly by SDS-PAGE and immunoblotting with antiphosphotyrosine-STAT1 (New England Biolabs, Beverly, MA). Immunoblots were developed using either horseradish peroxidase or enhanced chemiluminescence (ECL) or nitroblue tetrazolium chemistry, respectively.
RESULTS
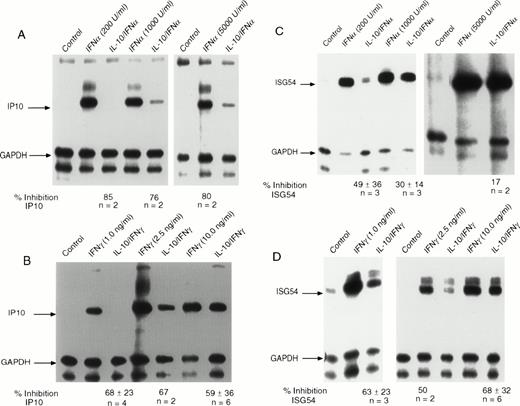
We initially addressed the question of which early response genes induced by IFNs might be inhibited by IL-10. Studies with STAT1-deficient mice showed that induction of IP-10 is dependent on STAT1 activation.28 The IP-10 gene encodes a chemokine of the cysteine-x-cysteine (CXC) type that is rapidly activated by IFNγ and IFNα. Recent data have shown that the IP-10 protein has strong anti-angiogenic properties and, in addition to inhibiting the development of carcinogenic tumors in several mouse models, has profound effects on chronic inflammatory conditions.29-31 We initially examined the effect of IL-10 on the induction of IP-10 in monocytes exposed to IFNα (Fig1A). We observed rapid induction of IP-10 expression that was markedly inhibited by IL-10 at all concentrations of IFNα tested. For some individuals (such as the one shown), nearly complete inhibition could be observed. Although the number of donors were only two per group, it was clear from the autoradiograph that at higher concentrations of IFNα, there was less inhibition by IL-10. IFNγ-stimulated IP-10 RNA expression was inhibited by pretreatment of monocytes with IL-10 (Fig 1B). At virtually all doses of IFNγ, over a range that occupies approximately 10% to 100% of the receptors for IFNγ, IL-10 suppressed IFNγ-induced IP-10 RNA expression by 59% to 68%. However, at the lower concentrations of IFNγ, inhibition tended to be greater. We went on to confirm that there was no change in receptor number of affinity by directly measuring the number of IFNγ receptors on monocytes after IL-10 treatment (data not shown).
IL-10 inhibits the induction of IP-10 and ISG54 by IFN and IFNγ. Monocytes were treated with 10 ng/mL of IL-10 for 60 minutes before treatment with the indicated doses of (A) IFN or (B) IFNγ for 90 minutes. RNA was isolated from the cells, and RNAse protection assays were performed. The arrows indicate the protected fragment corresponding to IP-10 and the control gene, GAPDH. IL-10 inhibits IFN-induced expression of ISG54 by (C) IFN or (D) IFNγ. The arrows indicate the protected gene fragment for ISG54 and the control gene, GAPDH. For all RNAse protection assays, the ratio of the values for the IFN-induced gene divided by those for the control (GAPDH) gene was computed, and for cells that had been pretreated with IL-10 followed by IFN. The values were calculated from Phospho-Imager data analysis. The percentage of inhibition was calculated by subtracting the values for cells treated with IL-10 from values for IFN-treated cells and dividing this number by the values for the cells treated with IL-10 and IFN. The values under the gel represent the mean of 2 to 6 experiments ± standard deviation at the various doses.
IL-10 inhibits the induction of IP-10 and ISG54 by IFN and IFNγ. Monocytes were treated with 10 ng/mL of IL-10 for 60 minutes before treatment with the indicated doses of (A) IFN or (B) IFNγ for 90 minutes. RNA was isolated from the cells, and RNAse protection assays were performed. The arrows indicate the protected fragment corresponding to IP-10 and the control gene, GAPDH. IL-10 inhibits IFN-induced expression of ISG54 by (C) IFN or (D) IFNγ. The arrows indicate the protected gene fragment for ISG54 and the control gene, GAPDH. For all RNAse protection assays, the ratio of the values for the IFN-induced gene divided by those for the control (GAPDH) gene was computed, and for cells that had been pretreated with IL-10 followed by IFN. The values were calculated from Phospho-Imager data analysis. The percentage of inhibition was calculated by subtracting the values for cells treated with IL-10 from values for IFN-treated cells and dividing this number by the values for the cells treated with IL-10 and IFN. The values under the gel represent the mean of 2 to 6 experiments ± standard deviation at the various doses.
In addition to IP-10, we also examined IFN activation of ISG54 (Fig1C). Although ISG54 was one of the first genes recognized as an IFN-inducible early response gene, virtually no information is available regarding the function of the protein that it encodes. IFNα-stimulated ISG54 is entirely dependent on the assembly of a complex of transcription factors, including STAT1, STAT2, and p48, that bind to the ISRE enhancer. Prior incubation of monocytes with IL-10 resulted in IFNα-induced inhibition of the accumulation of ISG54 RNA, compared with incubation with IFNα alone (Fig 1C). The observed inhibition by IL-10 was greater when cells were incubated with submaximal concentrations of IFNα (49% v 17%). There is some variability, and some donors clearly show inhibition of greater than 60%. Early reports discounted the ability of IFNγ to induce the ISG54 gene because of the lack of a GRR sequence within its 5′ regulatory region. However, IFNγ treatment of peripheral blood monocytes can clearly stimulate the accumulation of ISG54 RNA (Fig 1D). Incubation of monocytes with IL-10 showed a consistent suppressive effect on IFNγ-stimulated ISG54 RNA expression to a similar extent as that observed for the IP-10 gene. In many donors, the suppression reached levels of greater than 90% at lower doses of IFNγ (1 ng/mL, in this experiment; Fig 1D). These concentrations of IFNγ are generally at a level in which receptors are only 10% occupied, but genes are maximally expressed.27
In addition to IP-10 and ISG54 genes, we examined the ability of IL-10 to suppress the induction of several other IFN-inducible genes. These genes included ICAM-1 and IRF-1. Although the IRF-1 gene has GAS elements that drive its transcription by IFNγ, there was no perceptible inhibition by IL-10 in monocytes. For ICAM-1, only IFNγ was capable of inducing its expression; IFNα had no effect (data not shown). ICAM-1 expression is clearly important for immune cell adherence and tumor cell recognition.32,33 For those experiments in which this gene had increased expression of at least fourfold after exposure to IFNγ, pretreatment with IL-10 inhibited ICAM-1 RNA expression by 50% (over a range of IFNγ concentrations of 0.25 to 10 ng/mL; data not shown). These results are consistent with a recent report describing the inhibitory effects of IL-10 on IFNγ-induced ICAM-1 expression in monocytes.34
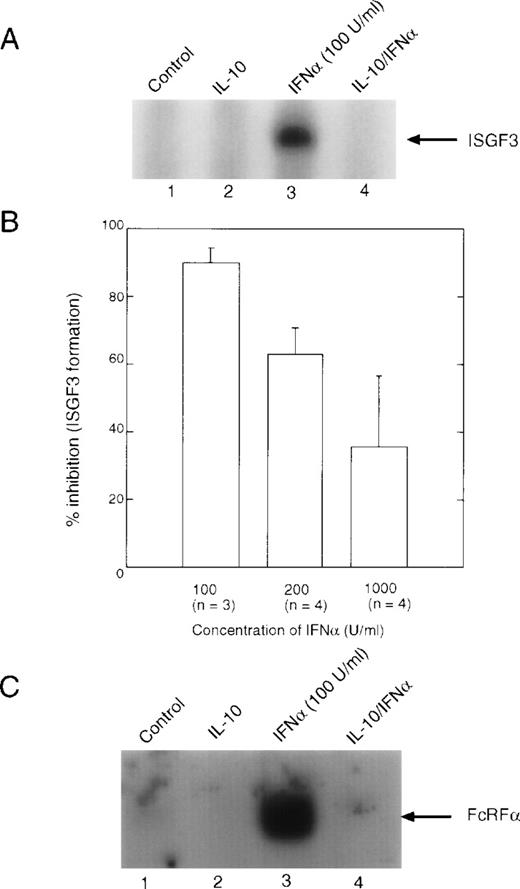
Both IFNα and IFNγ activate STATs, which activate gene expression by binding to ISRE and GAS elements in the promoters of IFN-inducible genes. IFNγ essentially activates only STAT1 in human peripheral blood monocytes, although one group has reported the activation of STAT5 in the histiocytic lymphoma cell line, U937.35 IFNα can activate several STATs, including STAT1, STAT2, STAT3, STAT4, and STAT5. Multimers of these STATs bind to ISRE or GAS enhancers to facilitate gene transcription. When we investigated the effect of IL-10 on the IFNα-induced formation of ISGF3 (Fig2A), we noted that there was a clear dose response with respect to IFNα concentrations, such that at 100 U/mL (0.5 ng/mL) of IFNα, inhibition by IL-10 was greater than 90%, yet at 1,000 U/mL of IFNα, inhibition by IL-10 had decreased to approximately 35% (Fig 2B). This result suggested that formation of ISGF3 was markedly inhibited by IL-10 at lower doses of IFNα. However, higher doses of IFNα could overcome to some extent this inhibition by increased receptor occupancy, suggesting that there may be a limiting amount of an inhibitory component induced by IL-10 whose function can be competed for when more IFNα binds to the cell. The addition of exogenous p48 to the reactions did not affect the ability of the complex to bind to the ISRE probe (data not shown). When a similar experiment was performed using the GRR probe, which is recognized by STAT1 dimers, the same result was observed (Fig 2C). Because binding to the GRR probe probably occurs through a mechanism different from that to the ISRE (ie, neither p48 nor STAT2 is involved), this result implies that STAT1 may potentially be a target of IL-10 suppression.
EMSA analysis of IFN-induced DNA binding complexes. Monocytes were treated with 10 ng/mL of IL-10 for 90 minutes, followed by treatment with the indicated doses of IFN for 30 minutes. Nuclear extracts were prepared, and EMSA were performed. (A) EMSA using the ISRE probe, which represents binding of the ISGF3 complex. (B) Summaries of the Phospho-Imager data from several experiments, such as those presented in (A), were quantitated using the Phospho-Imager. The value given is the mean ± standard deviation. (C) EMSA using the GRR probe, which predominantly measures binding of STAT1 dimers.
EMSA analysis of IFN-induced DNA binding complexes. Monocytes were treated with 10 ng/mL of IL-10 for 90 minutes, followed by treatment with the indicated doses of IFN for 30 minutes. Nuclear extracts were prepared, and EMSA were performed. (A) EMSA using the ISRE probe, which represents binding of the ISGF3 complex. (B) Summaries of the Phospho-Imager data from several experiments, such as those presented in (A), were quantitated using the Phospho-Imager. The value given is the mean ± standard deviation. (C) EMSA using the GRR probe, which predominantly measures binding of STAT1 dimers.
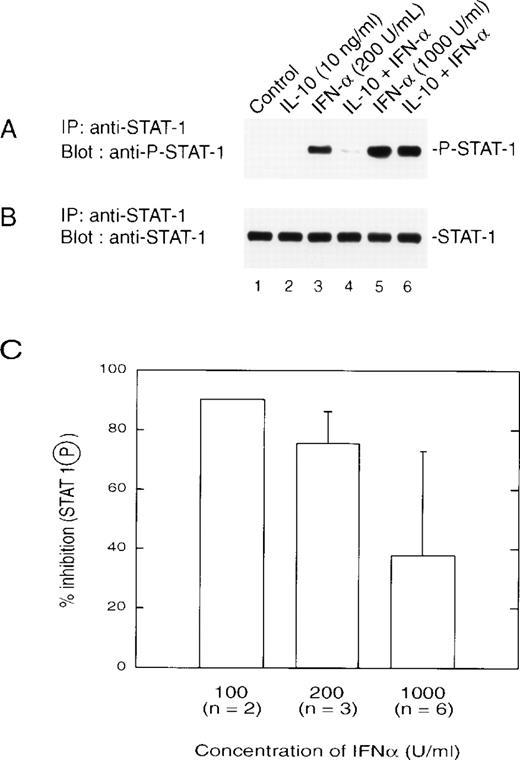
We next examined the effect of IL-10 on IFNα activation of STAT1 directly through the use of an antibody that recognizes only the tyrosine-phosphorylated form of STAT1 (Fig3A). IL-10 pretreatment of monocytes resulted in reduced levels of tyrosine-phosphorylated STAT1 when cells were treated with lower doses of IFNα (200 U/mL or lower). This result correlated with IL-10–mediated inhibition of IFN-induced gene expression and formation of ISGF3. All lanes expressed equal amounts of STAT1 loaded onto the gel (Fig 3B). With increasing doses of IFNα, there was a decrease in the inhibitory action of IL-10 on STAT1 tyrosine phosphorylation (Fig 3A, lanes 5 and 6), such that at 1,000 U/mL (5 ng/mL), inhibition was still present, but reduced to approximately 40% (Fig 3C). This was consistent with the results observed for gene induction and ISRE binding.
(A) IL-10 inhibits the IFN-induced tyrosine phosphorylation of STAT1. Monocytes were treated with 10 ng/mL of IL-10 for 60 minutes, followed by treatment for 30 minutes with the indicated doses of IFN at 200 U/mL (lanes 3 and 4) or 1,000 U/mL (lanes 5 and 6). Extracts were prepared, and equal amounts of protein were applied to an SDS-PAGE gel. Separated proteins were transferred and a Western blot was performed with an antibody that recognizes tyrosine-phosphorylated STAT1. (B) The blot was stripped and reprobed for STAT1. (C) The inhibitory effects of IL-10 on IFN-stimulated phosphorylation of STAT1 are dose-dependent. Monocytes were prepared as in (A), and the resulting autoradiographs were scanned for phosphorylated STAT1 using a Laser densitometer (LKB-Brommo, Piscataway, NJ). The numbers in parentheses refer to the number of experiments performed with different donors. The data are presented as the mean ± standard deviation.
(A) IL-10 inhibits the IFN-induced tyrosine phosphorylation of STAT1. Monocytes were treated with 10 ng/mL of IL-10 for 60 minutes, followed by treatment for 30 minutes with the indicated doses of IFN at 200 U/mL (lanes 3 and 4) or 1,000 U/mL (lanes 5 and 6). Extracts were prepared, and equal amounts of protein were applied to an SDS-PAGE gel. Separated proteins were transferred and a Western blot was performed with an antibody that recognizes tyrosine-phosphorylated STAT1. (B) The blot was stripped and reprobed for STAT1. (C) The inhibitory effects of IL-10 on IFN-stimulated phosphorylation of STAT1 are dose-dependent. Monocytes were prepared as in (A), and the resulting autoradiographs were scanned for phosphorylated STAT1 using a Laser densitometer (LKB-Brommo, Piscataway, NJ). The numbers in parentheses refer to the number of experiments performed with different donors. The data are presented as the mean ± standard deviation.
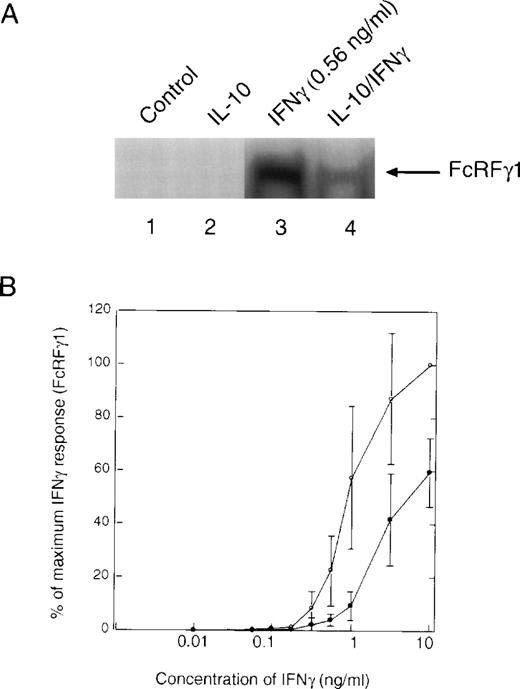
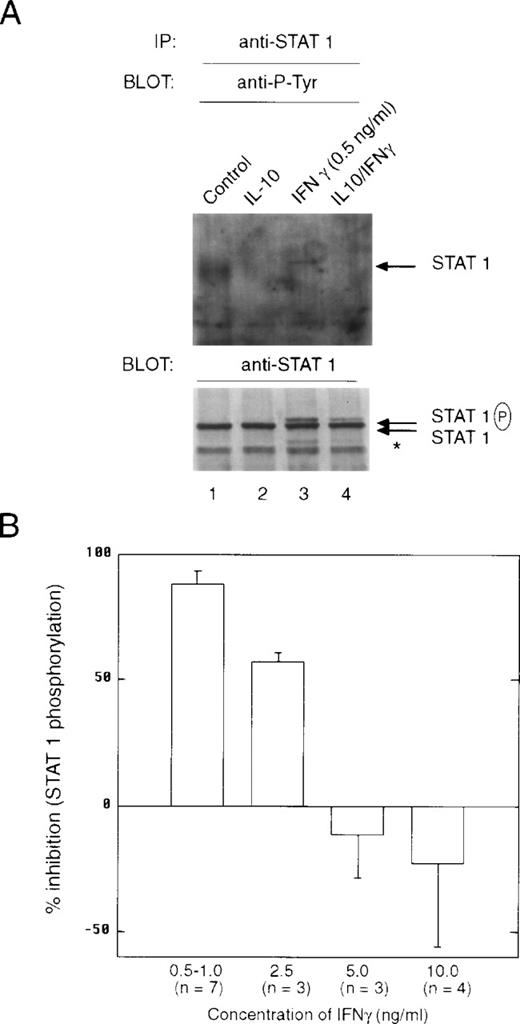
Pretreatment of cells with IL-10 also suppressed the ability of IFNγ-stimulated STAT1 dimeric complexes to bind to the GRR probe (Fig4A, lane 3 v 4). There was no evidence of IL-10–stimulated STAT binding to the GRR probe, because the time of treatment with IL-10 (t = 90 minutes) before IFNγ was sufficient for this binding to have dissipated. Under these conditions, IL-10 inhibited IFNγ activation of GRR-binding complexes at several concentrations of IFNγ (Fig 4B). The inability to repeatedly scan the same exact region in EMSA autoradiographs from different experiments leads to the variation observed in the plot. We also looked directly at the ability of IL-10 to block activation of STAT1 using phosphotyrosine immunoblotting. Monocytes were pretreated with IL-10 and then incubated with IFNγ for 15 minutes. The cells were lysed, and phosphorylation of STAT1 was assessed with an antiphosphotyrosine antibody. There was substantial inhibition of STAT1 tyrosine phosphorylation (Fig 5A, lanes 3 and 4). The levels of tyrosine phosphorylation of both the 91-kD and a lower molecular weight STAT1 (Fig 5A, asterisk) were markedly decreased by IL-10 pretreatment (Fig5A, lower panel). Because the preincubation of monocytes with vanadate, a phosphotyrosine phosphatase (PTP) inhibitor, had no effect on IL-10 inhibition (data not shown), it is unlikely that SHP1, a PTP, induces the effects of IL-10. Moreover, because only tyrosine-phosphorylated STAT1 translocates to the nucleus, a decrease in tyrosine phosphorylation implies an IL-10 event must occur at the receptor. Again, we observed a dose-response effect of the inhibition, such that at higher doses of IFNγ, the suppressive effects of IL-10 were much less evident (Fig 5B). However, in most inflammatory conditions, IFNγ concentrations are generally low, and the inhibitory effect of IL-10 is marked at concentrations of 1 ng/mL, in which activation of genes is generally at a near maximal level.
Effect of IL-10 on IFNγ-induced assembly of STAT1 dimers. (A) Monocytes were preincubated with 10 ng/mL of IL-10 for 90 minutes and then incubated with IFNγ for 20 minutes. Whole-cell extracts were prepared, and an EMSA was performed using the GRR probe, which is recognized by phosphorylated STAT1. The upper band of the shifted complex was presumed to be multimers of STAT1 binding to the probe. The autoradiograph represents that part of the total gel analyzed. (B) EMSA with GRR probe was performed as in (A), except that the indicated doses of IFNγ were used. The response was measured as a percentage of the maximum response of cells treated with IFNγ at 10 ng/mL. Three experiments were performed using individual donors, and the data are presented as the mean ± standard error. (○) Treated with IFNγ alone; (•) treated with IL-10 and IFNγ.
Effect of IL-10 on IFNγ-induced assembly of STAT1 dimers. (A) Monocytes were preincubated with 10 ng/mL of IL-10 for 90 minutes and then incubated with IFNγ for 20 minutes. Whole-cell extracts were prepared, and an EMSA was performed using the GRR probe, which is recognized by phosphorylated STAT1. The upper band of the shifted complex was presumed to be multimers of STAT1 binding to the probe. The autoradiograph represents that part of the total gel analyzed. (B) EMSA with GRR probe was performed as in (A), except that the indicated doses of IFNγ were used. The response was measured as a percentage of the maximum response of cells treated with IFNγ at 10 ng/mL. Three experiments were performed using individual donors, and the data are presented as the mean ± standard error. (○) Treated with IFNγ alone; (•) treated with IL-10 and IFNγ.
IL-10 inhibits the IFNγ-induced tyrosine phosphorylation of STAT1. (A) Monocytes were pretreated with 10 ng/mL of IL-10 for 60 minutes, and then treated with 0.5 ng/mL of IFNγ for 20 minutes. The upper panel represents an anti-STAT1 immunoprecipitation blotted with antiphosphotyrosine antibody. The lower panel represents the same membrane reprobed with anti-STAT1. The slower migrating band is tyrosine-phosphorylated STAT1 (STAT1P). The asterisk refers to a lower molecular weight band that is frequently observed in the immunoprecipitations and is related to STAT1 activation. (B) Monocytes were prepared as in (A), except that the indicated doses of IFNγ were used. Summary of laser densitometric scans of multiple antiphosphotyrosine immunoblots is shown. The values for percentage of inhibition were calculated, and the data are presented as the mean ± standard deviation for the indicated number of independent experiments.
IL-10 inhibits the IFNγ-induced tyrosine phosphorylation of STAT1. (A) Monocytes were pretreated with 10 ng/mL of IL-10 for 60 minutes, and then treated with 0.5 ng/mL of IFNγ for 20 minutes. The upper panel represents an anti-STAT1 immunoprecipitation blotted with antiphosphotyrosine antibody. The lower panel represents the same membrane reprobed with anti-STAT1. The slower migrating band is tyrosine-phosphorylated STAT1 (STAT1P). The asterisk refers to a lower molecular weight band that is frequently observed in the immunoprecipitations and is related to STAT1 activation. (B) Monocytes were prepared as in (A), except that the indicated doses of IFNγ were used. Summary of laser densitometric scans of multiple antiphosphotyrosine immunoblots is shown. The values for percentage of inhibition were calculated, and the data are presented as the mean ± standard deviation for the indicated number of independent experiments.
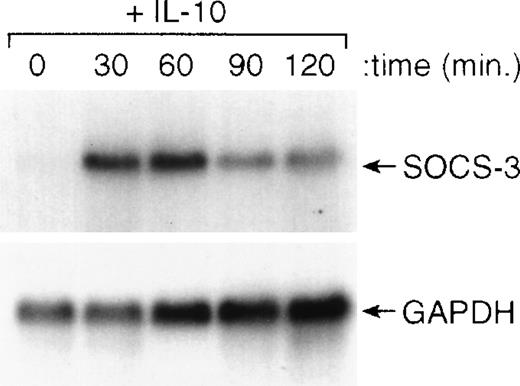
Recently, a series of genes have been cloned that are known to inhibit cytokine-induced activation of cells.36-39 To determine if IL-10 induced the expression of any of these genes, we treated cells with IL-10 and isolated RNA for Northern blot analysis. The results of this analysis showed that IL-10 could rapidly (within 30 minutes) induce the expression of the SOCS3 gene and maintain its expression for at least 120 minutes after the addition of IL-10 (Fig6). The kinetics of this induction of SOCS3 gene expression by IL-10 were consistent with its potential role in counterregulating IFN-induced STAT1 activation and IFN-induced gene expression. In preliminary experiments, we found that IL-10 did not upregulate expression of the related genes, CIS-1, SOCS-1, or SOCS-2, in monocytes (data not shown).
IL-10 induces expression of the SOCS3 gene in monocytes. Monocytes were incubated for the designated times with 10 ng/mL of IL-10 and then processed for RNA isolation. Northern blot analysis was performed as outlined in the Materials and Methods.
IL-10 induces expression of the SOCS3 gene in monocytes. Monocytes were incubated for the designated times with 10 ng/mL of IL-10 and then processed for RNA isolation. Northern blot analysis was performed as outlined in the Materials and Methods.
DISCUSSION
IL-10 suppresses the ability of LPS to elicit increases in mRNA of IL-1, tumor necrosis factor, IL-6, and other cytokines or cytokine receptors.13 In some of these studies, it has also been shown that the ability of IFNγ to enhance the effects of LPS can also be suppressed by IL-10.40 However, there has been no study addressing whether IL-10 inhibits IFNγ- and IFNα-stimulated early response genes in monocytes. Both IFNα and IFNγ induce the expression of these genes by activating STAT proteins that bind to either GAS or ISRE enhancers.16 In this report, we showed that IL-10 can inhibit the induction of IFN-induced genes by preventing the assembly of ISGF3 and GRR/GAS transcription complexes. The suppression of gene expression correlates with the IL-10–induced inhibition of the tyrosine phosphorylation of STAT1, which is required for this protein to bind DNA. Moreover, we have shown that the SOCS3 gene is induced by IL-10 and may be involved in the suppression of STAT1 phosphorylation.
IL-10 has numerous effects on hematopoietic cells, but most notable is its association with the development of Th2 lymphocytes. The polarization of T-cell development is thought to progress from a Th0 phase to either a Th1 or Th2 cell, depending on the presence or absence of various ILs.41 IL-10 is secreted by Th2 cells, and once secreted, it plays a major role in inhibition of monocyte/macrophage function, thereby suppressing the Th1 phenotype.42 Like IFNs, IL-10 activates STAT complexes.15,17,43,44 Incubation of monocytes with IL-10 activates primarily STAT3 and, to a lesser extent, STAT1.15 However, studies using STAT1-deficient mice indicated that STAT3 is important for IL-10 inhibition of LPS-induced monocyte activation.28 Because IL-10 activation of STATs is transient in monocytes (<30 minutes), by the time IFNs were added, activated STATs were not present (data not shown).
Because IL-10 treatment of monocytes does not alter the number or affinity of IFN cell surface receptors, another mechanism must be regulated by which IL-10 inhibits IFN-stimulated phosphorylation of STAT1. There has been recently discovered a new class of molecules referred to as SOCS proteins that seem to be induced by cytokines and then act as negative regulators to inhibit signal transduction.36-39 These molecules possess SH2 domains and can bind to phosphotyrosines on Jaks or cytokine receptors. Of interest, Jak1 is used by all three molecules: IFNα, IFNγ, and IL-10. There does seem to be some specificity with regard to the inhibitory actions of SOCS, because only SOCS1 and 3, but not CIS (an inhibitor of GM-CSF–induced STAT5), were able to inhibit growth hormone-induced STAT5 activation.45 Constitutive SOCS1 expression in transfected cells was shown to suppress IFN activation.36 No studies have shown whether IL-10 induces the expression of any SOCS genes. Experiments to detect association between Jaks and SOCS proteins have been very difficult using available antibodies. Only very high expression of a transiently transfected myc-JAB (CIS-like) gene and Jak2 (that is constitutively tyrosine phosphorylated) allowed coimmunoprecipitation of these two proteins.38 In this report, we do show that IL-10 can rapidly (30 minutes) induce the expression of SOCS3, possibly through its ability to induce tyrosine phosphorylation of STAT3. Whether STAT3-stimulated expression of SOCS3 accounts for the IL-10 inhibition of STAT1 tyrosine phosphorylation by IFNs is not yet known.
Another well-documented mechanism to inhibit cytokine signaling is the activation of tyrosine phosphatases that inactivate Jaks by dephosphorylation. Ligand-stimulated tyrosine phosphorylation of the erythropoietin receptor can be inhibited by targeting the phosphatase SHP1 (PTP-1C) to the receptor, resulting in dephosphorylation of Jak2.46 47 Although we have found that the amount of Jak1 is unchanged after IL-10 treatment, under the current culture conditions we have not been able to quantitate its degree of tyrosine phosphorylation, either by in vitro kinase assay or by immunoblotting with antiphosphotyrosine. When the cells were incubated with vanadate, a tyrosine phosphatase inhibitor, there was no evidence that the inhibitory actions of IL-10 were altered, suggesting that IL-10–regulated phosphatase activity is not involved (data not shown).
One common theme throughout the study has been the effect of the dose of IFN on the inhibitory action of IL-10. At high doses of either IFNα or IFNγ, the inhibitory action of IL-10 is reduced. At lower doses of IFN, less STAT1 is tyrosine phosphorylated in the presence of IL-10, suggesting the possibility that an event may be occurring at the plasma membrane that is regulated by IL-10. We and others have previously shown that Jaks are associated with IFN receptors, and STAT1 migrates to the receptor once IFN binds. Because the major event that determines the amount of tyrosine phosphorylation of STAT1 is the concentration of IFN binding to its receptor, it seems reasonable to hypothesize that the effect of IL-10 is to disrupt this multimolecular complex, such that the efficiency of STAT1 tyrosine phosphorylation is diminished. IL-10 induction of SOCS3 would be one such mechanism to account for the observed inhibitory actions of IL-10.
We have shown that IL-10 can inhibit the induction of certain IFN-induced genes. ICAM-1 was only induced by IFNγ and showed good IL-10–induced inhibition. The inhibition by IL-10 was fairly consistent among the genes, except for IRF-1. Although IRF-1 has a GAS motif that drives this gene, there was only a minimal effect by IL-10.48 This gene was readily induced by IFNγ in monocytes, but no induction was observed with IFNα (data not shown). There are several other elements in the promoter of this gene that may override IFNγ activation through its GAS element, if this enhancer is inhibited by incubation of cells with IL-10. These include Sp1 and NFκB binding sites. Recently, mice deficient in the IFN-induced-gene, double-stranded RNA-activated Ser/Thr protein kinase (PKR) were studied for their responsiveness to IFNγ.49 PKR is important for IFN-induced activation of NFκB. Cells from these mice showed no IFNγ-induced expression of IRF-1-promoter-reporter constructs. This response was restored on transfection with wild-type PKR, suggesting that NFκB may play an important role in mediating the expression of IRF-1.
It was recently shown that in human monocytes, there was no inhibition of GAS binding activity or STAT1 tyrosine phosphorylation after pretreatment with IL-10 in a study of ICAM-1 induction by IFNγ.34 One possible reason for the discrepancy between our results is the differences in the way the monocytes were prepared. In Song et al,34 the monocytes were passed over Ficoll-Hypaque and then allowed to adhere for 1.5 hours in medium containing 10% fetal bovine serum. This process allows for adherence of some B cells and possibly even some T cells. These monocyte preparations were generally only 85% pure. Moreover, Song et al34 used a concentration of IFNγ (10 ng/mL) that was 10-fold higher than the maximal concentrations of IFNγ we used to observe the effects described in this report.27 We observed a decrease in IFN-induced STAT binding to two distinct elements and a decrease in the corresponding STAT1 tyrosine phosphorylation for both IFNα and IFNγ. This suggests that, for cells isolated by our method with minimal handling and more physiological concentrations of IFN, incubation with IL-10 inhibits IFN-induced gene activation by preventing the phosphorylation of STAT1 that may occur through activation of a new set of genes that encode for proteins that suppress continued cytokine activation.24 The availability of antibodies to these molecules will enable further characterization of their mechanism of action. Our findings define an important mechanism by which IL-10 can antagonize IFN-induced gene expression in monocytes.
ACKNOWLEDGMENT
The authors thank Dr D. Hilton (Walter and Eliza Hall Institute, Parkville, Australia) for providing the SOCS cDNAs; Genentech, Inc (South San Francisco, CA) for IFNγ; Roche, Inc (Nutley, NJ) for IFNα; and Schering-Plough, Inc (Kenilworth, NJ) for IL-10.
Supported by the Oak Ridge Institute for Science and Education through an interagency agreement between the Department of Energy and the FDA (S.I., H.D., and N.V.).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to David S. Finbloom, MD, Division of Cytokine Biology, Center for Biologics Evaluation and Research, Food and Drug Administration, 29 Lincoln Dr (Bldg 29A, Room 2D20), Bethesda, MD 20892-4555.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal