Abstract
We have quantitated the relative contributions of the constitutively active 4β1 and 4β7 integrins and the domains embodying their cognate binding sites in mediating human B-cell lymphoma adhesion and chemotaxis on fibronectin. By cooperating, the central cell-binding and IIICS carboxy-terminal domains were entirely responsible for the adhesion activity displayed by fibronectin, and their relative contribution to this process was estimated to be 30% versus 70%. Assessment of the leukocyte-substrate binding strength (ie, dynes/cell) indicated a 10-fold higher avidity of the cell-IIICS domain interaction. The two integrins interchangeably recognized both domains, but differed quantitatively in their participation in the adhesive event, as well as in domain preference. The use of 3Fn (according to the nomenclature proposed by Bork and Koonin [Curr Opin Struct Biol 6:366, 1996] for the type III fibronectin modules) module-specific antibodies and recombinant polypeptides showed that 4 integrins recognized both the RGD sequence (3Fn10) and an apparently novel synergistic site located within the 3Fn8 module; even in this case, the integrins displayed a distinct binding site preference. Interleukin-1β (IL-1β)/IL-2–induced chemotaxis also involved cooperative function of the central cell-binding and IIICS domains, but the mechanisms regulating this phenomenon differed markedly from those controlling cell adhesion. First, the relative contribution of the individual domains was comparable, but neither of the individual domains promoted migration to the extent observed on intact fibronectin. Secondly, 4β1 and 4β7 integrins were both involved in the domain-binding necessary for initiation of migration, but the relative contribution of each receptor in the chemotactic process was less disparate than for initial cell adhesion. Thirdly, the mode by which chemotactic B-lymphoma movement was supported by the central cell-binding domain differed from that sustaining cell adhesion in that it involved independent recognition of either the 3Fn8 or the 3Fn9 module, which acted in synergy with the 3Fn10 module. Our data provide novel evidence concerning the relative importance of the constitutively active 4β1 and 4β7 integrins for the interaction of B-cell lymphoma cells with fibronectin, and they emphasize a multiple and diverse recognition of sites responsible for either anchorage or locomotion of tumor leukocytes on this matrix molecule.
THE INTERACTION OF normal and neoplastic leukocytes with fibronectin is thought to be critically involved in the regulation of leukocyte trafficking through tissues. The central cell-binding domain of fibronectin contains the RGD sequence, which is believed to be the primary cell adhesion site of this domain, and at least two additional, spatially well-separated cell-binding sites that are thought to operate synergistically.1-4 The carboxy-terminal IIICS variable region also contains two potential cell-binding sites designated CS-1 and CS-5 (connecting segment 1 and 5).5-8 We and other investigators have recently demonstrated that, although the RGD sequence is a critical determinant for the α5β1-mediated adhesion of neoplastic leukocytes to fibronectin, optimal interaction of the cells requires recognition of a cooperative cell adhesion site residing within 3Fn9 (according to the nomenclature proposed by Bork and Koonin9 for the type III fibronectin modules) and defined by PHSRN.8,10The arginine residue is the key amino acid of this site11and α5β1 integrin binding to it seems to be strongly influenced by the activity state of the receptor8 as well as the proper spatial orientation of the 3Fn9-10 ligand modules.12
The α4β1 and α4β7 integrins have been described as characteristic lymphocyte integrins reacting with the CS-1 cell attachment site within the IIICS region,7,13,14 in addition to a series of other cell surface and extracellular ligands, including vascular cell adhesion molecule-1 (VCAM-1), mucosal addressin cell adhesion molecule-1 (MadCAM-1), invasin, thrombospondin, and von Willebrand factor propeptide. However, several points remain unclear concerning this apparently rather broad interaction repertoire. First, it is not yet unambiguously determined whether all these ligands are truly functional in vivo. Secondly, it is not clear whether both integrins recognize all putative ligands equally well, simply by virtue of having in common the α4 subunit. Thirdly, the relative roles of the integrins in mediating adhesion and migration of lymphocytes on fibronectin remain to be firmly established. B-lymphoid cells, largely lacking α5β1, may alternatively use the α4β1 integrin to bind to the central cell-binding domain of fibronectin,15,16 but this putative RGD-recognition seems to be under strict control of the activation state of the receptor. This is believed because only integrin complexes in which the β1 subunit is artificially activated by ligation with the activating antibody TS2/16 are capable of exhibiting this binding specificity.15,16 Finally, although the α4β1 integrin may recognize multiple binding sites within the fibronectin molecule, the present evidence is that the human α4β7 integrin binds to the CS-1 site and a putative recognition sequence recently identified within 3Fn5.17-19
We have previously shown that two highly malignant cell lines, Ci-1 (lymphoblastic B-cell lymphoma) and Sc-1 (noncleaved Burkitt’s type lymphoma), bind constitutively to fibronectin and express high levels of α4 integrins and low to poorly detectable levels of α5β1.20 In this study, we have used these model cells to determine the relative contribution of the α4β1 and α4β7 integrins in the processes of cell attachment and cytokine-induced chemotaxis in response to the central versus IIICS cell-binding domains. Thus, we have addressed comparatively both the relative contributions of the two primary fibronectin domains in adhesion/migration phenomena and of the sites within these domains that are recognized by the two integrins.
MATERIALS AND METHODS
Antibodies and their specificities.
The specificity and sources of the monoclonal antibodies (MoAbs) used in this study were as follows: anti-β1 MoAb 4B4 was purchased from Coulter Scientifics, Ltd (Palo Alto, CA); activating anti-β1 antibody TS2/16 was provided by Dr Martin Hemler (Dana-Farber Cancer Research Institute, Harvard Medical School, Boston, MA); anti-α4 MoAb HP2-1 was kindly provided by Dr Francisco Sanchez-Madrid (Servicios de Immunologia, Hospital de la Princesa Università Autònoma de Madrid, Madrid, Spain); anti-α4 MoAb P4G2, anti-α5 MoAb P1D6, and anti-α5β1 complex MoAb JBS5 were purchased from Chemicon International (Temecula, CA); anti-α4β7 MoAb Act-1 was kindly provided by Dr Andrew Lazarovitz (Department of Nephrology, University of Ontario, London, Ontario, Canada); and anti-α4 MoAb PS2 (hybridoma) was purchased from ATCC (Rockville, MD). Antibodies against fibronectin included MoAb 333 directed against an epitope localized within the Fn3-10-11 module of fibronectin4; MoAb HFN-7 (which was purified from the corresponding hybridoma cells obtained from the ATCC) is directed against an epitope within the 3Fn9 module10; and MoAb IST-6 recognizes the 3Fn8 module.10 Antibody P3D4 (Chemicon International) was raised against a carboxy-terminal fragment of fibronectin and is known to block the interaction of cells with the 38-kD and 58-kD fragments of the A and B chains of fibronectin.21 MoAb FN15 (Sigma, St Louis, MO) reacts with the central cell-binding domain of fibronectin, but does not affect cell adhesion.
Fibronectin fragments, recombinant polypeptides, and synthetic peptides.
Bacterial expression proteins of molecular weight (Mr) 110,000 (110 kD) and Mr120,000 (120 kD), the latter one including the EDIIIBalternatively spliced 3Fn module, correspond to the entire central cell-binding domain of fibronectin and were produced essentially according to the procedures described in Moyano et al.19These recombinant proteins were indistinguishable in their attachment- and migration-promoting activity, indicating that the EDIIIB module did not contribute to the cell biological activities of the protein in our system. We have therefore used these proteins interchangeably throughout the study, and for simplicity, we consistently refer to the 110-kD polypeptide when describing the data related to these polypeptides. The Mr 38,000 (38 kD) tryptic fragment of fibronectin, comprising the Hep II domain and the putative cell attachment sites CS-1 through CS-3 of the IIICS region, was a generous gift from Dr Angeles Garcia-Pardo (Department of Immunology, Centro de Investigationes Biologicas, Madrid, Spain). A recombinant protein of Mr 52,600 and encompassing the carboxy-terminal 3Fn12-15 modules and the complete IIICS domain (ie, CS-1 through CS-5)22 was kindly provided by Dr Martin Humphries (Department of Biochemistry and Molecular Biology, University of Manchester, Manchester, UK). Because the two carboxy-terminal fragments supported cell adhesion to equivalent extents, they were used interchangeably throughout the study and described as the 38-kD fragment. Synthetic peptides with the composition RGDNP, GPenGRGDSPCA, and GDRGDSPASSK were purchased from GIBCO BRL, Life Technologies (Paisley, UK), and peptides with the composition GRGDSP, GRGESP, DELPQLVTLPHNLHGPEILDVPST (CS-1), GEEIQIGHIPREDVDYHLYP (CS-5), and DRVPHSRNSIT (B peptide) were synthesized on an automated Milligen 9050 peptide synthesizer (Millipore, Bedford, MA) using Fmoc chemistry and were purified by reverse-phase high-performance liquid chromatography (HPLC) according to standard procedures and analyzed for purity by amino acid analysis (identified active recognition sequences of these peptides are underlined). GRGD-containing peptides were found to be equally active and were used interchangeably throughout the study. Recombinant bacterial polypeptides corresponding to the fibronectin modules 3Fn4-6, 3Fn7-9, 3Fn9, and 3Fn10 were produced according to the procedures described in Moyano et al.19 These polypeptides are arbitrarily denoted R-3Fn4-6, R-3Fn7-9, R-Fn9, and R-Fn10. Another series of recombinant polypeptides was produced from previously generated cDNA clones by reamplification with adapter primers and cloning into the pQE-30 high expression vector (Qiagen, Hilden, Germany), in which the recombinant protein is fused to an N-terminal 6-His affinity tag. The positive clones were sequenced to rule out Taq polymerase errors, and the recombinant fragments were expressed by standard protocols. To improve refolding of these recombinant polypeptides, the fusion products were bound to a Ni2+ chromatography matrix, urea was removed, and the bound proteins were eluted with phosphate-buffered saline (PBS). The purity of the final products was ascertained by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). This protocol was used to generate the following polypeptides. R-3Fn3/8R (where R refers to the critical arginine of the recognition PHSRN motif) encompassing the 3Fn8 and 3Fn10 modules, but lacking the 3Fn9 module and including the PHSRN sequence derived from the 3Fn9 module, was generated. This sequence was inserted in the most homologous region identified between the 3Fn8 and 3Fn9 modules.23 R-3Fn8/10D is identical to R-3Fn8/10R, with the exception that the arginine residue in PHSRN was substituted by an aspartic acid residue (ie, PHSDN).23 R-3Fn6-10SPSDN encompasses 3Fn6-10, but carries a 16-amino acid mutation including a substitution of PHSRN → SPSDN of the 3Fn9 synergy sequence.11
Cell adhesion assay (centrifugal assay for fluorescence-based cell adhesion [CAFCA]).
The cell adhesion assay CAFCA (TECAN Group)23,24 used in this study is based on differential centrifugation25 and represents an extensive refinement of an assay previously described by our group.10,20 Major innovations include the use of commercially available six-well strips of flexible polyvinyl chloride (PVC) denoted CAFCA miniplates (TECAN Austria, Salzburg, Austria), covered with double-sided tape (bottom units), and fluorescent cell labeling with the vital fluorochrome calcein AM (Molecular Probes, Inc, Eugene, OR) for 20 minutes at 37°C. Labeled cells were rinsed extensively in Ca2+- and Mg2+-free PBS followed by one rinse in 50 mmol/L Tris-HCl, pH 7.2, containing 150 mmol/L NaCl (TBS) and 1 mmol/L EDTA, and then aliquoted into the bottom CAFCA miniplates at a density of 1 to 3 × 104 cells/well. Cell adhesion to substrates was assayed in TBS containing 0.1% bovine serum albumin (BSA), 1 mmol/L MgCl2, and 2% India ink as a fluorescence quencher. In some instances, cells were preincubated with the anti-integrin antibodies (1 to 3 μg/106 cells) anti-β1 (MoAb 4B4; inhibitory), anti-β1 (MoAb TS2/16; stimulatory), anti-α4 (MoAb HP2-1), anti-α4β7 (MoAb Act-1), or anti-α5 (MoAb P1D6) or with 1 to 100 μg of recombinant R-3Fn fragments in PBS for 20 minutes at 37°C and then plated onto the substrates. Synthetic peptides were added directly to the cell adhesion medium at final concentrations of 0.01 to 1 mg/mL. CAFCA miniplates were placed on specifically devised hard plastic holders (TECAN Austria) and centrifuged at 142gfor 5 minutes at 37°C to synchronize the contact of the cells with the substrate. The miniplates were then incubated for 30 minutes at 37°C and subsequently mounted together with a similar CAFCA miniplate lacking double-sided tape (top unit) to create communicating chambers for subsequent reverse centrifugation. Tests were initially performed to determine the minimal reverse centrifugal force required to detach cells nonspecifically adhering to a BSA-coated substrate, but yet sufficiently low to detect low-avidity binding to a matrix substrate. This force was established to be 12g when applied for 5 minutes at 37°C for the human B-cell lymphoma lines examined herein. Other sets of experiments, in which the miniplate assemblies were centrifuged at 12g, 45g, 100g, 177g, 399g, and 710g, were also performed to determine the relative strength of cell adhesion to the substrate. Higher centrifugation forces could not be used because they caused detectable damage to the cells. The relative number of cells bound to the substrate (ie, remaining bound to the bottom miniplates) and of unbound cells (in wells of the top miniplates) was estimated by top/bottom fluorescence detection in a computer-interfacedSPECTRAFluor microplate fluorometer (TECAN Austria). Fluorescence values were analyzed with the CAFCA software (TECAN Austria) to determine the percentage of adherent cells of the total cell population analyzed according to a previously devised formula.24-26 Relative strengths of cell adhesion in dynes/cell (Afd) were calculated as previously described24-26 according to the formula: Afd = (Dc − Dm) × Vc × Fc, where Dc is the specific density of the cell, previously established to be 1.07 g/mL25,26; Dm is the specific density of the medium equal to 1.00 g/mL; Vc is the volume of the cell; and Fc is the centrifugal force exerted on the bound cell. The average cell diameter of Sc-1 and Ci-1 cells was comparable and was established to be 9 μm. If the substrate contact area of cells binding to a specific ligand is estimated,Afd may also be converted to the recently proposed adhesion constant ι, ie, force-length2(dynes/cm2).27 Statistical significance, as determined using the Student’s t-test, was set at P< .001.
Cell motility assays.
Transmigration experiments involving chemotactic movement of the cells through a porous membrane were performed according to a novel fluorescence-based motility assay denoted fluorescence-assisted transmigration invasion and motility assay (FATIMA; TECAN Austria).26 Specifically devised Unicell-24 plates containing Transwell-like units in which a fluorescence-shielding polycarbonate membrane (Polyfiltronics, Inc, Boston, MA; TECAN Austria) with 5-μm pores was mounted were coated with 5 to 20 μg/mL of intact fibronectin or fibronectin-derived polypeptides (control wells had uncoated membranes) as described for CAFCA (50 μL/well), followed in some cases by incubation with various antifibronectin antibodies and extensive washing with RPMI. At the time of cell plating, the wells of the lower tray of the Unicell-24 plates were filled with 600 μL of RPMI with or without 5 ng/mL interleukin-1β (IL-1β) or 50 ng/mL IL-2 (ImmunoKontakt, Frankfurt, Germany), and 2 × 105cells suspended in 100 μL of RPMI were added to the top of each transwell. Optimal cytokine concentrations were determined in separate tests.
In perturbation experiments with anti-integrin antibodies, cells were preincubated for 30 minutes at 37°C with the various antibodies before being aliquoted into the Unicell-24 plates. Pilot tests indicated that a minimum of 24 hours of incubation was needed to observe substantial transmigration; consequently, all of the measurements were performed at 24 to 48 hours. Transwell units containing cells that had not transmigrated were removed, and the bottom trays containing the transmigrated cells were gently centrifuged for 5 minutes at 8g. Subsequently, 20 μmol/L calcein AM was added, and cells were further incubated for 20 to 30 minutes at 37°C, followed by fluorescence detection with theSPECTRAFluor fluorometer. The numbers of cells transmigrated were derived from the fluorescence values by extrapolation from parallel dilution curves of labeled cells. The percentage of transmigrated cells varied from 22% to 26% of the total added to the system, and nonspecific passage through membranes in the absence of surface coating was consistently less than 30% of the total migration observed. Statistical significance was determined as for CAFCA.
RESULTS
Constitutive B-cell lymphoma interaction with the central and IIICS fibronectin cell-binding domains.
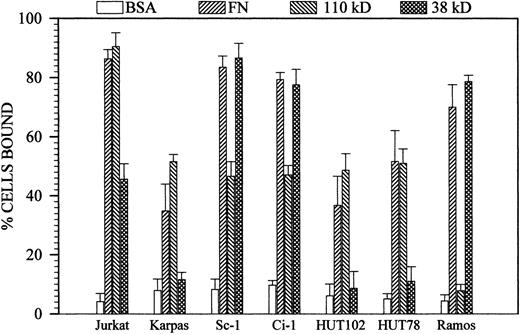
Sc-1 and Ci-1 B-lymphoma cells constitutively expressing high levels of α4β1 and α4β7 (as confirmed by flow cytometry; data not shown) adhered to both the carboxy-terminal, IIICS-containing 38-kD and to the 110-kD polypeptide corresponding to the entire central cell-binding domain (Fig 1A). Stable attachment to 38 kD and 110 kD did not require any prior chemical or immunological activation of the cells; accordingly, preincubation of the cells with the β1-integrin activating antibody TS2/16 did not alter the adhesion pattern of the cells (Fig 1A). Cell adhesion to both domains, as well as to intact fibronectin, was entirely cation-dependent and was equally promoted by physiological concentrations of either Mg2+ or Mn2+ (1 to 3 mmol/L or 30 to 100 μmol/L, respectively). However, Ca2+ failed to stimulate cell adhesion, even when used at concentrations up to 10 mmol/L (not shown). The simultaneous presence of Mg2+ and Mn2+ did not potentiate cell binding, and the addition of Ca2+ to cells binding in the presence of the former ions similarly did not affect the extent of cell adhesion (not shown). Cell adhesion to the 38-kD fragment largely paralleled that seen on intact fibronectin, when compared in molar equivalents (Fig 1A) and corrected for the independently established relative coating efficiencies of the proteins. The 110-kD and 120-kD fragments also supported substantial cell attachment that, in molar equivalents, corresponded to up to 30% of the maximal attachment seen on intact fibronectin (Fig 1A). These results indicate that the relative contributions of the two cell-binding domains to the promotion of Sc-1/Ci-1 cell adhesion to fibronectin were 70% versus 30%.
(A) Sc-1 and Ci-1 cell adhesion to 38-kD fragment and 110-kD polypeptide with and without addition of the anti-β1 activating antibody TS2/16. Data are presented as the ratio of cells bound to each of the two proteins when compared with cells adhering to the intact fibronectin molecule, after subtraction of nonspecific binding to BSA (5% to 10%). The extent of cell attachment was analyzed at various concentrations of the fragment/polypeptide and is expressed as molar equivalents of intact fibronectin. (B) Relative avidity of Sc-1 and Ci-1 cell binding to the 38-kD fragment and 110-kD polypeptide assessed by applying increasing centrifugal forces to detach the adherent cells.
(A) Sc-1 and Ci-1 cell adhesion to 38-kD fragment and 110-kD polypeptide with and without addition of the anti-β1 activating antibody TS2/16. Data are presented as the ratio of cells bound to each of the two proteins when compared with cells adhering to the intact fibronectin molecule, after subtraction of nonspecific binding to BSA (5% to 10%). The extent of cell attachment was analyzed at various concentrations of the fragment/polypeptide and is expressed as molar equivalents of intact fibronectin. (B) Relative avidity of Sc-1 and Ci-1 cell binding to the 38-kD fragment and 110-kD polypeptide assessed by applying increasing centrifugal forces to detach the adherent cells.
Although fewer cells adhered to the 110-kD polypeptide than to the 38-kD fragment, the application of CAFCA using differential centrifugation forces to determine cell-substratum binding avidities10,20 24-26 showed that Sc-1 cells attaching to the two fragments did so with roughly equal avidity (∼2.5 × 10−4 dynes/cell; Fig 1B). This avidity was comparable to that detected on intact fibronectin. Ci-1 cells, on the other hand, differed in the force of attachment to the two fragments, binding with estimated forces of 1.4 × 10−4 dynes/cell to the 38-kD fragment and 2.0 × 10−5 dynes/cell to the 110-kD polypeptide.
Comparative assays with a number of well-known B and T neoplastic leukocytes expressing one, two, or all three of the integrins α5β1, α4β1, and α4β7 clearly demonstrated that cells expressing high levels of α5β1 or medium/high levels of α4β1 (ie, Karpas 299, HUT78, and HUT102) strongly bound to the 110-kD polypeptide, whereas Ramos cells exclusively expressing α4-integrins bound solely to the 38-kD fragment (Fig 2). Jurkat cells expressing all three receptors at high levels, ie, α5β1, α4β1, and α4β7, characteristically showed more extensive binding to the 110-kD polypeptide than to the 38-kD fragment, and this behavior markedly differed from that of Sc-1 and Ci-1 cells (Fig 2). The observation that Ramos cells failed to recognize the central cell-binding domain in a constitutive manner28 is in agreement with previous studies showing that this ability is conferred to these cells only after immuno-mediated β1 integrin subunit activation.15 16
Comparison between the constitutive attachment profiles of Sc-1/Ci-1 cells and various neoplastic human T and B leukocytes adhering to intact fibronectin, the 110-kD polypeptide, or the 38-kD fragment. Integrin expression as determined by FACS was as follows: Karpas, HUT78 and HUT102 5β1high/4β1low; Ramos 4β1high; and Jurkat 5β1high/4β1high/4/β7high. Each substrate molecule was coated at a concentration of 10 μg/mL (not in molar equivalents). In all cases, binding to the control BSA substrates was less than 10%.
Comparison between the constitutive attachment profiles of Sc-1/Ci-1 cells and various neoplastic human T and B leukocytes adhering to intact fibronectin, the 110-kD polypeptide, or the 38-kD fragment. Integrin expression as determined by FACS was as follows: Karpas, HUT78 and HUT102 5β1high/4β1low; Ramos 4β1high; and Jurkat 5β1high/4β1high/4/β7high. Each substrate molecule was coated at a concentration of 10 μg/mL (not in molar equivalents). In all cases, binding to the control BSA substrates was less than 10%.
Identification of the sites responsible for the tethering of Sc-1 and Ci-1 cells to fibronectin.
A synthetic peptide containing the RGD cell-recognition motif did not affect Sc-1 and Ci-1 cell attachment to either the 110-kD polypeptide or 38-kD fragment, whereas it entirely blocked α5β1-mediated K562 (reference α5β1-positive/RGD-dependent cells) cell adhesion to the 110-kD polypeptide (Fig 3). Peptides containing the previously identified synergistic sequence PHSRN (B peptide) of the 3Fn9 module, and peptides corresponding to the penultimate segment (CS-5) of the IIICS domain, similarly failed to affect B-cell attachment to both the 110-kD polypeptide and 38-kD fragment (Fig 3). In contrast, the CS-1 peptide substantially blocked cell adhesion to both proteins (Fig 3). Preincubation of fibronectin substrates with the function-blocking antibodies 333 (occluding the RGD site) or P3D4 (blocking cell interaction with the CS-1 site) did not alter Sc-1 and Ci-1 cell adhesion to intact fibronectin when used alone (Fig 4). In contrast, these antibodies entirely abrogated cell binding when added in combination (not shown), confirming that, at least in this system, these two domains accounted for all cell adhesion activity exerted by the fibronectin molecule. Preincubation of 110-kD substrates with MoAb 333 resulted in a partial inhibition of cell adhesion (55%), as did similar preincubations with MoAb IST-6 directed against the 3Fn8 module. The scenario was different for MoAb HFN-7 reacting with the 3Fn9 module, which failed to perturb cell adhesion (not shown). The combination of MoAbs 333 and IST-6 entirely blocked cell binding to the 110-kD fragment, whereas the combination of MoAbs 333 and HFN-7 did not yield an additive effect (Fig 4). This latter finding indicated that abrogation of cell binding by the combination of MoAbs 333 and IST-6 was a consequence of the blockade of cooperating binding sites in 3Fn8 and 3Fn10 or possibly a site residing within the overlapping region between 3Fn8 and 3Fn9 in addition to the RGD site of 3Fn10.
Effects of synthetic peptides corresponding to known cell-binding sites on cell adhesion to the 110-kD polypeptide and 38-kD fragments. K562 cells, expressing constitutively high levels of 5β1 integrin, were used as reference cells to confirm the inhibitory activity of RGD-containing peptides and to determine the optimal blocking concentration. The optimum for these cells was established to be 50 μg/mL of GRGDSP peptides, and raising the concentration to 550 μg/mL still did not inhibit Sc-1/Ci-1 cell binding. Dose-dependent inhibition curves with CS-1 peptides showed a concentration optimum of 150 μg/mL peptide. B peptide and CS-5 were tested at concentrations up to 850 μg/mL.
Effects of synthetic peptides corresponding to known cell-binding sites on cell adhesion to the 110-kD polypeptide and 38-kD fragments. K562 cells, expressing constitutively high levels of 5β1 integrin, were used as reference cells to confirm the inhibitory activity of RGD-containing peptides and to determine the optimal blocking concentration. The optimum for these cells was established to be 50 μg/mL of GRGDSP peptides, and raising the concentration to 550 μg/mL still did not inhibit Sc-1/Ci-1 cell binding. Dose-dependent inhibition curves with CS-1 peptides showed a concentration optimum of 150 μg/mL peptide. B peptide and CS-5 were tested at concentrations up to 850 μg/mL.
Inhibitory effects of antifibronectin antibodies on Sc-1 and Ci-1 cell adhesion to intact fibronectin and the 110-kD polypeptide. Antibody P3D4 was used as a control antibody. Antibodies were used at concentrations of 1 to 10 μg/mL.
Inhibitory effects of antifibronectin antibodies on Sc-1 and Ci-1 cell adhesion to intact fibronectin and the 110-kD polypeptide. Antibody P3D4 was used as a control antibody. Antibodies were used at concentrations of 1 to 10 μg/mL.
Sc-1/Ci-1 cell adhesion to fibronectin is cooperatively mediated by α4β1 and α4<giGb7 integrins with diverse cell-binding site preferences.
The interaction of the Sc-1 and Ci-1 cells with intact fibronectin was unaffected by anti-α5 integrin subunit antibodies (MoAb P1D6; Fig 5A and B) and an antibody against the α5β1 complex (MoAb JBS5; data not shown). In contrast, adhesion of the cells to the intact molecule was entirely abrogated by MoAb HP2-1 against the α4 integrin subunit (Fig 5A and B). When added alone, the anti-β1 MoAb 4B4 partially blocked cell adhesion to intact fibronectin, and the anti-α4β7 MoAb Act-1 only had a marginal, but significant, effect. Simultaneous addition of the two antibodies strongly reduced the ability of cells to tether to fibronectin (Fig 5A and B). Cell binding to both the 110-kD polypeptide and 38-kD fragment was also entirely abrogated after the addition of MoAb HP2-1 (Fig 5A and B). However, cell adhesion to the 110-kD polypeptide was altered to an equivalent extent by MoAbs 4B4 and Act-1 (Fig 5A and B).
Inhibitory effects of anti-integrin antibodies, used alone or in combination, on Sc-1 (A) and Ci-1 (B) cell adhesion to intact fibronectin and its cell-binding domains: P1D6, anti-5 (identical results were obtained with the anti-5β1 MoAb JBS5); HP2-1, anti-4; 4B4, anti-β1; and Act-1, anti-4β7. To determine the optimal inhibitory concentration of each antibody, antibodies were individually titrated in independent tests and found to be optimally active at 1 to 3 μg/106 cells. (C) Relative strength of substratum adhesion estimated for Sc-1 cells binding to the 38-kD fragment or 110-kD polypeptide in the presence or absence of either MoAb 4B4 or MoAb Act-1, as determined by exposing bound cells to the centrifugal forces indicated on the abscissa.
Inhibitory effects of anti-integrin antibodies, used alone or in combination, on Sc-1 (A) and Ci-1 (B) cell adhesion to intact fibronectin and its cell-binding domains: P1D6, anti-5 (identical results were obtained with the anti-5β1 MoAb JBS5); HP2-1, anti-4; 4B4, anti-β1; and Act-1, anti-4β7. To determine the optimal inhibitory concentration of each antibody, antibodies were individually titrated in independent tests and found to be optimally active at 1 to 3 μg/106 cells. (C) Relative strength of substratum adhesion estimated for Sc-1 cells binding to the 38-kD fragment or 110-kD polypeptide in the presence or absence of either MoAb 4B4 or MoAb Act-1, as determined by exposing bound cells to the centrifugal forces indicated on the abscissa.
Exposure of Sc-1 cells that had attached to either the 38-kD fragment or 110-kD polypeptide to progressively incremented centrifugal forces in the alternative presence of MoAb 4B4 or Act-1 demonstrated that binding to the 110-kD polypeptide was strongly dependent on the activity of α4β7, whereas the opposite was true for cell attachment to the 38-kD fragment (Fig 5C). Assessment of the relative adhesive forces responsible for the cell-substratum interaction in the presence of the antibodies yielded an Adf of 5.35 × 10−5 dynes/cell for cells attached to the 38-kD fragment in the presence of MoAb Act-1 versus an Adf of 1.40 × 10−5dynes/cell in the presence of MoAb 4B4, when extrapolated to a centrifugal force retaining 50% of the cells originally bound. On the other hand, an Adf of 2.25 × 10−5 dynes/cell versus an Adf of 1.20 × 10−5 dynes/cell was estimated for cells attaching to the 110-kD polypeptide in the presence of MoAbs 4B4 and Act-1, respectively.
Localization of the sites responsible for the α4β1- and α4β7-mediated adhesion of Sc-1 cells to the central cell-binding domain of fibronectin.
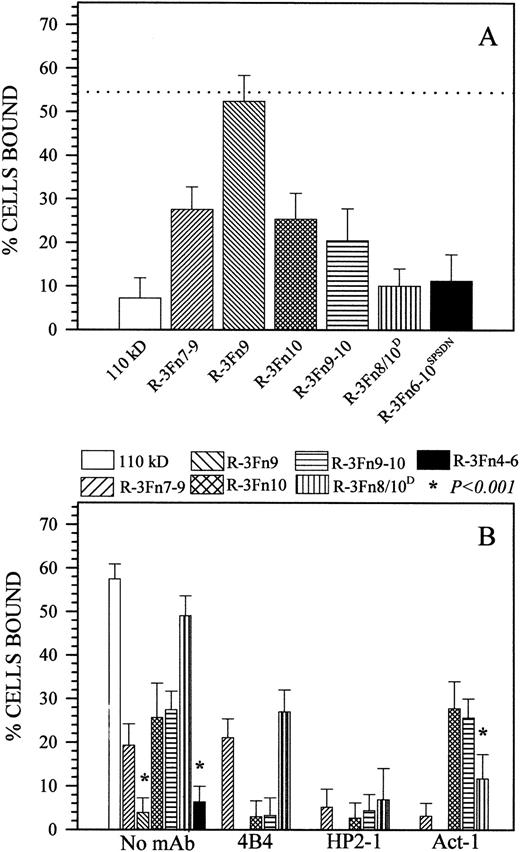
All R-3Fn polypeptides composed of the 3Fn8, the 3Fn10, or both modules strongly inhibited Sc-1 cell binding to the 110-kD polypeptide (Fig 6A), whereas polypeptides R-3Fn9 and R-3Fn4-6 (not shown) were completely inactive. A marked difference was also observed between the inhibitory efficiency of polypeptides R-3Fn6-10SPSDN and R-3Fn8/10D and the other polypeptides, with the former displaying competing activity comparable to the larger 110-kD polypeptide (Fig 6A). Sc-1 cell attachment to the immobilized recombinant polypeptides R-3Fn7-9, R-3Fn9-10, and R-3Fn10 was significantly (38% to 61%; P < .001) less pronounced than that to the intact 110-kD fragment (even when the polypeptides were coated onto plastic at a >10-fold molar excess in comparison with the parent fragment), whereas fragments R-3Fn9 and R-3Fn4-6 were completely inactive (Fig 6B). On the other hand, cell attachment to polypeptide R-3Fn8/10D was not significantly distinguishable (P > .01) from that seen on the 110-kD fragment when assayed at a 1:5 molar ratio (Fig 6B). However, in contrast to the findings with immobilized polypeptides, the inhibitory effect of soluble polypeptides R-3Fn6-10SPSDN and R-3Fn8/10D was not distinguishable from that of polypeptides R-3Fn7-9, R-3Fn9-10, and R-3Fn10 (Fig 6A). This observation indicated that, in liquid phase, the putative cell recognition sites exposed in both the R-3Fn8/10D and R-3Fn10 polypeptides were of equal importance for the anchorage of cells to the central cell-binding domain.
(A) Sc-1 cell binding to the 110-kD polypeptide after the addition of 110-kD or R-3Fn polypeptides. The dotted line indicates cell adhesion in the absence of competing polypeptide. Inhibition resulting from 110 kD, R-3Fn7-9, R-3Fn9-10, R-3Fn10, R-3Fn6-10SPSDN, and R-3Fn8/10D are significantly different. (B) Sc-1 cell adhesion to immobilized recombinant polypeptides in the presence or absence of anti-integrin antibodies at their optimal blocking concentrations. The amount of R-3Fn polypeptide coated onto plastic, or added in solution, was not in molar equivalents, because it is well known that the biological activity of such fragments is rather low3,11 23 and must be compensated for by using an excess of added protein. Fragments were therefore used at concentrations up to 100 μg/mL, based on independent dose-dependency tests, which established that the fragments were not significantly more active above the indicated concentrations. Antibody inhibition assays reported here do not include immobilized 110 kD, R-3Fn9, or R-3Fn4-6.
(A) Sc-1 cell binding to the 110-kD polypeptide after the addition of 110-kD or R-3Fn polypeptides. The dotted line indicates cell adhesion in the absence of competing polypeptide. Inhibition resulting from 110 kD, R-3Fn7-9, R-3Fn9-10, R-3Fn10, R-3Fn6-10SPSDN, and R-3Fn8/10D are significantly different. (B) Sc-1 cell adhesion to immobilized recombinant polypeptides in the presence or absence of anti-integrin antibodies at their optimal blocking concentrations. The amount of R-3Fn polypeptide coated onto plastic, or added in solution, was not in molar equivalents, because it is well known that the biological activity of such fragments is rather low3,11 23 and must be compensated for by using an excess of added protein. Fragments were therefore used at concentrations up to 100 μg/mL, based on independent dose-dependency tests, which established that the fragments were not significantly more active above the indicated concentrations. Antibody inhibition assays reported here do not include immobilized 110 kD, R-3Fn9, or R-3Fn4-6.
In accordance with data obtained using the larger 110-kD polypeptide, cell attachment to polypeptides R-3Fn7-9, R-3Fn9-10, R-3Fn10, and R-3Fn8/10D was completely blocked by the anti-α4 MoAb HP2-1 (Fig 6B). In the case of the R-3Fn9-10 and R-3Fn10 polypeptides, Sc-1 cell adhesion was also completely blocked by the anti-β1 MoAb 4B4, whereas cell attachment to the R-3Fn7-9 and R-3Fn8/10Dpolypeptides was either unaffected or only partially (45%) affected (Fig 6B). The anti-α4β7 MoAb Act-1, on the other hand, eliminated entirely cell binding to R-3Fn7-9 and reduced cell adhesion to the R-3Fn8/10D polypeptide by 75% (Fig 6B).
Differential role of the central cell-binding and IIICS fibronectin domains in cytokine-induced chemotaxis.
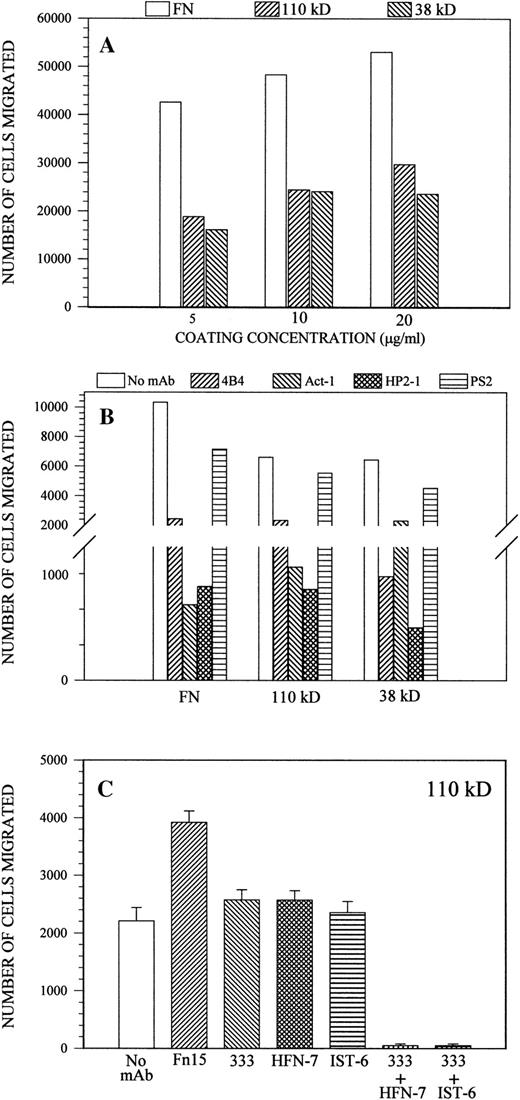
Both IL-1β and IL-2 independently stimulated the chemotactic movement of Sc-1 cells in response to the 110-kD and 38-kD substrates, but in contrast to what was observed for cell adhesion, these proteins did so to a largely comparable extent. However, none of the individual molecules was capable of promoting migration to an extent similar to that observed with intact fibronectin (Fig7A). Thus, assuming a comparable protein coating efficiency of the polycarbonate membranes as for plastic, the results indicated that the motility-promoting activity of fibronectin induced by the chemoattractive stimuli involved a 50% relative contribution of each of the central cell-binding and IIICS domains, and optimal leukocyte movement was only attained when both domains exerted their promoting effects in a cooperative fashion.
(A) Dose-related motility response of Sc-1 cells to intact fibronectin, 110-kD polypeptide, or 38-kD fragment under the chemoattractive stimuli of IL-1β/IL-2, as monitored FATIMA (see Materials and Methods). (B) IL-1β/IL-2–elicited chemotaxis of Sc-1 cells on intact fibronectin (10 μg/mL), 120-kD polypeptide, or 38-kD fragment (20 μg/mL) in the presence of antibodies to 4 (HP2-1 and PS2), β1 (4B4), or 4β7 (Act-1) integrins, added at their maximal inhibitory concentration. (C) Effects of antibodies against the central cell-binding domain of fibronectin on IL-1β/IL-2–induced Sc-1 cell chemotaxis. The membrane to be transmigrated was coated with the 110-kD polypeptide (20 μg/mL) followed by incubation with the different antibodies as described for CAFCA. FN15 corresponds to a cell-binding domain-directed MoAb with no reported functional effects.
(A) Dose-related motility response of Sc-1 cells to intact fibronectin, 110-kD polypeptide, or 38-kD fragment under the chemoattractive stimuli of IL-1β/IL-2, as monitored FATIMA (see Materials and Methods). (B) IL-1β/IL-2–elicited chemotaxis of Sc-1 cells on intact fibronectin (10 μg/mL), 120-kD polypeptide, or 38-kD fragment (20 μg/mL) in the presence of antibodies to 4 (HP2-1 and PS2), β1 (4B4), or 4β7 (Act-1) integrins, added at their maximal inhibitory concentration. (C) Effects of antibodies against the central cell-binding domain of fibronectin on IL-1β/IL-2–induced Sc-1 cell chemotaxis. The membrane to be transmigrated was coated with the 110-kD polypeptide (20 μg/mL) followed by incubation with the different antibodies as described for CAFCA. FN15 corresponds to a cell-binding domain-directed MoAb with no reported functional effects.
The IL-1β/IL-2–induced chemotaxis of Sc-1 cells, both in response to intact fibronectin and to the isolated 110-kD and 38-kD polypeptide/fragment, was in all cases effectively blocked by MoAb 4B4, HP2-1, or Act-1. As expected, the antimurine α4 integrin antibody PS2, known to display only a marginal cross-reactivity with the human integrin homologue, had only a minor effect (Fig 7B). The substantially less effective inhibition caused by addition of the anti-α4β7 MoAb Act-1 on cell movement on the 38-kD fragment, when compared with the 110-kD polypeptide and intact fibronectin (Fig 7B), was largely consistent with its effect on initial cell adhesion. However, in contrast to cell adhesion, estimation of the relative contribution of the two integrin receptors indicated 40% versus 60% in the contribution of the α4β7 and α4β1, respectively, in the binding to the central cell-binding domain. None of the recombinant R-3Fn polypeptides supporting cell adhesion was able to promote chemotactic migration of Sc-1 cells significantly above background levels (ie, on uncoated membranes; data not shown). Yet another difference between the requirements for initial cell adhesion and chemotaxis was that the single MoAbs 333 and IST-6 significantly blocked the chemotactic movement elicited by the cytokines and supported by either intact fibronectin or the 110-kD polypeptide (Fig 7C). In contrast, dual blockade with MoAbs 333 and IST-6 completely impeded Sc-1 cell chemotaxis (Fig 7C). Finally, an additional discrepancy observed between the inhibitory effects exerted by the anti-central cell-binding domain antibodies on cell adhesion and cytokine-induced motility was the ability of the anti-3Fn9 antibody HFN-7 to abrogate chemotaxis when used in combination with MoAb 333 (Fig 7C). This finding indicated the participation of a 3Fn9 binding site in the regulation of α4 integrin-mediated leukocyte movement on fibronectin.
DISCUSSION
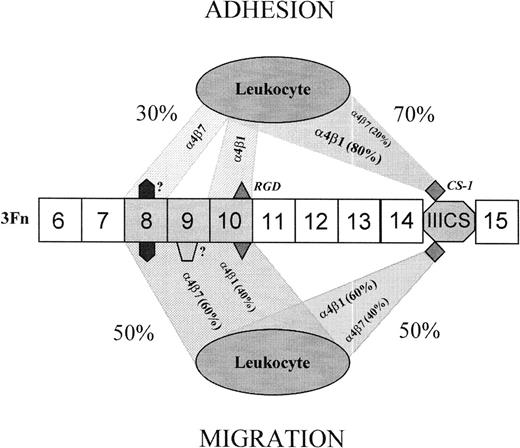
The present study shows that certain highly malignant, α4β1high/α4β7high/α5β1lowhuman B-lymphoma cells can recognize the central cell-binding region of fibronectin constitutively, ie, without any prior artificial cellular activation or immunological manipulation of their integrin receptor complexes. In fact, Sc-1 and Ci-1 cells, but not a number of other neoplastic leukocytes, were found to use this region of the molecule to functionally complement the alternatively spliced carboxyl-terminal IIICS region, as summarized in Fig 8. In the case of initial cell tethering, a tentative assessment of this complementarity indicated that the relative contribution of these two cell-binding domains was 70% to 30% in favor of the IIICS (when based on a quantification of the binding activity in molar equivalents). The corresponding relative contribution of the α4β1 and α4β7 integrins in this adhesion phenomenon was estimated to be 80% to 20%, as suggested by the results obtained after differential antibody blockade of the two integrins. Although acting in concert, the receptors’ capability to mediate cell interaction with the two domains seemed to differ in that the α4β1 integrin preferentially mediated binding to the CS-1 site, whereas the α4β7 was particularly important for the recognition of the central cell-binding domain. Both integrin receptors mediated binding to at least two sites within this latter domain: the RGD site and a synergistic site distinct from the previously reported PHSRN and likely to reside within the 3Fn8 module or at boundary region between 3Fn8 and 3Fn9. Even in this case, there was a differential preference for binding sites in that the α4β1 preferentially reacted with the RGD sequence and the α4β7 proved to be the pivotal receptor in the recognition of the putative 3Fn8 synergistic site (Fig 8).
Suggested model for the relative contributions of the cooperating 4β1 and 4β7 integrins and their cognate binding sites in the process of cell adhesion and migration on fibronectin. Both receptors interchangeably recognize at least 4 different binding sites residing within the central cell-binding and IIICS alternative spliced domains and acting in concert. Within the former, three 3Fn modules are the ones primarily implicated, ie, 3Fn8-10. Each of these modules embodies at least one binding site, which is differentially recognized by the integrins during the process of leukocyte adhesion and chemotactic migration. The RGD site is the active one of 3Fn10, whereas the synergistic sites residing within 3Fn8/3Fn9 remain unidentified. The CS-1 site seems to be the only active one of the IIICS domain. Both the relative contributions of the two domains and of the integrin receptors recognizing the corresponding sites within these domains differ in the processes of adhesion and migration. In the case of cell adhesion, 4β7 shows a bias towards the putative novel synergistic site within 3Fn8, whereas 4β1 preferentially binds to the RGD site of 3Fn10.
Suggested model for the relative contributions of the cooperating 4β1 and 4β7 integrins and their cognate binding sites in the process of cell adhesion and migration on fibronectin. Both receptors interchangeably recognize at least 4 different binding sites residing within the central cell-binding and IIICS alternative spliced domains and acting in concert. Within the former, three 3Fn modules are the ones primarily implicated, ie, 3Fn8-10. Each of these modules embodies at least one binding site, which is differentially recognized by the integrins during the process of leukocyte adhesion and chemotactic migration. The RGD site is the active one of 3Fn10, whereas the synergistic sites residing within 3Fn8/3Fn9 remain unidentified. The CS-1 site seems to be the only active one of the IIICS domain. Both the relative contributions of the two domains and of the integrin receptors recognizing the corresponding sites within these domains differ in the processes of adhesion and migration. In the case of cell adhesion, 4β7 shows a bias towards the putative novel synergistic site within 3Fn8, whereas 4β1 preferentially binds to the RGD site of 3Fn10.
In contrast to initial cell adhesion, the relative contribution of the two fibronectin cell-binding domains in IL-1β/IL-2–induced chemotaxis was virtually equal, and none of the cell- binding domains alone was capable of supporting levels of cell movement paralleling that sustained by intact fibronectin. In the chemotactic motility process, the integrins seemed equally important in mediating the migratory response, exhibiting a fully interchangeable binding to the IIICS and central cell-binding domains. However, the mode by which the cytokine-induced motility was supported by the central cell-binding domain of fibronectin differed from the initial cell adhesion event. In fact, the former involved multiple synergizing binding sites within the 3Fn8-10 modules (Fig 8).
The ability of B lymphomas, notably certain Burkitt’s type lymphomas such as Ramos and Daudi cells, to recognize the central cell-binding domain of fibronectin through their α4 integrins has previously been documented.15,16 However, this activity was demonstrated to require the currently debated, experimental antibody-mediated hyperactivation of β1 integrin receptors.29 It is now evident that a rather nonspecific mechanism of 3Fn module recognition can be elicited in cultured cells by antibody activation or by Mn2+ binding to the integrin subunits, irrespectively of the site or matrix molecule in which these modules are presented to the cells.30 Thus, we have identified here a novel physiological mechanism of α4-integrin interaction with the central cell-binding domain of fibronectin that may operate in untreated neoplastic B leukocytes. Moreover, our data establish that recognition of the RGD-containing region of fibronectin by constitutively active α4 integrins cannot be perturbed by soluble RGD-containing peptides, suggesting that, in contrast to α5β1 and αv integrins, α4 integrins require structural integrity of the RGD loop and its surrounding sequences. The discrepancies observed between constitutive versus artificially activated α4 integrins may also be due to differences in the ligand-induced, conformation-dependent modulation of the activity of these receptors.30 Alternatively, the divergent binding properties may be related to cell phenotype-specific regulation of these integrins, which confer on the receptors disparate intrinsic capacities to bind with high avidity to the various sites in the central cell-binding domain of fibronectin.
Our data suggest that α4 integrins display higher affinity for a sequence(s) contained within the CS-1 peptide (when provided in soluble form) than for the RGD sequence, when presented comparably in solution phase. This finding is in accord with a recent study showing that interconversion of the RGD cell-binding motif to LDV (contained by the CS-1 peptide) retains a potent antagonistic effect on α4β1 integrins.31 Moreover, binding of constitutively active α4β1 integrins to CS-1 and VCAM-1 can be perturbed by a cyclic peptide containing the RGD sequence.32,33These binding interactions can further be inhibited up to 1,000-fold more effectively by LDV-containing peptides synthesized in an analogous cyclic configuration.31-33 Thus, taken together, these previous observations and our results suggest that the actual activation state of α4 integrins may determine their relative affinity for synthetic peptides with different amino acid compositions and/or tertiary structures in solution, and they imply the following order of affinity: cyclic LDV > linear CS-1 > cyclic RGD > linear RGD peptides. Finally, because of the apparent high affinity of the α4 integrin subunit for the LDV sequence, it may not be surprising that the experimentally activated receptors can bind efficiently to KLDAPT peptides,19 in which the valine to alanine amino acid substitution may cause little or no change in the structural configuration of the binding site.
In analogy with the well-studied α5β1-mediated leukocyte interaction with fibronectin,8,10 we report that α4 integrin-dependent B-lymphoma binding to the central cell-binding domain of fibronectin seems to involve at least two independent binding sites. The latter conclusion derives from the observation that both MoAb 333, reacting with an epitope localized within the 3Fn10/3Fn11 modules,3 and the 3Fn8-directed MoAb IST-6 partially blocked cell adhesion to the central cell-binding domain when used alone, but completely abrogated the cell-fibronectin interaction when used in combination. The inhibitory effect of MoAb IST-6 identifies an ancillary synergistic cell adhesion site within the 3Fn8 module or possibly within the amino-terminal portion of the 3Fn9 module in the region adjacent to the 3Fn8. Experiments involving the use of antibodies against the α4 and β1 integrin subunits, and against the α4β7 heterodimer, showed that both receptor complexes were cooperatively responsible for the B-cell lymphoma-fibronectin interaction. However, their relative participation in this interaction differed. In fact, the contribution of α4β7 to the initial adhesion of cells to the central cell-binding domain was proportionally greater than that to the carboxyl-terminal IIICS region, whereas the corresponding relative contribution of the α4β1 integrin to the binding to the two separate domains was equivalent. This relationship was demonstrated by the twofold to fourfold difference in the force required to break the bonds between α4β1/α4β7 integrins and the 38-kD or 110-kD fragment/polypeptide in the presence of anti-integrin antibodies. Thus, although strongly supporting other investigators’ suggestions of a concerted action of the α4β1 and α4β7 integrins in mediating lymphocyte-fibronectin interaction, our observations establish quantitatively for the first time their relative contributions and provide new evidence that both integrin receptors can interact with multiple binding sites within the central cell-binding domain of the fibronectin molecule.
Our present data emphasize that the α4β7 integrin can display a particular preference for a putative synergistic site located within the 3Fn8 module, which is distinct from the previously identified PHSRN synergy site of the 3Fn9 module.8,23 Considering that α4 integrins may also recognize the recently identified KLDAPT site present within the 3Fn5 module,19 the physiological relevance of the versatile reactivity of these receptors with at least 5 disparate fibronectin sequences4-7,15,16 19 remains to be clarified. We speculate that the reason for needing two interaction sites within the central domain versus one in the carboxyl-terminal domain may be that, in cases in which the high-affinity CS-1 site is missing in fibronectin due to alternative splicing of the IIICS region, the synergizing low-affinity sites within the central cell-binding domain may cooperatively compensate for that loss. Naturally, this does not preclude transduction of different signals by the different sites to the cells in both situations.
Both fibronectin cell-binding domains were found to be capable of sustaining significant chemotactic cell movement in response to IL-1β and IL-2. However, in contrast to initial cell adhesion, the motility-promoting effect of the individual domains remained significantly lower than that exerted by intact fibronectin. These findings establish two important points: (1) differences in the involvement of the central cell-binding and IIICS domains in human B-lymphoma cell adhesion versus migration; and (2) an independent capability of the two domains to promote migration, with the central one being pivotal in the cytokine-induced chemotactic movement of the cells. Several interpretations can be put forward for the apparently diverse roles of the two primary cell-binding domains of fibronectin in mediating B-lymphoma adhesion versus migration. One possibility is simply that cell motility may be favored by low-affinity binding of the cells to multiple sites, whereas stable substrate anchorage may be preferentially promoted by high-affinity binding sites, even if fewer in number. Experiments involving antibody blockade of the RGD-containing 3Fn10 module and the upstream 3Fn8 and 3Fn9 modules, encompassing the putative synergistic sites, showed that leukocytes required free access to at least one of these latter synergistic sites to accomplish chemotactic migration. Either of these sites appeared to be equally effective in cooperating with the RGD site of the 3Fn10 module. Nonetheless, prolonged contact of the cells with the central cell-binding domain in which the RGD recognition site was occluded still permitted some locomotion of the cells, presumably due to the formation of transient contacts with the alternative binding sites within the 3Fn8 and 3Fn9 modules. This finding would preclude an absolute requirement for the RGD sequence in α4β1/α4β7-mediated chemotaxis. Another intriguing observation is that the isolated 3Fn9-10 and 3Fn8/10 modules (ie, polypeptides R-3Fn9-10 and R-3Fn8/10) failed to promote chemotactic movement of the cells, despite containing both the RGD and one each of the identified putative synergistic sites involved in the promotion of cell motility. Thus, we conclude that an effective cell locomotory process requires the presence and proper configuration of at least sites within the central cell-binding domain.
ACKNOWLEDGMENT
The authors are indebted to Drs Angeles Garcia-Pardo, Martin Hemler, Andrew Lazarovitz, and Francisco Sanchez-Madrid for providing various fibronectin fragments and anti-integrin antibodies. Maria Teresa Mucignat is thanked for her technical assistance.
Z.Y. and E.G. have contributed equally to this work.
Supported by grants Ricerca Finalizzata from the Fondo Sanitario Nazionale (FSN 1994-1995), Associazione Italiana Ricerca sul Cancro (AIRC), and Consiglio Nazionale delle Ricerche (CNR; Grant No. ACRO).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Roberto Perris, PhD, Division for Experimental Oncology 2, Centro di Riferimento Oncologico Aviano, Istituto Nazionale Centroeuropeo, Aviano (PN) 33081, Italy; e-mail:rperris@ets.it.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal