Abstract
In an effort to characterize molecular events contributing to lineage commitment and terminal differentiation of stem/progenitor cells, we have used differential display reverse transcription polymerase chain reaction (DDRT-PCR) and cell lines blocked at two distinct stages of differentiation. The cell lines used were EML, which is representative of normal multipotential primitive progenitors (Sca-1+, CD34+, c-Kit+, Thy-1+) able to differentiate into erythroid, myeloid, and B-lymphoid cells in vitro, and MPRO, which is a more committed progenitor cell line, with characteristics of promyelocytes able to differentiate into granulocytes. One clone isolated by this approach was expressed in MPRO but not in EML cells and contained sequence identical to the 3′ untranslated region of D3, a gene cloned from activated peritoneal macrophages of unknown function. We have observed a novel pattern of D3 gene expression and found that D3 is induced in EML cells under conditions that promote myeloid cell differentiation (interleukin-3 [IL-3], stem cell factor [SCF], and all-trans-retinoic acid [atRA]) starting at 2 days, corresponding to the appearance of promyelocytes. D3 RNA expression reached a maximum after 5 days, corresponding to the appearance of neutrophilic granulocytes and macrophages, and decreased by day 6 with increased numbers of differentiated neutrophils and macrophages in vitro. Induction of D3 RNA in EML was dependent on IL-3 and was not induced in response to SCF or atRA alone or SCF in combination with 15 other hematopoietic growth factors (HGF) tested. Similarly, D3 was not expressed in the normal bone marrow cell (BMC) counterpart of EML cells, Linlo c-Kit+Sca-1+ progenitor cells. D3 RNA expression was induced in these cells when cultured for 7 days in IL-3 plus SCF. A comparison of the expression of D3 RNA in cell lines and normal BMC populations demonstrated that D3 is induced during macrophage and granulocyte differentiation and suggests a potential physiological role for D3 in normal myeloid differentiation.
HEMATOPOIETIC development results in the formation of eight morphologically and functionally distinct blood cell lineages from pluripotential hematopoietic stem cells (PHSC). Mature cells have a limited life span and are continuously replenished from a pool of PHSC throughout an individual’s lifetime. Significant progress has been made in characterizing the PHSC and in understanding the growth factors/cytokines required for their survival, growth, and differentiation. Although less is known about the molecular events involved in PHSC differentiation, progress has been made in identifying lineage- and stage-specific genes and transcription factors contributing to the development and differentiation of normal PHSC.1-3 Genes involved in hematopoiesis have been identified using a variety of techniques. For example, studies of naturally occurring mutations in the mouse and oncogene-induced malignancy have elucidated critical roles for c-myb, c-myc, c-kit, and c-fos in hematopoiesis.3 Other genes have been identified using molecular approaches such as polymerase chain reaction (PCR) or low stringency hybridization to isolate genes with sequence similarity to known genes (eg, MZF-1) or by using subtractive hybridization techniques. Other genes, for example, AML1 and PLZF, have been identified by mapping sites of chromosome translocations or by their aberrant expression in leukemias.3
Several properties of stem cells have made the isolation and characterization of genes involved in stem cell development difficult. First, PHSC are present at low frequency in the bone marrow (0.01% to 0.005% of nucleated bone marrow cells).4 Second, even while these cells can be greatly enriched, they may still be heterogeneous. Therefore, to circumvent these difficulties, we used a murine lymphohematopoietic progenitor cell line, EML,5 to identify developmentally regulated genes. This cell line was generated by insertion of a retrovirus carrying a dominant negative retinoic acid receptor gene (RARα403) into bone marrow cells. EML cells are phenotypically similar to primitive murine hematopoietic stem cells, defined by Li and Johnson6 as Linloc-Kit+ Sca-1+ cells that are highly enriched for PHSC. EML cells are stem cell factor (SCF)-dependent and can differentiate into erythroid, myeloid, megakaryoctye, mast, and B-lymphoid cells in vitro. Myeloid differentiation is blocked in EML due to the expression of RARα403, but the block can be overcome by treatment with supraphysiological concentrations of all-trans-retinoic acid (atRA). A similarly derived cell line, MPRO,7 is granulocyte-macrophage colony-stimulating factor (GM-CSF)–dependent and is blocked at a more committed, promyelocyte stage of differentiation. MPRO cells possess morphology (enlarged cytoplasm) and cell surface marker expression (Mac-1 and 8C5 positive) characteristic of promyelocytes. Thus, the EML and MPRO cell lines are temporally related (separated by approximately 48 hours of differentiation) and provide a unique model to examine differences in gene expression between two distinct stages of myeloid differentiation.
In this study, we determined (1) whether the EML and MPRO cell lines could be used as a model system to isolate novel and potentially important genes involved in stem/progenitor cell growth and differentiation and (2) the expression pattern of such isolated genes in various lineages and whether these genes were regulated during myeloid cell differentiation. To accomplish this, we used differential display reverse transcription-polymerase chain reaction (DDRT-PCR). This method was designed to analyze changes in gene expression by amplifying short cDNA sequences representative of mRNAs present in cell populations. The amplified cDNAs are displayed on a sequencing gel, allowing both comparative analysis of gene expression during differentiation and the reamplification and cloning of differentially expressed cDNAs.8 Two clones isolated by this procedure have novel 3′ sequence: one is expressed in EML but not MPRO, and one is expressed at higher levels in EML than MPRO. A third clone, expressed in MPRO but not in EML, was identified as the previously described gene, D3.9 The function of D3 is unknown, and its potential role in myeloid differentiation has not been characterized; therefore, we chose to focus on the regulation of this gene during EML cell and normal hematopoietic cell differentiation.
MATERIALS AND METHODS
Cell lines and cytokines.
EML cells were maintained in Iscove’s modified Dulbecco’s medium (IMDM), 20% horse serum (GIBCO-BRL, Gaithersburg, MD), 2 mmol/L L-glutamine, and 1% penicillin-streptomycin supplemented with 8% to 15% BHK/MLK conditioned media (CM), as a source of SCF, or 100 ng/mL recombinant SCF (Peprotech, Rocky Hills, NJ) (EML complete medium). To induce the differentiation of EML cells, complete media was supplemented with 30 ng/mL interleukin-3 (IL-3; Peprotech) and 10−5 mol/L atRA (Sigma, St Louis, MO). MPRO and EPRO5 cells were maintained in DMEM, 10% fetal calf serum (FCS; Gemini Bio-Products, Calabasas, CA), 2 mmol/L L-glutamine, and 1% penicillin-streptomycin, supplemented with 20 ng/mL recombinant murine GM-CSF (Peprotech), and were differentiated with addition of atRA (10−5 mol/L). The following cell lines were maintained in RPMI 1640, 10% FCS, 2 mmol/L L-glutamine, and 1% penicillin-streptomycin, and supplemented as indicated: FDC-P1 (IL-3 dependent),10 32D-Cl3,1132D-Cl23,11 DA-3,12 and M-NFS-60 (ATCC, Manassas, VA), with 10% Wehi-3b conditioned media; FDC-P1 (IL-4 dependent) with 100 ng/mL IL-4 (Peprotech); and CTLL-213 with 20 ng/mL IL-2 (Cetus, San Francisco, CA). HCD-5714 was maintained in IMDM, 30% FCS, 5 × 10−5 mol/L 2-mercaptoethanol, 2 mmol/L L-glutamine, 1% penicillin-streptomycin, and 5 U/mL recombinant human erythropoietin (EPO; R.W. Johnson Pharmaceutical Research Institute, Raritan, NJ). NIH 3T3 (ATCC) cells were maintained in DMEM, 10% calf serum (GIBCO-BRL), 2 mmol/L L-glutamine, and 1% penicillin-streptomycin.
Isolation and purification of normal bone marrow cells (BMC).
Animal care was provided in accordance with the procedures outlined in the “Guide for the Care and Use of Laboratory Animals,” NIH Publication No. 86-23, 1985. Normal murine BMC were obtained by aspirating the femurs of 12-week-old female Balb/c mice. Light-density BMC (LDBMC) were separated by centrifugation on lymphocyte separation medium (Organon Teknika Corp, Durham, NC). Cells were washed twice in IMDM and resuspended in IMDM supplemented with 10% FCS (complete IMDM). LDBMC were purified to remove committed hematopoietic cells according to a previously described protocol.4 Briefly, LDBMC were resuspended in complete IMDM and incubated at 4°C for 30 minutes with a cocktail of antibodies, Ly-6G (Gr-1), CD4 (L3T4), CD45R (B220), CD11b (Mac-1), and CD8 (all from Pharmingen, San Diego, CA). Cells were washed twice and resuspended in complete IMDM and magnetic beads (Dynal, Great Neck, NY) were added at a ratio of 40:1 (beads/cell). The mixture was incubated for 30 minutes at 4°C and the cells were separated using a magnetic particle concentrator resulting in a lineage-low (Linlo) cell population.
To isolate Linlo c-Kit+ progenitors, Linlo cells were directly labeled with phycoerythrin (PE)-conjugated monoclonal antibodies (MoAbs) that recognize c-Kit or isotype-matched control antibodies (Pharmingen). Cells were incubated for 30 minutes at 4°C with the recommended concentration of antibodies, washed twice in complete IMDM, resuspended in the same medium to approximately 2 to 5 × 107 cells/mL, and separated by fluorescence-activated cell sorting (FACS; Becton Dickinson Co, San Jose, CA). To isolate Linloc-Kit+ Sca-1+ progenitor cells, Linlo cells were further purified by two-color FACS using fluorescein isothiocyanate (FITC)-conjugated c-Kit antibodies and PE-conjugated Ly6A/E (Sca-1) MoAbs (Pharmingen).
Normal B220+ B cells were purified by flow cytometry from a fresh suspension of mouse spleen cells using FITC-conjugated anti-B220 MoAbs or isotype-matched control antibodies (Pharmingen). GR-1+ granulocytes were isolated by flow cytometry from freshly aspirated cell suspensions of unfractionated mouse BMC using FITC-conjugated anti-8C5 MoAbs (Pharmingen). To isolate Ter119+ erythroid cells, unfractionated mouse BMC were labeled with biotinylated MoAbs that recognize Ter119 or isotype-matched control, stained with avidin-conjugated PE, and separated by FACS.
Soft agar colony formation.
To measure colony formation of EML cells in vitro, cells were suspended in 1 mL of EML complete media and 0.3% sea plaque agarose (FMC, Rockland, ME) with cytokines and plated in 35-mm Lux petri dishes (Miles Scientific, Naperville, IL), incubated at 37°C in 5% CO2 for 7 to 10 days, and scored for colony growth (>50 cells).
Proliferation assays.
To measure the proliferative response of EML cells to cytokines, cells were washed three times in complete media and plated at 1 × 104 cells/mL in microtiter plates in triplicate with single cytokines: 100 ng/mL SCF, 30 ng/mL IL-3, 50 ng/mL GM-CSF, 100 ng/mL IL-7 (Peprotech), 100 ng/mL IL-6 (Peprotech), 5 U/mL EPO, 50 ng/mL granulocyte colony-stimulating factor (G-CSF; Amgen, Thousand Oaks, CA), 100 ng/mL macrophage colony-stimulating factor (M-CSF; Peprotech), 20 ng/mL IL-1α (Hazleton, Vienna, VA), 500 U/mL interferon-γ (IFN-γ; Biosource Int, Camarillo, CA), and 20 ng/mL transforming growth factor β (TGFβ; Oncogene Corp, Seattle, WA). Cells were incubated for 48 hours and pulsed with 1 μCi3H-thymidine (6.7 Ci/mmol/L; NEN, Boston, MA) for the last 6 to 8 hours of incubation and then harvested (Tomtech Harvester 96; Tomtech, Inc, Orange, CT) onto glass-fiber filter paper (Filtermat A; Wallac Oy, Turku, Finland). Filter strips were dried and counted by liquid scintillation.
RNA isolation, electrophoresis, and Northern blotting.
Total RNA was purified using RNeasy (Qiagen, Chatsworth, CA) as outlined by the manufacturer. Separation of RNA samples (10 to 15 μg) by electrophoresis was performed on 1% agarose, 5.2% formaldehyde (37% solution), 1× MOPS gels. The running buffer was 1× MOPS, 5% formaldehyde. RNA was transferred to nylon membrane (MSI, Westboro, MA) and hybridized at 60°C in Church-Gilbert hybridization solution15 (0.5 mol/L NaPO4, pH 7.0, 1 mmol/L EDTA, 1% bovine serum albumin [BSA], 7% sodium dodecyl sulfate [SDS]). cDNA probes were random prime labeled using Prime-It II (Stratagene, La Jolla, CA) following the manufacturer’s instructions. To quantitate the levels of expression, Northern blots were stripped and reprobed with either hu-β-actin or G3PDH (Clontech, Palo Alto, CA). Signals were quantitated, relative to controls, by scanning densitometry.
DDRT-PCR and cDNA sequencing.
Total RNA from EML and MPRO was isolated as described above and treated with DNase I to remove DNA using MessageClean (GenHunter Corp, Brookline, MA). DDRT-PCR was performed using RNAimage (GenHunter) as described by the manufacturer. First, in three separate reactions using one of three primers (H-T11G, H-T11A, and H-T11C), RNA samples were reverse transcribed. Each of the first-strand cDNA pools was then amplified in the presence of α-35S-dATP in 4 separate reactions with either H-AP1 (5′-AAGCTTGATTGCC-3′), H-AP2 (5′-AAGCTTCGACTGT-3′), H-AP3 (5′-AAGCTTTGGTCAG-3′), or H-AP4 (5′-AAGCTTCTCAACG-3′) in combination with the primer used to reverse transcribe the sample. PCR products were separated on 6% denaturing polyacrylamide gels that were dried onto 3M paper (3MM Whatman, Maidstone, UK) and exposed to film. Bands that consistently appeared to be differentially expressed in two separate PCR reactions were isolated, reamplified, and cloned into the vector, pCR II (Invitrogen, Carlsbad, CA). The inserts were sequenced by automated sequencing (ABI Model 373, Foster City, CA) using M13 primers.
RT-PCR.
Total RNA was isolated as described, and 100 ng was reverse transcribed in 5 mmol/L MgCl2, 1× PCR buffer, 1 mmol/L dCTP, 1 mmol/L dGTP, 1 mmol/L dATP, 1 mmol/L dTTP (Perkin Elmer Core Reagents; Roche, Branchburg, NJ), 1 U/μL RNase Inhibitor (Promega, Madison, WI), and 2.5 U/μL Moloney murine leukemia virus reverse transcriptase (GIBCO-BRL). cDNA from 4 to 5 ng of total RNA input was then amplified by PCR using Perkin Elmer Core reagents as specified by the manufacturer. For amplification of actin, the primer sequences used were as follows: 5′ GAC ATG GAG AAG ATC TGG CAC 3′ and 5′ GAT CTT CAT GGT GCT AGG AG 3′. Standard conditions for PCR were denaturation at 94°C for 2 minutes; 25 cycles of 94°C for 1 minute, 60°C for 1 minute, and 72°C for 1 minute; and extension at 72°C for 7 minutes. For amplification of D3/204, the primers used were as follows: 5′ TCG GCT AAG AAC CAA AAA TCA CA 3′ and 5′ CAC TCC CCA CAA CTT CTA TCC TTC 3′. The full-length cDNA clones for D3 and 204 were generous gifts from Dr Thomas Hamilton (Research Institute, Cleveland Clinic Foundation, Cleveland, OH) and Dr Peter Lengyel (Department of Biophysics and Biochemistry, Yale University, New Haven, CT), respectively. Standard PCR conditions were as indicated above, except that cycle number was increased to 30.
RESULTS
Isolation of cDNA clones by DDRT-PCR.
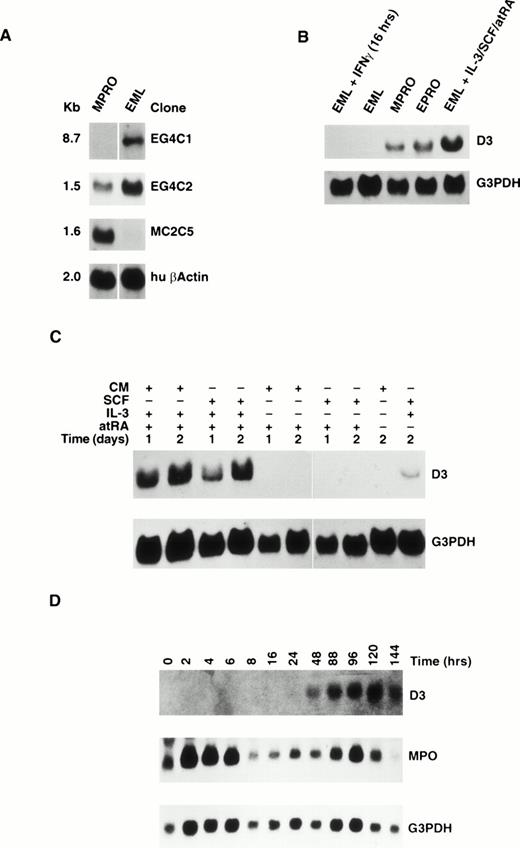
To isolate and characterize genes that are regulated during myeloid differentiation, DDRT-PCR was employed using EML and MPRO cell lines. Using 12 different primer pairs for comparison, 15 differentially expressed cDNA clones were obtained, 3 of which were confirmed to be differentially expressed in the EML and MPRO cell lines by Northern blot analysis (Fig 1A). One cDNA clone, designated EG4C1, hybridized to an 8.7-kb transcript expressed in EML cells that was absent in MPRO cells. The second cDNA clone, EG4C2, hybridized to transcripts of 2.7 and 1.5 kb that are downregulated in MPRO cells compared with EML cells. The third cDNA clone, MC2C5, hybridized to a 1.6-kb transcript expressed in the MPRO cell line that was absent in EML cells.
(A) Northern blot analysis of RNA extracted from the EML and MPRO cell lines. The blots were probed with partial cDNA clones isolated by DDRT-PCR and reprobed with human β-actin. (B) Northern blot analysis of RNA extracted from MPRO, EPRO, and EML cell lines and EML cells treated with IFNγ for 16 hours or IL-3/SCF/atRA for 6 days. The blot was probed with the 3′ UTR of the D3 gene and reprobed with G3PDH. (C) Northern blot analysis of RNA extracted from EML cells treated for 1 or 2 days with (+) or without (−) CM, SCF, IL-3, and atRA. The blot was probed with the 3′ UTR of D3 gene and reprobed with G3PDH. (D) Northern blot analysis of RNA extracted from EML cells cultured for 0 to 144 hours in IL-3/BHK CM/atRA. The blot was probed with D3 (top), MPO (center), and G3PDH (bottom).
(A) Northern blot analysis of RNA extracted from the EML and MPRO cell lines. The blots were probed with partial cDNA clones isolated by DDRT-PCR and reprobed with human β-actin. (B) Northern blot analysis of RNA extracted from MPRO, EPRO, and EML cell lines and EML cells treated with IFNγ for 16 hours or IL-3/SCF/atRA for 6 days. The blot was probed with the 3′ UTR of the D3 gene and reprobed with G3PDH. (C) Northern blot analysis of RNA extracted from EML cells treated for 1 or 2 days with (+) or without (−) CM, SCF, IL-3, and atRA. The blot was probed with the 3′ UTR of D3 gene and reprobed with G3PDH. (D) Northern blot analysis of RNA extracted from EML cells cultured for 0 to 144 hours in IL-3/BHK CM/atRA. The blot was probed with D3 (top), MPO (center), and G3PDH (bottom).
The nucleotide sequences of the three differentially expressed cDNAs were determined and compared with sequences in gene databases. No significant homology was found for the EG4C1 and EG4C2 clones, whereas the nucleotide sequence of the MC2C5 clone was identical to the 3′ untranslated region (UTR) of the previously described gene, D3, of unknown function. The D3 gene was initially shown to be induced during macrophage activation in response to IFN-β, IFN-γ, and lipopolysaccharide (LPS)16 and is a member of a family of genes called the 200 family.
D3 gene expression is induced in response to IL-3/SCF/atRA in EML.
To determine whether D3 is induced during myeloid cell differentiation, EML cells were induced to differentiate by culturing with SCF, IL-3, and atRA. D3 gene expression was analyzed by Northern blots with RNA from MPRO, EPRO, EML, and EML cells cultured for 6 days in IL-3, BHK conditioned media (CM) as a source of SCF, and atRA that were probed with the 3′ UTR specific to D3 (Fig 1B). The EPRO cell line is similar to MPRO, except that it was derived directly from the EML cell line by inducing EML differentiation with atRA for 2 days, generating cells blocked at the promyelocyte stage.5 As predicted, EPRO cells express approximately the same level of D3 RNA as MPRO cells, whereas EML cells show no detectable expression. However, D3 RNA is induced in EML cells cultured in IL-3, SCF (BHK CM), and atRA for 6 days. Furthermore, because D3 was originally shown to be rapidly induced by IFN-γ, EML cells were also cultured in SCF and IFN-γ for 16 hours. Lane 1 of Fig 1B demonstrates that, in EML cells, D3 is not induced in response to IFN-γ.
To determine what combinations of SCF, IL-3, and atRA were required to induce D3 expression, EML cells were cultured in recombinant SCF (rSCF) in the presence and absence of IL-3 and atRA. First, Northern blot analysis confirmed that D3 gene expression was induced with rSCF plus IL-3 and atRA to the same extent and with the same kinetics (27% between days 1 and 2) as with CM plus IL-3 and atRA (28% between days 1 and 2; Fig 1C). Second, neither CM nor rSCF alone or in combination with atRA was able to induce D3 expression. Third, IL-3 alone was not able to induce D3 expression within 48 hours of IL-3 stimulation (data not shown). Finally, low levels of D3 gene expression were observed when EML cells were cultured in a combination of IL-3 and rSCF for 2 days. Thus, IL-3 in combination with SCF is required to induce D3 RNA expression in EML cells. This effect is greatly enhanced in the presence of atRA.
To investigate whether other hematopoietic growth factors (HGF), with positive and negative effects on proliferation of EML cells and normal progenitor cells, could induce D3 expression, EML cells were cultured in rSCF in the presence or absence of atRA in combination with other HGF (GM-CSF, EPO, G-CSF, M-CSF, IL-7, IL-6, IL-1, IFN-γ, or TGFβ) for 2 days. To determine whether EML cells have biologically active receptors for these HGF, proliferation assays were performed with single HGF. Table 1 shows that EML cells respond to SCF, IL-3, GM-CSF, IL-6, IL-7, and TGFβ, indicating the presence of functional receptors for these HGF. Only the combination of IL-3 and SCF was sufficient to activate D3 gene expression, with much higher levels observed in the presence of atRA.
Cytokine-Induced D3 Expression in EML Cells
| Cytokine . | Receptor . | D3 Expression . | |
|---|---|---|---|
| +atRA . | −atRA . | ||
| SCF | + | − | − |
| IL-3 | + | +++ | + |
| GM-CSF | + | − | − |
| EPO | − | − | − |
| G-CSF | − | − | − |
| M-CSF | − | − | − |
| IL-7 | + | − | − |
| IL-6 | + | − | − |
| IL-1α | − | − | − |
| IFNγ | − | − | − |
| TGFβ | + | − | − |
| Cytokine . | Receptor . | D3 Expression . | |
|---|---|---|---|
| +atRA . | −atRA . | ||
| SCF | + | − | − |
| IL-3 | + | +++ | + |
| GM-CSF | + | − | − |
| EPO | − | − | − |
| G-CSF | − | − | − |
| M-CSF | − | − | − |
| IL-7 | + | − | − |
| IL-6 | + | − | − |
| IL-1α | − | − | − |
| IFNγ | − | − | − |
| TGFβ | + | − | − |
RNA was extracted from EML cells that were treated with SCF plus the indicated HGF in the presence or absence of atRA according to the procedures described in Materials and Methods. Northern blots of EML RNA were probed with D3 resulting in no expression (−), low expression (+), or moderate/high expression (+++). Northern blots were reprobed with G3PDH to verify equal loading in all samples. The presence (+) or absence (−) of biologically active receptors was determined in proliferation assays with EML cells using single cytokines as described in Materials and Methods.
Kinetics of D3 gene expression in EML during myeloid differentiation.
To determine the kinetics of D3 gene expression, RNA was obtained from EML cells treated with SCF, IL-3, and atRA for 0 to 6 days. Northern blot analysis demonstrated that D3 expression was induced beginning at day 2 (48 hours), with maximal expression observed at day 5 (70% increase in expression between days 2 and 5) that decreased after 6 days (Fig 1D). When D3 gene expression was compared with myloperoxidase (MPO), a granule protein known to be expressed in maturing neutrophils, MPO gene expression showed two peaks of expression. First, there was a rapid increase in MPO RNA levels 2 to 4 hours after treatment that returned to baseline levels of expression after 8 hours of treatment in culture. Then, a second peak of MPO RNA expression occurred at day 4, which decreased at day 5, and further decreased to levels below background by day 6 in culture. Thus, D3 is not induced rapidly (within 8 hours) in response to growth and differentiation signals, but rather corresponds to the second peak of MPO expression that correlates with the appearance of maturing granulocytes in culture5 (see below).
D3 expression in hematopoietic cell lines.
To determine whether D3 expression is lineage restricted, RNA was obtained from a panel of hematopoietic cell lines of various lineages and Northern blots were hybridized with D3 specific probes. First, seven myeloid progenitor cell lines were tested, each of which is blocked at a different stage of differentiation, although all are thought to be blocked at stages more primitive than promyelocytes. Of the seven, only NSF-58 contained D3 RNA expression levels comparable to MPRO cells (Table 2). Low but detectable expression was observed in FDC-P1 cultured in IL-3. Second, two T-cell lines (CTLL-2 and EL4) showed no D3 RNA expression, and no expression was detected in an erythroid cell line (HCD-57). Finally, one (Wehi-3b) of two myelomonocytic leukemia cells tested expressed D3 RNA. As a positive control for D3 expression, the macrophage cell line GG2EE was stimulated with IFN-γ to induce D3 mRNA expression. Thus, the expression pattern in cell lines indicates that D3 is restricted to cells of the myeloid lineage and that expression is absent in cells that represent more primitive stages of myeloid differentiation.
Expression of D3 RNA in Hematopoietic Cell Lines
| Cell Line . | Lineage . | D3 . |
|---|---|---|
| EML | Multipotential progenitor, SCF dependent | − |
| MPRO | Promyelocyte, GM-CSF dependent | ++ |
| EPRO | Promyelocyte, GM-CSF dependent | ++ |
| FDC-P1 (IL-3) | Myeloid progenitor, IL-3 dependent | + |
| 32D.Cl3 | Myeloid progenitor, IL-3 dependent | − |
| 32D.Cl23 | Myeloid progenitor, IL-3 dependent | − |
| DA-3 | Myeloid progenitor, IL-3 dependent | − |
| NFS-58 | Myeloid progenitor, IL-3 dependent | ++ |
| FDC-P1 (IL-4) | Myeloid progenitor, IL-4 dependent | − |
| M-NFS-60 | Myeloid progenitor, M-CSF dependent | − |
| EL-4 | T lymphoma, factor independent | − |
| CTLL-2 | T cytotoxic, IL-2 dependent | − |
| HCD-57 | Erythroleukemia, EPO dependent | − |
| M1 | Myelomonocytic leukemia | − |
| Wehi-3b | Myelomonocytic leukemia | +++ |
| GG2EE | Macrophage | − |
| GG2EE + IFNγ | + | |
| NIH 3T3 | Fibroblast | − |
| Cell Line . | Lineage . | D3 . |
|---|---|---|
| EML | Multipotential progenitor, SCF dependent | − |
| MPRO | Promyelocyte, GM-CSF dependent | ++ |
| EPRO | Promyelocyte, GM-CSF dependent | ++ |
| FDC-P1 (IL-3) | Myeloid progenitor, IL-3 dependent | + |
| 32D.Cl3 | Myeloid progenitor, IL-3 dependent | − |
| 32D.Cl23 | Myeloid progenitor, IL-3 dependent | − |
| DA-3 | Myeloid progenitor, IL-3 dependent | − |
| NFS-58 | Myeloid progenitor, IL-3 dependent | ++ |
| FDC-P1 (IL-4) | Myeloid progenitor, IL-4 dependent | − |
| M-NFS-60 | Myeloid progenitor, M-CSF dependent | − |
| EL-4 | T lymphoma, factor independent | − |
| CTLL-2 | T cytotoxic, IL-2 dependent | − |
| HCD-57 | Erythroleukemia, EPO dependent | − |
| M1 | Myelomonocytic leukemia | − |
| Wehi-3b | Myelomonocytic leukemia | +++ |
| GG2EE | Macrophage | − |
| GG2EE + IFNγ | + | |
| NIH 3T3 | Fibroblast | − |
RNA was extracted from the indicated cell lines for Northern blots that were probed with D3 according to the procedures outlined in Materials and Methods. Blots were reprobed with G3PDH to verify equal loading in all samples. No expression (−), low expression (+), and moderate/high expression (++/+++) of D3 are indicated.
D3 expression in normal tissues and normal hematopoietic cells.
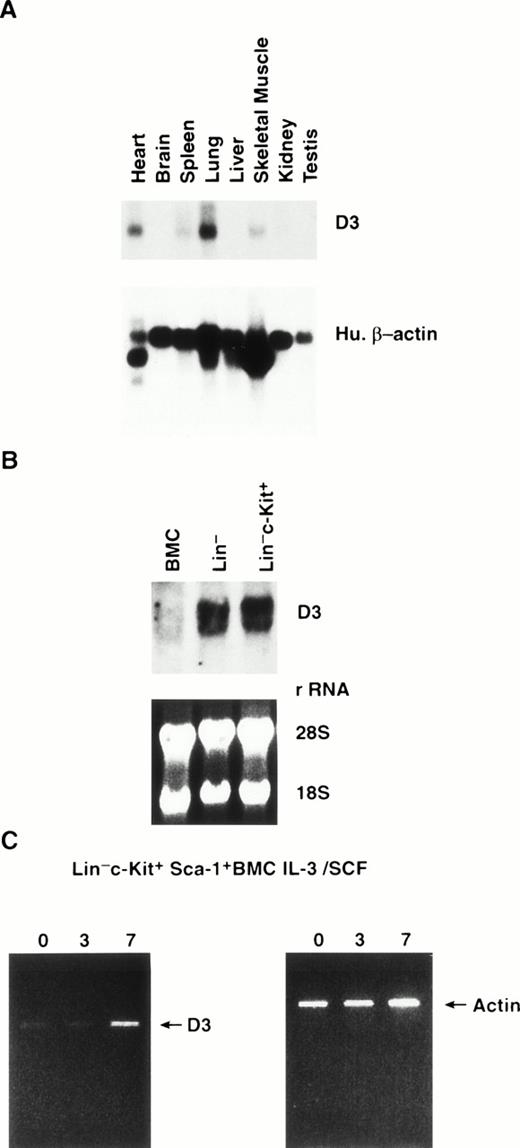
This analysis was extended to determine the expression of D3 in normal murine tissues. On multiple tissue Northern blots probed with a D3 specific probe, the highest mRNA levels were observed in the lung, with lower levels of expression detected in heart, spleen, and skeletal muscle (Fig 2A). D3 message was not detected in the brain, liver, kidney, or testis.
(A) Northern blot analysis of poly A+ RNA extracted from normal mouse tissues. The blots were probed with a D3 specific probe (top) and reprobed with human β-actin (bottom). (B) Northern blot analysis of RNA extracted from BMC, lineage-depleted BMC (Lin−), and Lin− cells purified by flow cytometry for expression of c-Kit (Lin−c-Kit+). The blots were probed with the D3 specific probe (top) and equal loading of samples was verified by ethidium bromide staining of rRNA (bottom). (C) RT-PCR analysis of D3 expression (left) in Lin− c-Kit+ Sca-1+ BMC cultured 0, 3, and 7 days in IL-3/SCF. Amplification of actin from each cDNA sample is shown (right).
(A) Northern blot analysis of poly A+ RNA extracted from normal mouse tissues. The blots were probed with a D3 specific probe (top) and reprobed with human β-actin (bottom). (B) Northern blot analysis of RNA extracted from BMC, lineage-depleted BMC (Lin−), and Lin− cells purified by flow cytometry for expression of c-Kit (Lin−c-Kit+). The blots were probed with the D3 specific probe (top) and equal loading of samples was verified by ethidium bromide staining of rRNA (bottom). (C) RT-PCR analysis of D3 expression (left) in Lin− c-Kit+ Sca-1+ BMC cultured 0, 3, and 7 days in IL-3/SCF. Amplification of actin from each cDNA sample is shown (right).
To determine the expression of D3 in normal hematopoietic cells, we isolated RNA from normal unseparated BMC, Linlo progenitor enriched BMC, and Linlo BMC further purified by flow cytometry for cells that express c-Kit. Little or no D3 RNA was detected in unseparated BMC; however, higher RNA levels were obtained in the progenitor purified Linlo and in Linloc-Kit+ cells (Fig 2B). On normal BMC Northern blots probed with the D3 specific 3′ UTR probe, two bands (∼1.6 and 1.8 kb) were detected as opposed to the single, 1.6-kb transcript detected on the cell line Northern blots.
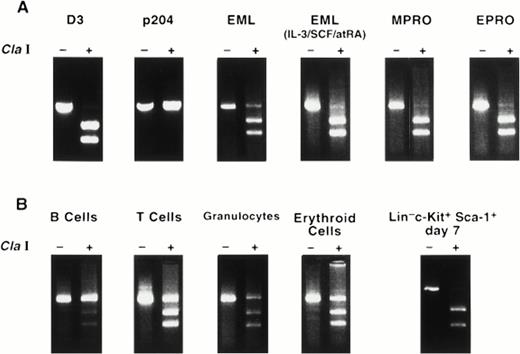
We further purified the Linlo, c-Kit+population using antibodies that recognize Sca-1 and flow cytometry to enrich for the most primitive progenitor cells, which phenotypically represent the normal counterparts of EML cells. Because low numbers of highly purified cells were obtained by this purification process, a PCR approach was developed to detect D3 message. The primer pairs used for the analysis amplify both D3 and the highly homologous family member, 204. However, the amplification products can be distinguished by restriction enzyme digestion with Cla I, which digests D3 once but does not cut within the 204 cDNA (demonstrated using plasmids containing cDNAs for either gene; see Fig 4A). Linloc-Kit+ Sca-1+ cells were cultured for 0, 3, or 7 days in IL-3 plus SCF, after which RNA was isolated and reverse transcribed with oligo (dT) and cDNA was amplified with the D3/204 primers (Fig 2C). To verify that equal quantities of cDNA were used in each PCR reaction, cDNAs were titrated and amplified with actin-specific primers and demonstrated to be equal within a linear range of amplification (data not shown). Low levels of amplified product were observed at days 0 and 3 in Linloc-Kit+ Sca-1+ cells, with a significant increase detected after 7 days in culture using the D3/204 amplification primers. Similarly, low levels of D3 expression were observed in EML cells at day 0, even though no D3 RNA can be detected by Northern blot analysis. Next, to determine whether the amplification product was D3 or 204, the amplification product from day 7 RNA was reamplified in a second round of PCR to obtain adequate amounts of material for the digests. The purified PCR fragments were then digested with Cla I. Figure 3B shows that the message amplified from Linlo c-Kit+Sca-1+ day 7 was completely digested with Cla I, indicating that the amplification product was D3 and not 204. In summary, little or no D3 expression is detected in primitive Linlo c-Kit+ Sca-1+ cells, but treatment of cells with the combination of SCF plus IL-3 induces D3 RNA expression, supporting the view that the EML cell line provides a model for this progenitor cell stage.
RT-PCR analysis of D3 expression in cell lines and in normal hematopoietic cells. (A) Plasmids containing the full-length cDNAs for D3 and 204 were purified and amplified using D3/204 primers, which amplifies a 505-bp band visualized by ethidium bromide staining. The amplification products were then purified and treated with (+) or without (−) Cla I. RNA extracted from EML, EML cultured in IL-3/SCF/atRA for 6 days, MPRO, and EPRO cell lines were reverse transcribed with oligo (dT), amplified with D3/204 primers, and treated with (+) or without (−) Cla I. (B) RNA from normal hematopoietic cells was purified and analyzed for D3 expression by RT-PCR as indicated above.
RT-PCR analysis of D3 expression in cell lines and in normal hematopoietic cells. (A) Plasmids containing the full-length cDNAs for D3 and 204 were purified and amplified using D3/204 primers, which amplifies a 505-bp band visualized by ethidium bromide staining. The amplification products were then purified and treated with (+) or without (−) Cla I. RNA extracted from EML, EML cultured in IL-3/SCF/atRA for 6 days, MPRO, and EPRO cell lines were reverse transcribed with oligo (dT), amplified with D3/204 primers, and treated with (+) or without (−) Cla I. (B) RNA from normal hematopoietic cells was purified and analyzed for D3 expression by RT-PCR as indicated above.
Using the PCR-based detection method for D3, we examined RNA obtained from normal T cells (thymocytes), B220+ B cells, 8C5+ granulocytes, and Ter-119+ erythroid cells. Figure 3 shows that the PCR product amplified in MPRO and EPRO cell lines is exclusively D3 (completely digested by Cla I), whereas normal B220+ B cells exclusively express the 204 RNA species (completely undigested by Cla I). In contrast, 8C5+ granulocytes, T cells, and Ter-119E+erythroid cells express both D3 and 204 RNAs.
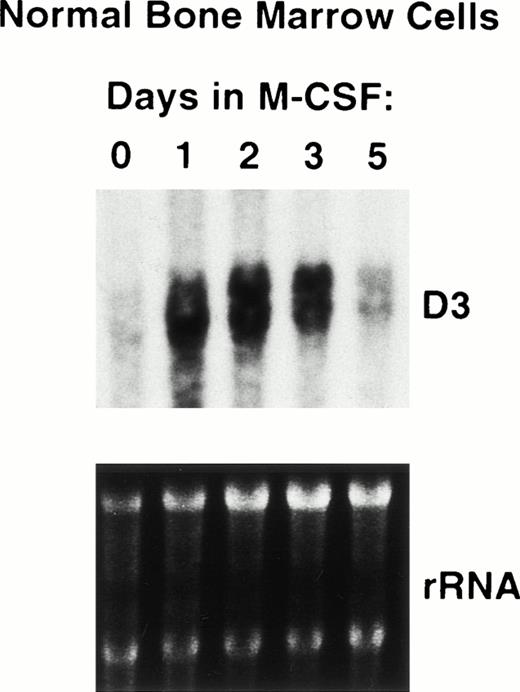
Finally, to determine whether D3 is expressed and/or induced during macrophage differentiation, we isolated total RNA from unseparated normal BMC and BMC cultured in M-CSF for 0 to 5 days (conditions that promote terminal unilineage macrophage differentiation in vitro). As shown in Fig 2, Northern blots probed with D3 detected little or no expression in unseparated normal BMC (Fig 4, lane 1). In addition, high levels of D3 RNA expression were observed after 1 day in cultures supplemented with M-CSF that were sustained until day 5, when D3 RNA expression levels decreased to the level observed in unfractionated BMC (Fig 4). This correlated with the appearance of mature differentiated macrophages (data not shown). Thus, D3 is upregulated during macrophage differentiation and is downregulated upon maturation. As previously reported, D3 expression can be induced again in mature macrophages when they are activated with interferons.9
Induction of D3 expression in differentiating bone marrow macrophages. RNA was extracted from normal BMC treated 0 to 5 days in M-CSF in vitro as described in Materials and Methods. RNA was analyzed on Northern blots, which were probed with D3 (top), and equal loading of RNA samples was verified by ethidium bromide staining of rRNA (bottom).
Induction of D3 expression in differentiating bone marrow macrophages. RNA was extracted from normal BMC treated 0 to 5 days in M-CSF in vitro as described in Materials and Methods. RNA was analyzed on Northern blots, which were probed with D3 (top), and equal loading of RNA samples was verified by ethidium bromide staining of rRNA (bottom).
Kinetics of morphological changes and colony formation in EML cells during differentiation.
To determine whether there was a correlation between biological events associated with EML cell differentiation and D3 gene expression, we performed a kinetic analysis of morphological changes and the production of more differentiated HGF-responsive colony-forming cells in differentiating EML cell cultures. Morphological changes in EML cells undergoing myeloid differentiation were assessed by culturing cells for 0 to 6 days in SCF and atRA with or without IL-3. On days 0, 2, 4, and 6, cells were cytospun and stained with Wright-Giemsa stain. Before treatment, approximately 95% of the EML cells were undifferentiated blasts (myeloblast) and promyelocytes (Table 3). By 2 days in IL-3, SCF, and atRA, the percentage of undifferentiated blasts/promyelocytes decreased to 78%, with an increase in granulocytic neutrophils with segmented or banded nuclei and macrophages with enlarged vacuolated cytoplasm (to 13% and 9%, respectively). After 4 days, 50% of the EML cells appear as undifferentiated blasts/promyelocytes, whereas 35% of cells are granulocytic neutrophils and 15% of EML are macrophages. On day 6, 34% of EML cells were undifferentiated blasts/promyelocytes, 44% were granulocytic neutrophils, and 22% were macrophages. These morphological changes are dependent on the addition of IL-3. GM-CSF in combination with SCF is unable to promote morphological changes associated with atRA-induced myeloid differentiation (data not shown).
Morphology of EML Cells Treated 0 to 6 Days in SCF/atRA in the Presence and Absence of IL-3
| Day . | IL-3 . | Morphology . | ||
|---|---|---|---|---|
| Blasts/ Promyelocytes . | Myelocytes Segs/Bands . | Monocyte/ Macrophage . | ||
| 0 | − | 94% | 0 | 6% |
| + | 94% | 0 | 6% | |
| 2 | − | 94% | 0 | 6% |
| + | 78% | 13% | 9% | |
| 4 | − | 94% | 0 | 6% |
| + | 50% | 35% | 15% | |
| 6 | − | ND | ND | ND |
| + | 34% | 44% | 22% | |
| Day . | IL-3 . | Morphology . | ||
|---|---|---|---|---|
| Blasts/ Promyelocytes . | Myelocytes Segs/Bands . | Monocyte/ Macrophage . | ||
| 0 | − | 94% | 0 | 6% |
| + | 94% | 0 | 6% | |
| 2 | − | 94% | 0 | 6% |
| + | 78% | 13% | 9% | |
| 4 | − | 94% | 0 | 6% |
| + | 50% | 35% | 15% | |
| 6 | − | ND | ND | ND |
| + | 34% | 44% | 22% | |
EML cells were induced to undergo myeloid differentiation in the presence of SCF/IL-3 and atRA as described in Materials and Methods. The morphology of the cells was determined by examination of cytocentrifuge preparations of cells obtained at the indicated days that were stained with Giemsa. Percentages were determined by counting 100 to 500 cells per slide.
Abbreviation: ND, not determined.
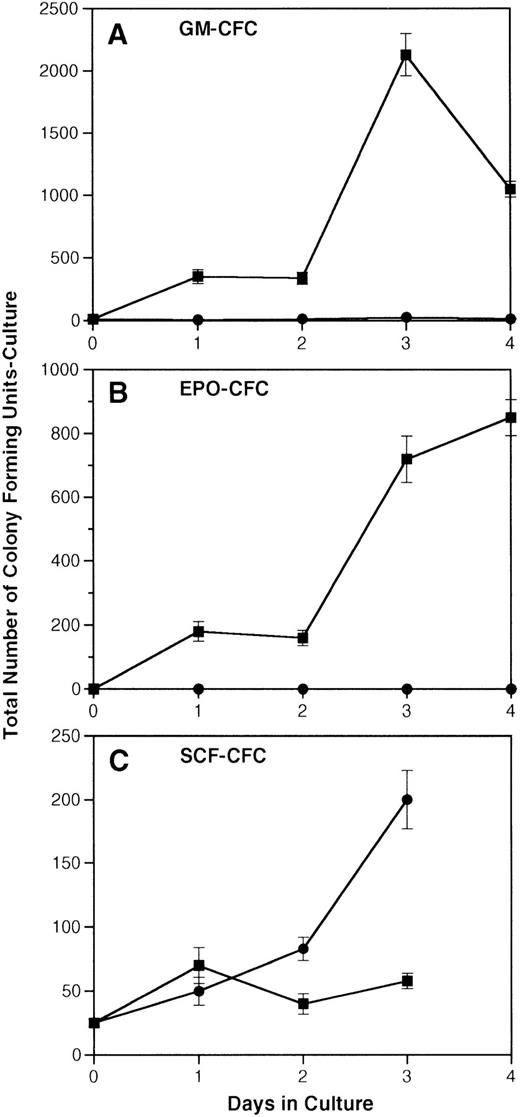
Over the same time course, production of more differentiated HGF-responsive colony-forming units-culture (CFU-c) was assessed by taking aliquots of cells cultured in SCF and atRA with or without IL-3 and replating them in soft agar colony assays containing either GM-CSF, EPO, or SCF (Fig 5A through C). The number of GM-CSF–responsive CFU-c generated from 1 × 105EML cells cultured in SCF, IL-3, and atRA went from 12 on day 0 to a maximum of 2,000 on day 3. In contrast, the maximum number of GM-CSF–responsive colonies produced in cultures containing only the combination of SCF and atRA was 26 on day 3. The number of EPO-responsive colonies produced in cultures of EML in IL-3, SCF, and atRA reached a maximum of 850 on day 4 versus none generated by culture with SCF and atRA alone. Lastly, the number of SCF-responsive colonies remained fairly constant over time in cultures containing SCF/IL-3 and atRA, whereas there was an increase in SCF-responsive colonies from 2.5 × 104 to 2 × 105 generated with SCF plus atRA (the same increase was seen in cultures with SCF alone). Taken together, using two different assays to measure differentiation (morphology and generation of CFU-c), D3 RNA expression in EML cells correlates with the appearance of morphologically differentiated neutrophils and the generation of more mature cells responsive to GM-CSF and EPO.
Induction of HGF-responsive CFU-c progenitors from EML cells. EML cells were induced to undergo myeloid differentiation according to the procedures described in Materials and Methods. (A) The total number of GM-CSF–responsive colonies generated from 1 × 105 EML cells cultured in SCF/atRA with or without IL-3. (B) The total number of EPO-responsive colonies generated from 1 × 105 EML cells cultured in SCF/atRA with or without IL-3. (C) The total number of SCF-responsive colonies generated from 1 × 105 EML cells cultured in SCF/atRA with or without IL-3. (▪) SCF + RA + IL-3; (•) SCF + RA.
Induction of HGF-responsive CFU-c progenitors from EML cells. EML cells were induced to undergo myeloid differentiation according to the procedures described in Materials and Methods. (A) The total number of GM-CSF–responsive colonies generated from 1 × 105 EML cells cultured in SCF/atRA with or without IL-3. (B) The total number of EPO-responsive colonies generated from 1 × 105 EML cells cultured in SCF/atRA with or without IL-3. (C) The total number of SCF-responsive colonies generated from 1 × 105 EML cells cultured in SCF/atRA with or without IL-3. (▪) SCF + RA + IL-3; (•) SCF + RA.
DISCUSSION
It is becoming increasingly clear that hematopoiesis is regulated, in part, by proteins that regulate growth, commitment, and differentiation and that aberrant expression of these regulatory proteins can contribute to leukemogenesis. In this study, our goal was to address whether the EML cell line could be used as a model to isolate and characterize novel genes involved in normal hematopoiesis. To this end, we performed DDRT-PCR using RNA from the EML and MPRO cell lines and demonstrated that these cell lines are particularly suited to differential screening techniques, because they represent clonal populations of cells arrested at distinct, temporally related stages of myeloid differentiation. The utility of these cell lines in allowing us to isolate molecules involved in normal myeloid differentiation can be quickly confirmed by comparing expression in normal stem/progenitor cell populations. Also, because EML can be induced to differentiate into other lineages depending on the culture conditions used, we predict that the system described here may be similarly useful in isolating and characterizing genes involved in erythroid, megakaryocyte, mast, and lymphoid cell differentiation.
In this report, we demonstrated the isolation of two novel cDNAs and a cDNA identical to a previously described gene, D3, of unknown function. The D3 gene was originally isolated from peritoneal macrophages treated with macrophage activating agents, including interferons and LPS.16 D3 is a member of a multigene cluster of interferon-inducible genes (200 family). There are at least four and possibly six members in the mouse (p202, p204, p203, and D3) and three human homologues (MNDA, IFI16, and AIM2) that map to a conserved linkage group in human band region 1q21-32 and distal mouse chromosome 1.17-22 However, there is currently no known biological function for any of the 200 family members of interferon-inducible genes.
We have demonstrated here that D3 is induced during myeloid differentiation by using both cell line models and normal cells allowing us to determine (1) the growth factors/cytokines involved in the induction of D3 expression, (2) the kinetics of this expression during myeloid differentiation, and (3) the lineage restriction of D3 gene expression. D3 RNA is not expressed in the multipotential progenitor cell line, EML; however, it is expressed in the promyelocytic cell line MPRO and can be induced in EML cells in culture conditions that promote myeloid cell differentiation (IL-3 plus rSCF plus atRA). We have demonstrated that, in EML cells, the induction appears to be independent of interferons and that D3 is not induced in response to atRA alone, distinguishing it from retinoic acid-inducible genes. In light of the lack of effect observed on hematopoiesis in IL-3–deficient mice,23 it is interesting to note that IL-3 appears to be required for EML cells to differentiate and for the induction of D3, which cannot be substituted with GM-CSF or other HGF tested to date. This suggests that either there are uncharacterized factors in vivo with activities that overlap with IL-3 or that dependence on IL-3 for differentiation may be a unique property of the EML cell line.
The relevance of D3 expression to normal hematopoietic cell differentiation was demonstrated using Linloc-Kit+ Sca-1+ BMC (EML cell equivalent) and Linlo cells (MPRO cell equivalent). Like the EML cells, Linlo c-Kit+ Sca-1+ cells do not express D3; however, expression can be induced by culturing these cells in medium containing IL-3 plus SCF. We are currently testing whether D3 RNA can be induced in normal Linlo c-Kit+Sca-1+ primitive BMC in response to HGF other than the combination of IL-3 and SCF. Like the MPRO and EPRO cell lines, Linlo cells express high levels of D3 RNA. Our results demonstrate that the EML and MPRO cell lines can be successfully used to isolate novel and potentially important genes involved in normal myeloid differentiation and that these cell lines are at least partially representative of their normal cellular counterparts in terms of gene expression patterns.
EML cells undergoing myeloid differentiation begin to express D3 RNA after 24 to 48 hours; expression peaks after 5 days and decreases by day 6 with the appearance of increasing numbers of terminally differentiated neutrophils and macrophages. These relatively slow kinetics of IL-3/SCF–induced D3 RNA expression in EML cells (24 to 48 hours by Northern blot analysis) suggest that the regulation of D3 gene expression is a complex process requiring multiple downstream events from receptor engagement. For example, IL-3 in combination with SCF may promote EML cells to synthesize transcription factors, receptors, and/or cytokines that, in turn, induce D3 gene expression. Whereas D3 RNA expression was clearly observed on Northern blots after 48 hours (Fig 1), with peak expression after 4 to 5 days in EML cells, the onset of D3 expression in normal BMC treated with M-CSF occurred 24 hours earlier than in EML cells (Fig 4). Maximum D3 expression in M-CSF–treated BMC cultures was observed between days 1 and 2, with low levels of expression detected by day 5 when the culture consisted of greater than 90% terminally differentiated macrophages. We believe that differences in the kinetics of induction of D3 could be explained, in part, by the state of differentiation of EML progenitor cells (primitive) and M-CSF–induced BMC cultures (committed). Thus, D3 RNA expression is temporally regulated during granulocyte and macrophage differentiation.
In an effort to understand the tissue and lineage restriction of D3 expression, we have examined panels of tissue blots, cell lines, and purified normal cells. Analysis of RNA obtained from normal mouse tissues showed no D3 expression in brain, liver, kidney, or testis. Modest levels of D3 were detected in the lung, with lower levels observed in spleen, skeletal muscle, and heart. Because lung tissue contains activated macrophages, we predicted this tissue to be positive for D3. Also, because spleen contains a small percentage of myeloid cells, this could account for the low expression seen in that site. The expression of D3 in heart and skeletal muscle suggests a potential role for this gene in these tissues.
Our survey of RNA expression in a wide range of hematopoietic cell lines showed that D3 RNA expression is highly restricted to the myeloid lineage. D3 gene expression was not detected in two T-cell lines tested (EL4 and CTLL-2), and no expression was detected in an erythroid cell line (HCD-57). Furthermore, D3 was not expressed in six of seven myeloid progenitor cell lines. Although all of these cell lines are blocked in differentiation at slightly distinct stages, they represent myeloid cells developmentally more primitive than promyelocytes. This suggests that not only is D3 expression restricted to cell lines of the myeloid lineage, but also to myeloid cells at or beyond the promyelocyte stage.
To determine whether D3 showed lineage restriction in normal hematopoietic cells, RNA was obtained from purified cell populations of various lineages and analyzed for D3 gene expression by RT-PCR. Using this highly sensitive, nonquantitative analysis, we were able to determine that splenic B220+ B cells do not express D3 RNA; however, expression was detected in Gr-1+ granulocytes, Ter 119+ erythroid cells, and thymic T cells. Although D3 gene expression could be detected by RT-PCR in thymocytes, we believe that the level of expression is extremely low, because D3 was not detected by Northern blot analysis of thymocyte RNA (data not shown). Finally, D3 RNA expression was induced in nomal BMC cultured in M-CSF, which promotes monocyte/macrophage lineage differentiation. Taken together, these results indicate that D3 is expressed in differentiating granulocytes and macrophages and possibly in erythroid cells.
There is currently no known biological function for D3 or any of the 200 family members of interferon-inducible genes. All of the family members are structurally related and contain one or two copies of a highly homologous 200 amino acid domain. Like MNDA, D3 contains one of the 200-amino acid homology domains, whereas the other family members contain two of these domains.9 Although the subcellular localization of D3 is unknown, other members of this family, including p202, p204, MNDA, and IFI 16, localize to the nucleus,24,25 and the open reading frame of D3 encodes a highly basic protein of 425 amino acids containing putative nuclear localization sequences.9 Interestingly, p202 has been demonstrated to bind a variety of proteins involved in both cell cycle regulation and transcription, including retinoblastoma protein (pRB),26 p53-binding protein 1 (53BP-1),27NF-κB, and AP-1.28 The p202 protein has also been demonstrated to inhibit E2F-mediated transcription.29 30These results suggest that the 200 family proteins may mediate pleotropic effects on growth and differentiation by binding DNA directly or, more likely, through protein-protein interactions. We are currently testing whether D3 has DNA and/or protein binding activity.
The interferon family of cytokines has been shown to mediate a large number of biological effects, including antiviral, antibacterial, immunomodulatory, and antiproliferative activity.31 These effects are the result of receptor-mediated signaling events that lead to the induction of gene expression of a variety proteins, including the 200 family. For example, D3 can be induced in terminally differentiated macrophages by exposure to interferons.9 In addition to the immunomodulatory effects, interferons have been shown to modulate cellular differentiation.31 In this regard, we have shown here for the first time that D3 expression is induced during normal myeloid cell growth and differentiation by HGF other than interferons, including SCF and IL-3. It is currently not known what role D3 and the other 200 family members play in regulating these diverse effects, which include the immunomodulatory actions of the interferons and the growth and differentiation effects induced by SCF/IL-3. We are currently investigating whether the relationship between D3 expression and myeloid differentiation is causal.
ACKNOWLEDGMENT
The authors are grateful to Drs Sally Spence, Joost Oppenheim, Peter Donovan, Peter Johnson, and Simon Williams for their review of this manuscript.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Jonathan R. Keller, PhD, Intramural Research and Support Group, Science Applications International Corp-Frederick, National Cancer Institute-Frederick Cancer Research and Development Center, Frederick, MD 21702-1201.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal