Abstract
In this study, we analyzed the regulation of NF-Y expression during human monocyte to macrophage maturation. NF-Y is a ubiquitous and evolutionarily conserved transcription factor that binds specifically to the CCAAT motif present in the 5′ promoter region of a wide variety of genes. We show here that in circulating monocytes, NF-Y binding activity is not detected on the CCAAT motif present in the promoters of genes such as major histocompatibility complex (MHC) class II, gp91-phox, mig, and fibronectin, whereas during macrophage differentiation, a progressive increase in NF-Y binding activity is observed on these promoters. Analysis of NF-Y subunit expression indicates that the absence of NF-Y activity in circulating monocytes is caused by a lack of the A subunit. Furthermore, addition of the recombinant NF-YA subunit restores NF-Y binding. We show that the lack of NF-YA protein is due to posttranscriptional regulation and not to a specific proteolytic activity. In fact, NF-YA mRNA is present at the same level at all days of monocyte cultivation, whereas the protein is absent in freshly isolated monocytes but is progressively synthesized during the maturation process. We thus conclude that the NF-YA subunit plays a relevant role in activating transcription of genes highly expressed in mature monocytes. In line with this conclusion, we show that the cut/CDP protein, a transcriptional repressor that inhibits gpc91-phox gene expression by preventing NF-Y binding to the CAAT box, is absent in monocytes.
THE CCAAT box IS A common motif present in direct or reverse orientation in many eukaryotic promoters. It usually resides around 80 bp upstream of the transcription start site and may be present in one or few copies. Several studies have shown the importance of the CCAAT box for the expression of genes constitutively expressed, as well as of genes active only in cells fully differentiated.1-5 To date, a large number of proteins able to bind this motif have been described: some are tissue-specific, whereas others are expressed ubiquitously.6 Although CCAAT elements can be found elsewhere in the promoter, a survey of 502 unrelated promoter sequences shows that most CCAAT boxes, located to the −60 to −130 region of the promoter, precisely reflect the NF-Y target sequence, indicating that NF-Y is the major CCAAT box–recognizing transcription factor.7
NF-Y is one of the best characterized CCAAT binding proteins, and its unique structure and evolutionary conservation suggest that it plays a crucial role in transcription of eukaryotic genes.8 It is a ubiquitous heteromeric transcription factor, composed of three subunits, NF-YA, NF-YB, and NF-YC, all necessary for DNA binding.9-11 The close association of NF-YB and NF-YC is a prerequisite for NF-YA binding and sequence-specific DNA interaction.10 NF-Y is unable on its own to activate transcription, but is able to increase the activity of neighboring enhancer motifs as well as to participate in the correct positioning of other transcription factors at the transcription start site.12-14 Although NF-Y is ubiquitous and constitutive, it also participates in the regulation of some promoters by controlling gene expression in a lineage- and activation-specific manner (eg, albumin and major histocompatibility complex [MHC] class II).2,15 It remains to be proven whether NF-Y contributes to this restricted pattern of expression. However, recent data suggest a modulation of NF-Y activity during serum starvation, depletion of intracellular calcium, differentiation, and cell proliferation.16-19 We previously reported18that a modulation of NF-Y activity is responsible for the transcriptional regulation of the ferritin H-chain gene, both in heme-treated erythroleukemic cells and during monocyte to macrophage differentiation.
Hematopoietic stem cells and differentiated progenitors have been extensively analyzed and represent an important model system for the study of cell differentiation. Monocytes originate from bone marrow hematopoietic progenitors called colony-forming units–monocyte (CFU-M) that proliferate and differentiate in the presence of some cytokines, including colony-stimulating factor (CSF-1), granulocyte-macrophage colony-stimulating factor (GM-CSF), and interleukin-3 (IL-3). Monocytes isolated from peripheral blood are committed cells not yet fully differentiated that undergo terminal differentiation to macrophages after migration into extravascular tissues.20 These cells spontaneously mature in vitro: in fact, upon in vitro cultivation, monocytes adhere to the plastic surface and in a few days undergo a spontaneous, time-dependent differentiation process21 22 that highly mimics their in vivo maturation.
In this study, we have analyzed the regulation of NF-Y expression during monocyte to macrophage maturation. We report that the progressive increase in the expression of genes highly expressed in mature monocytes under the control of a NF-Y binding CCAAT box, correlates with an increase in NF-Y binding activity. Moreover, the absence of NF-Y binding activity in freshly isolated monocytes is due to lack of the NF-YA subunit. This activity can be restored by the addition of this recombinant subunit. This observation indicates that NF-YA seems to be a limiting factor in the transcriptional activation of these genes. We show that a posttranscriptional mechanism is responsible for the regulation of NF-YA subunit expression during monocytic maturation. We also show that the CCAAT displacement protein (cut/CDP), implicated as a transcriptional repressor of the gp91-phox gene in immature myeloid cells and downmodulated during myeloid maturation,23 24 is absent in monocytes. This finding suggests that, in these cells, the transcriptional control of gp91-phox gene expression may be regulated by NF-Y through modulation of NF-YA subunit synthesis.
MATERIALS AND METHODS
Cell cultures.
Human myelomonocytic U937 cells were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS). The cells were induced to monocyte differentiation with 250 ng/mL 1α25OH-vitamin D3 (kindly provided by Roche, Basel, Switzerland) for 2, 3, and 4 days. HeLa cells were grown in Dulbecco’s modified Eagle’s medium (D-MEM) supplemented with 5% FCS. For isolation and culture of monocytes/macrophages, peripheral blood mononuclear cells (PBMC) were obtained from 18- to 40-year-old healthy male and female donors and cultivated as previously described.25 Briefly, 1.5 × 107 total PBMC cells were seeded in 75-cm2culture flasks in Iscove’s medium containing 15% FCS (0.22 μm filtered). After 1 hour at 37°C, the adherent cells (96% CD14+) were extensively washed with Ca2+ and Mg2+ free-phosphate-buffered saline (PBS; Flow Laboratories, Irvine, CA) to remove nonadherent cells and then incubated with Iscove’s medium and cultured at 37°C. All reagents used for monocyte isolation and culture were endotoxin free, as evaluated by the Limulus amebocyte lysate assay (PBI, Milano, Italy). At different culture days, both nonadherent and adherent cells were recovered and analyzed. Adherent macrophages were recovered with a cell scraper after 30 minutes of incubation at 4°C in the presence of Ca2+ and Mg2+ free-PBS. This procedure did not affect cell viability, and both adherent and nonadherent cells terminally differentiated to macrophages. Nonadherent and adherent cells were mixed together and then processed for preparation of whole cell extracts and mRNA. In some experiments, monocytes were isolated by a three-step density centrifugation procedure on Ficoll and Percoll gradients.26 Only monocyte preparations containing greater than 95% CD14+ cells were used for the experiments reported here.
Flow cytometry analysis of HLA-DR expression.
HLA-DR expression was investigated on monocytes at different day of culture by flow cytometry analysis using fluorescein isothiocyanate (FITC)-labeled antihuman HLA-DR monoclonal antibody (MoAb; Becton Dickinson, Mountain View, CA). The cells were processed for flow cytometry analysis as previously reported.22
Preparation of cell extracts.
Monocytes and macrophages were washed twice in cold PBS and then collected by centrifugation. The pellet (1 × 107) was resuspended in 100 μL of lysis buffer containing 20 mmol/L HEPES, pH 7.9, 50 mmol/L NaCl, 10 mmol/L EDTA, 2 mmol/L EGTA, 0.5% (vol/vol) NP-40, supplemented with 0.5 mmol/L dithiothreitol (DTT), 10 mmol/L sodium molybdate, 10 mmol/L sodium orthovanadate, 100 mmol/L NaF, 10 μg/mL leupeptin, and 0.5 mmol/L phenylmethylsulfonyl fluoride (PMSF). After incubation for 30 minutes on ice, the suspension was centrifuged at 10,000g for 10 minutes. The supernatants were aliquoted and stored at −80°C.
Cell extracts for the analysis of the cut/CDP protein were prepared by resuspending the pellet (1 × 107) in 100 μL of lysis buffer containing 50 mmol/L HEPES, pH 7.9, 400 mmol/L KCl, 0.2 mmol/L EDTA, 0.2 mmol/L EGTA, 0.1% NP-40, 10% glycerol complemented with 1 mmol/L PMSF, 10 μg/mL leupeptin, 1 μg/mL aprotinin, 1 mmol/L di DTT, 4 mmol/L NaF, and 4 mmol/L sodium orthovanadate. Nuclear cell extracts were prepared as reported in Coqueret et al.27Briefly, cell pellets were resuspended in 10 mmol/L HEPES, pH 7.9, 1.5 mmol/L MgCl2, 10 mmol/L KCl, 1 mmol/L PMSF, 1 μg/mL leupeptin, 1 μg/mL aprotinin, and 1 mmol/L DTT. After three freeze-thaw cycles, the suspension was centrifuged at 10,000gfor 1 minute, and the supernatant was recovered. Nuclear pellets were resuspended in 20 mmol/L HEPES pH 7.9, 1.5 mmol/L MgCl2, 420 mmol/L KCl, 0.2 mmol/L EDTA, 25% glycerol, 1 mmol/L PMSF, 1 μg/mL leupeptin, 1 μg/mL aprotinin, and 1 mmol/L DTT. After incubation for 30 minutes on ice, nuclear extracts were spun down at 10,000g for 5 minutes, and the supernatants were recovered.
DNA electrophoretic mobility shift assay (EMSA).
To measure the association of DNA-binding proteins with different DNA sequences, the synthetic double stranded oligonucleotides, prepared on an Applied Biosystems DNA synthesizer (Applied Biosystems, Foster City, CA), were end-labeled using the T4 polynucleotide kinase or the Klenow polymerase. Binding reaction mixture (20 μL final volume) contained labeled oligonucleotide probes (20,000 cpm) in binding buffer (75 mmol/L KCl, 20 mmol/L Tris-HCl, pH 7.5, 1 mmol/L DTT), 5 μg/mL bovine serum albumin (BSA) and 14% (vol/vol) glycerol, and 3 μg poly(dl)-poly(dC). Binding reaction mixture for cut/CDP EMSA was performed according to Coqueret et al.27 Whole (15 μg) or nuclear cell extracts (10 μg) were added, and the reaction mixture was incubated for 20 minutes at room temperature. For reconstitution experiments, the recombinant proteins were incubated with whole cell extracts for 30 minutes at 4°C before addition to the EMSA reaction mixture. Samples were electrophoresed in 5% polyacrylamide gel in 0.5 × Tris-borate/EDTA (TBE) buffer for 2 hours at 200 V at 18°C. Gels were then dried and autoradiographed. The DNA sequences of the oligonucleotides used in these studies were as follows: MHC class II Ea, CCAAT 5′-GTCTGAAACATTTTTCTGATTGGTTAAAAGTTGAGTGCT-3′; gp91-phox, 5′-GCAAGCT TTTCAGTTGACCAATGATTATTAGCCAATTC-3′; mig, 5′-GGTCAGCTGAGGAGACCAGCCAATCAGAGACGGGAAGG-3′; fibronectin, 5′-CGTCACCCGGAGCCCGGGCCAATCGGGCGCGGTCGGCTG-3′; CDPα, 5′-CTTTTCAGTTGACCAATGATTATTAGCCAATTTC TGATAAAAAGAAAAGGAAACCGATTGC-3′; Cut, 5′-AAAAGAAGCT TATCGATACCGT-3′; PU.1, 5′-TGCCTAGCTAAAAGGGGAAGAAGAG GATCAGCCCAAGGAG-3′; and PU.1 mut, 5′-TGCCTAGCTAAAAGGGATCGTAGCGGATCAGCCCAAGGAG-3′.
Western blot assay.
For Western blot analysis, 30 μg of whole cell extracts prepared from fully differentiated macrophages was added to 30 or 60 μg of extracts prepared from freshly isolated monocytes. Both of these extracts were prepared without adding proteinase inhibitors. The mixture was incubated for 40 minutes at room temperature and then denatured and separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred onto nitrocellulose paper, incubated with rabbit anti–NF-YA, and detected by the Enhanced Chemiluminescence System using antirabbit horseradish peroxidase-coupled secondary antibody (Amersham International Plc, Little Chalfont, Buckinghamshire, UK). Incubation and washes were performed as previously described.2 For cut/CDP protein, blots were incubated with polyclonal antibody raised against the N-terminal half of CDP (a generous gift of A. Nepveu, McGill University, Montreal, Quebec, Canada) and performed as described.27
RNase protection experiments.
Total RNA was isolated from monocytes and macrophages at different stages of differentiation by the guanidium cesium chloride method.28 Total RNA (5 μg) was hybridized for 18 hours to the RNA probes (3 × 105 cpm) at 55°C in 25 μL of 80% formamide, 400 mmol/L NaCl, 40 mmol/L piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 6.8), and 1 mmol/L EDTA. Subsequently, samples were incubated with RNase A (40 μg/mL) and RNase T1 (2 μg/mL) for 1 hour at 33°C and then subjected to proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation. Gel electrophoresis was performed on standard 8% polyacrylamide 8 mol/L urea sequencing gel. For the construction of the human NF-YB riboprobe, the Pst IAcc I complementary DNA (cDNA) fragment was subcloned in theSma I site of the pBluescript KS (pBsKS). To generate a32P labeled 263 nucleotide (nt) long antisense RNA probe, this fragment was digested with Dde I and transcribed by T7 polymerase. A pBsSK plasmid containing the fragment from +240 nt to +640 nt of the human NF-YC cDNA was linearized withBamHI present in the polycloning site. To generate the antisense RNA of 400 nt, the linearized template was in vitro transcribed using T7 polymerase. To prepare the riboprobe for the human NF-YA subunit, the Bgl I fragment of 256 nt (from +122 nt to +378 nt) was subcloned in pBsKS Sma plasmid and linearized with HindIII present in the polycloning site. This riboprobe is able to evidentiate both the NF-YA mRNA isoforms. The 434-bp long pTRI- glyceraldehyde-3-phosphate dehydrogenase (GaPDH) mouse antisense control template (AMBION, Austin, TX) was used as an internal standard to establish the relative amount of RNA loaded. The probe was synthesized by in vitro transcription from linear template using SP6 polymerase.
RESULTS
Increasing NF-Y binding to the CCAAT box of promoters of genes upregulated during monocyte to macrophage differentiation.
We previously reported that freshly isolated monocytes (day 0) do not show any binding activity to the CCAAT box present in the promoter of the H-chain ferritin gene.18 However, starting at day 3 of culture and thereafter, a consistent NF-Y binding activity is induced, associated with an accumulation of ferritin mRNA.
Macrophages are physiologically involved in iron storage, and ferritin represents a functional molecule, whose expression is tightly regulated also at transcriptional level. To verify whether a similar mechanism is operative in transcription regulation of other genes upregulated during monocytic maturation, we analyzed the NF-Y binding to the functional CCAAT box present in the promoter of the MHC class II, gp91-phox, mig, and fibronectin genes. With the exception of mig,29,30 the importance of NF-Y in the transcription of these genes has been well established in functional studies.14,15,23 31
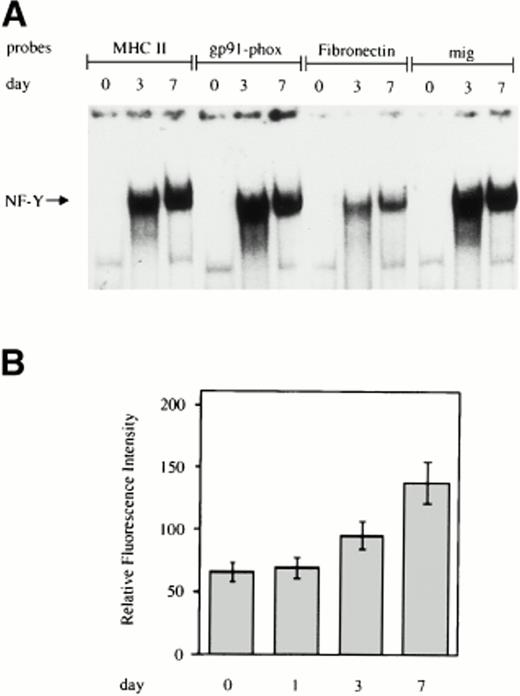
EMSA was performed on labeled double-stranded oligonucleotides derived from the sequences surrounding the CCAAT box present in the promoter of the indicated genes and incubated with whole cell extracts from monocytes at different days of cultivation. As shown in Fig1A, for all four of the CCAAT boxes examined, no NF-Y binding activity was present in freshly isolated monocytes, but a progressive increase in NF-Y activity was evident during macrophage differentiation. In line with these results, we observed a significant increase in the surface expression of MHC class II HLA-DR antigens during monocyte to macrophage maturation, as assessed by flow cytometric analysis (Fig 1B).
Increasing NF-Y binding to the CCAAT box region of the promoters of genes upmodulated during monocyte to macrophage differentiation. (A) Whole cell extracts obtained from human primary monocytes at day 0, 3, and 7 of culture were incubated with labeled oligonucleotides, corresponding to sequences surrounding the CCAAT box present in the promoters of the MHC class II, gp91-phox, fibronectin, and mig and analyzed by EMSA. (B) HLA-DR expression on the cell surface of in vitro grown monocytes. Monocytes were grown for the indicated time and processed for membrane fluorescence using anti–HLA-DR monoclonal antibody, as described in Materials and Methods.
Increasing NF-Y binding to the CCAAT box region of the promoters of genes upmodulated during monocyte to macrophage differentiation. (A) Whole cell extracts obtained from human primary monocytes at day 0, 3, and 7 of culture were incubated with labeled oligonucleotides, corresponding to sequences surrounding the CCAAT box present in the promoters of the MHC class II, gp91-phox, fibronectin, and mig and analyzed by EMSA. (B) HLA-DR expression on the cell surface of in vitro grown monocytes. Monocytes were grown for the indicated time and processed for membrane fluorescence using anti–HLA-DR monoclonal antibody, as described in Materials and Methods.
Exogenous addition of NF-YA to cell extracts from freshly isolated monocytes is able to reconstitute NF-Y binding.
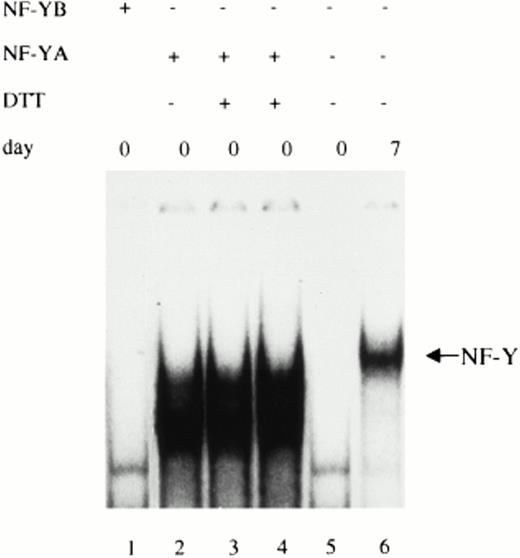
In our previous studies on the regulation of ferritin H-chain gene by NF-Y,18 it was hypothesized that the lack of NF-Y binding activity observed in cell extracts derived from freshly isolated monocytes may be caused by the absence of the NF-YA subunit. Consistent with this hypothesis, Western blot analysis indicated that the NF-YB subunit is equally expressed in freshly isolated monocytes (day 0), maturing (day 3), and fully differentiated macrophages (day 7). Conversely, the NF-YA subunit is undetectable in freshly isolated monocytes, appears on day 3, and further increases on day 7 of culture. To show that the lack of NF-Y binding to the CCAAT motifs shown in Fig1 was determined by the absence of the NF-YA subunit, we tested the ability of the recombinant protein to restore the binding when added to cell extracts prepared from freshly isolated monocytes. As shown in Fig2 (lane 2), the addition of 100 ng of the short form of NF-YA subunit to cell extracts derived from circulating monocytes was able to restore the binding of NF-Y to the CCAAT box present in the MHC class II promoter. The faster migrating complex obtained is probably a result of partial instability of the recombinant protein. Binding was not restored when the purified NF-YB subunit was added to monocyte cell extracts (lane 1). Similarly, the addition of NF-YB together with NF-YA did not modify the NF-Y binding activity (data not shown).
Exogenous addition of purified NF-YA subunit is able to reconstitute NF-Y binding activity in freshly isolated monocytes. Whole cell extracts obtained from human freshly isolated monocytes were incubated with the 32P-labeled oligonucleotide corresponding to the MHC class II CCAAT box. Additions of 100 ng of the recombinant NF-YB subunit (lane 1) or 100 ng of the recombinant short form of NF-YA subunit (lane 2) were made to the reaction mix before EMSA where indicated. In lanes 3 and 4, 10 or 20 mmol/L DTT was added to the reaction mix. Whole cell extracts prepared from freshly isolated monocytes (d0) and fully differentiated macrophages (d7) (lanes 5 and 6) were used as control of NF-Y binding activity.
Exogenous addition of purified NF-YA subunit is able to reconstitute NF-Y binding activity in freshly isolated monocytes. Whole cell extracts obtained from human freshly isolated monocytes were incubated with the 32P-labeled oligonucleotide corresponding to the MHC class II CCAAT box. Additions of 100 ng of the recombinant NF-YB subunit (lane 1) or 100 ng of the recombinant short form of NF-YA subunit (lane 2) were made to the reaction mix before EMSA where indicated. In lanes 3 and 4, 10 or 20 mmol/L DTT was added to the reaction mix. Whole cell extracts prepared from freshly isolated monocytes (d0) and fully differentiated macrophages (d7) (lanes 5 and 6) were used as control of NF-Y binding activity.
A recent report indicates32 that NF-YB needs to be reduced to allow interaction with NF-YC and efficient binding activity. Thus, band shift assays were also performed in the presence of either 10 or 20 mmol/L DTT. As shown in Fig 2 (lanes 3 and 4), the addition of this reducing agent to the reaction mixture did not increase the DNA binding activity of NF-Y, suggesting that, in freshly isolated monocytes, NF-YB is already in an active form and does not contribute to the impaired NF-Y binding activity.
Analysis of NF-Y subunits mRNA during monocyte to macrophage differentiation.
The results described above indicate that the absence of NF-YA subunit is the limiting factor for NF-Y binding activity in cell extracts prepared from freshly isolated monocytes.
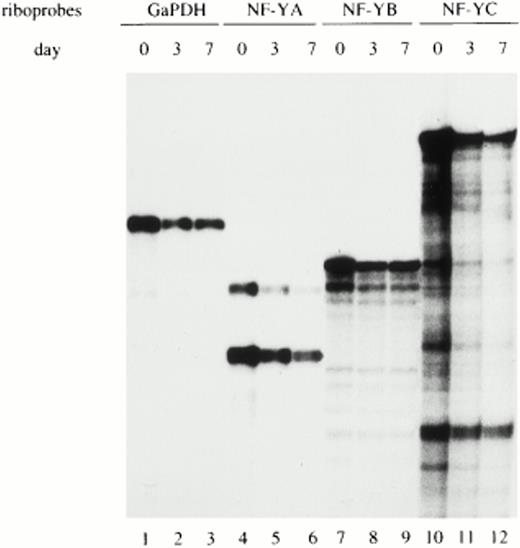
To determine whether the regulation of NF-YA subunit expression in monocytes/macrophages occurs at the transcriptional and/or posttranscriptional level, NF-YA, NF-YB, and NF-YC mRNAs were evaluated in monocytes at different days of maturation. RNase protection experiments were performed on total RNA prepared from freshly isolated (d0), maturing (d3), and fully differentiated monocytes (d7) and hybridized with specific riboprobes for the three NF-Y subunit transcripts. The GaPDH riboprobe was used as an internal control to standardize the amount of hybridized mRNA (lanes 1 and 3). Figure3 shows that similar amounts of mRNA for all three NF-Y subunits were present at all days of analysis. The specific riboprobe for NF-YA was able to recognize both the short- and long-NF-YA isoforms (lanes 4 through 6), arising from differential splicing events.33 Both isoforms are able to bind DNA and are equally active in transcriptional activation. NF-YA mRNA was present at comparable levels at all days of cultivation, with the short form being the most abundant.
Analysis of NF-Y subunits (A, B, and C) mRNA during monocyte to macrophage differentiation. Total RNA prepared from freshly isolated (d0), maturing (d3), and fully differentiated (d7) macrophages was hybridized with specific riboprobes for human NF-YA, NF-YB, and NF-YC subunits and analyzed by RNase protection. The two mRNA species of NF-YA mRNA correspond to the two isoforms derived by alternative splicing. GaPDH riboprobe was used as an internal control.
Analysis of NF-Y subunits (A, B, and C) mRNA during monocyte to macrophage differentiation. Total RNA prepared from freshly isolated (d0), maturing (d3), and fully differentiated (d7) macrophages was hybridized with specific riboprobes for human NF-YA, NF-YB, and NF-YC subunits and analyzed by RNase protection. The two mRNA species of NF-YA mRNA correspond to the two isoforms derived by alternative splicing. GaPDH riboprobe was used as an internal control.
These results indicate that in circulating monocytes, the NF-YA mRNA is present in discrete amounts, comparable with those of the other NF-Y subunits. Therefore, the lack of NF-YA protein is determined by posttranscriptional mechanisms.
The lack of NF-YA in cell extracts from freshly isolated monocytes is not due to a degradation of the protein.
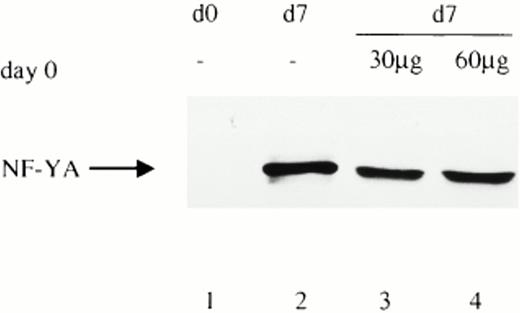
Because the NF-Y mRNA subunits are equally present at all days of monocyte maturation, we investigated if the absence of the NF-YA subunit in freshly isolated monocytes could be attributed to a specific degradation process operative in monocytes but not in macrophages. To test this hypothesis, increasing concentrations (30 to 60 μg) of cell extracts from freshly isolated monocytes were added to 30 μg of cell extracts from mature macrophages (Fig 4, lanes 3 and 4). Both of these extracts were prepared as indicated in Materials and Methods, except that protease inhibitors were not added to the lysis buffer. After incubation for 40 minutes at room temperature, samples were analyzed by Western blotting using specific affinity purified anti–NF-YA antibodies. As shown in Fig 4, a high amount of the protein is present at day 7 even in the absence of protease inhibitors during the extraction procedures. The amount of NF-YA present in macrophages (day 7) is not affected by the addition of whole cell extracts from freshly isolated monocytes (day 0). These results, although not conclusive, strongly suggest that NF-YA is a stable protein and that a specific protease activity is not present in immature cells.
The lack of NF-YA subunit in cell extracts from freshly isolated monocytes is not due to degradation. Western blot analysis (30 μg) from freshly isolated monocytes (d0) and fully differentiated macrophages (d7) (lanes 1 and 2) was performed with an affinity-purified antibody against NF-YA. Extracts were prepared, as indicated in Materials and Methods, but without the addition of protease inhibitors. In lanes 3 and 4, increasing amounts of whole cell extract from freshly isolated monocytes (d0) were added to 30 μg of cell extracts from d7 cultured monocytes, incubated for 40 minutes at room temperature, and then subjected to Western blot analysis.
The lack of NF-YA subunit in cell extracts from freshly isolated monocytes is not due to degradation. Western blot analysis (30 μg) from freshly isolated monocytes (d0) and fully differentiated macrophages (d7) (lanes 1 and 2) was performed with an affinity-purified antibody against NF-YA. Extracts were prepared, as indicated in Materials and Methods, but without the addition of protease inhibitors. In lanes 3 and 4, increasing amounts of whole cell extract from freshly isolated monocytes (d0) were added to 30 μg of cell extracts from d7 cultured monocytes, incubated for 40 minutes at room temperature, and then subjected to Western blot analysis.
The lack of NF-Y binding activity to the CCAAT box present on the gp91-phox gene promoter is not due to the repressor cut/CDP protein.
The gp91-phox gene, which encodes the cytochrome b heavy chain required for the microbicidal activity of phagocytic cells, is expressed nearly exclusively in terminally differentiating myelomonocytic cells.23 34
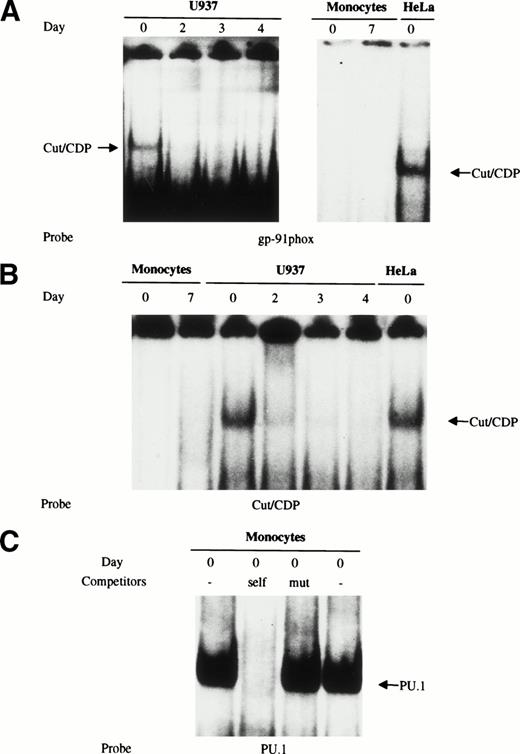
Several studies have shown that the cut/CDP is involved in the regulation of the gp91-phox gene, playing a repressive role by preventing, at least in part, the binding of the transcriptional activating factor NF-Y in immature myeloid cells; on the contrary, the DNA binding activity of cut/CDP is downregulated during terminal differentiation of phagocytic cells.1,23,24 This downmodulation correlates with an increase in the amount of gp91-phox expressed in macrophages.23,34 The low amount of gp91-phox protein present in freshly isolated monocytes35 could be due either to a repressor mechanism controlled by the cut/CDP protein or, as our data suggest, to the absence of an active NF-Y complex. To detect the amount and the activity of the cut/CDP protein in human primary monocytes, both EMSA experiments and Western blot analysis were performed. Both U937 myeloid cells and HeLa cells were used as a control for cells expressing the cut/CDP protein. Nuclear cell extracts were prepared from HeLa cells, freshly isolated monocytes, and fully differentiated macrophages, as well as from undifferentiated or 1α25OH-vitamin D3–induced U937 cells. These extracts were incubated with a labeled double-stranded oligonucleotide derived from the gp91-phox promoter known to have a great affinity binding site for the cut/CDP protein (CDPα oligonucleotide)23 (Fig5A) and with a specific consensus Cut binding site27 (Fig 5B). As shown in Fig 5A and B, the cut/CDP binding activity was undetectable in nuclear cell extracts from monocytes and macrophages. Conversely, cut/CDP binding activity was clearly detectable on both probes in extracts from undifferentiated U937 and HeLa cells. According to literature data,1 23 the complex between the cut/CDP protein and oligonucleotides disappears in U937 induced to differentiate with 1α25OH-vitamin D3 for 2, 3, and 4 days.
Cut/CDP binding activity is undetectable in human primary monocytes/macrophages. (A) Nuclear cell extracts from freshly isolated monocytes and fully differentiated macrophages were incubated with CDP-labeled oligonucleotide derived from the gp91-phox promoter and analyzed by EMSA. Nuclear cell extracts were from HeLa cells and U937 cells undifferentiated or induced to monocyte differentiation with 125OH-vitamin D3 for 2, 3, and 4 days. (B) The same nuclear extracts were incubated with a labeled specific consensus Cut binding site.27 (C) Whole (lane 1) and nuclear cell (lane 4) extracts prepared from freshly isolated monocytes were incubated with a labeled PU.1 oligonucleotide to test extract integrity. Competition assay with wild-type (lane 2) and mutated oligonucleotide (lane 3) was also performed on whole cell extracts.
Cut/CDP binding activity is undetectable in human primary monocytes/macrophages. (A) Nuclear cell extracts from freshly isolated monocytes and fully differentiated macrophages were incubated with CDP-labeled oligonucleotide derived from the gp91-phox promoter and analyzed by EMSA. Nuclear cell extracts were from HeLa cells and U937 cells undifferentiated or induced to monocyte differentiation with 125OH-vitamin D3 for 2, 3, and 4 days. (B) The same nuclear extracts were incubated with a labeled specific consensus Cut binding site.27 (C) Whole (lane 1) and nuclear cell (lane 4) extracts prepared from freshly isolated monocytes were incubated with a labeled PU.1 oligonucleotide to test extract integrity. Competition assay with wild-type (lane 2) and mutated oligonucleotide (lane 3) was also performed on whole cell extracts.
A positive control to assess extract integrity was included, because no EMSA complex was detected in day 0 monocyte extracts for either NF-Y or cut/CDP proteins, despite the presence of a large number of protease inhibitors. For this purpose, nuclear cell extracts from freshly isolated monocytes were incubated with a labeled oligonucleotide containing a specific binding site for the PU.1 transcription factor, a master regulator of myeloid genes. As shown in Fig 5C, a major DNA-protein complex was obtained using either whole (lane 1) or nuclear (lane 4) cell extracts prepared from freshly isolated monocytes. The binding of PU.1 was competed for by a 100-fold molar excess of cold PU.1 oligonucleotide, whereas the oligonucleotide containing a mutated PU.1 site was unable to compete (lanes 2 and 3).
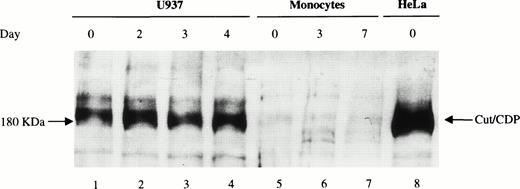
The amount of cut/CDP protein was also evaluated. Western blot analysis (Fig 6) was performed on total cell extracts from freshly isolated monocytes (d0), maturing (d3), and fully differentiated macrophages (d7) (lanes 5 through 7) and from HeLa (lane 8) and U937 cells (lanes 1 through 4) as positive controls. As expected,24 high levels of intact cut/CDP protein (180 kD) were detectable in HeLa cells and in U937 cells. Conversely, no cut/CDP protein was detected in monocytes at any day of maturation. This result indicates that the cut/CDP activity is not present in circulating monocytes, and thus, the negative transcriptional control, mediated by this protein, is probably not operative in these primary cells. We suggest that one of the mechanisms controlling the low expression of the gp91-phox in monocytes35 may be the lack of NF-YA subunit that hampers the transcriptional activity of NF-Y.
Analysis of cut/CDP protein expression during monocyte to macrophage maturation. Western blot analysis was performed on whole cell extracts (50 μg) from day 0, 3, and 7 monocyte culture (lanes 5 through 7) incubated with a specific anti–cut/CDP antibody. Whole cell extracts (50 μg) from U937 cells undifferentiated or induced to monocyte differentiation with 125OH-vitamin D3 for 2, 3, and 4 days (lanes 1 through 4) and HeLa cells (lane 8) were used as positive control for cut/CDP expression.
Analysis of cut/CDP protein expression during monocyte to macrophage maturation. Western blot analysis was performed on whole cell extracts (50 μg) from day 0, 3, and 7 monocyte culture (lanes 5 through 7) incubated with a specific anti–cut/CDP antibody. Whole cell extracts (50 μg) from U937 cells undifferentiated or induced to monocyte differentiation with 125OH-vitamin D3 for 2, 3, and 4 days (lanes 1 through 4) and HeLa cells (lane 8) were used as positive control for cut/CDP expression.
DISCUSSION
In this study we investigated the molecular mechanisms responsible for the specific expression of the NF-Y transcription factor during maturation/activation of human primary monocytes. Significant progress has recently been made in the characterization of several tissue-specific transcription factors such as PU.1, components of CCAAT/Enhancer-binding protein family, and acute myelogenous leukemia 1 (AML1), which play important roles in controlling myeloid specific gene expression (reviewed in Tenen et al36). In addition, it has been suggested that ubiquitous transcription factors may be relevant during myeloid differentiation. In this respect, Sp1 can mediate responses to several inducers of myeloid differentiation, and several myeloid promoters are dependent on a functional Sp1 site.37-42 Moreover, substantial variations in Sp1 expression have been found in some cell types at different stages of differentiation.43 It is thus conceivable that the control of cell differentiation is governed at the molecular level by the coordinate functional interaction between cell-specific and ubiquitous transcription factors.
We previously showed that the expression of the constitutive and ubiquitous transcription factor NF-Y is modulated during monocyte maturation/activation18: this conclusion was based on the analysis of the binding of NF-Y to the CCAAT box present in the ferritin promoter.
NF-Y plays a crucial role in regulating the expression of several genes, including genes upregulated during monocyte differentiation, such as MHC class II,2,14,15 Invariant Chain,44,45 and gp91-phox.1,23 The specificity for NF-Y activation can be provided in different tissues: (1) by the combined action with other cell specific transcription factors, (2) by the type of NF-YA isoform expressed in different cell types as recently shown by studies of the CD10/neutral endopeptidase 24.11 (NEP) promoter,46 and (3) by modulating its activity through the differential expression of one of its subunits as suggested by the present data. In freshly isolated monocytes, we show that the NF-YA subunit is absent, but its expression progressively increases during the maturation process.
This increase correlates with the pattern of NF-Y binding to MHC class II, gp91-phox, mig, and fibronectin promoters and with the increased expression of these genes during maturation. Moreover, addition of recombinant NF-YA protein to cell extracts prepared from circulating monocytes is able to completely restore the NF-Y binding. Although a formal analysis of NF-YC protein was not possible because anti–NF-YC antibodies are still not available, this result indicates that, in contrast to NF-YA, the B and C subunits are constantly expressed, thus suggesting that NF-YA is the limiting factor in the activation of NF-Y binding.
We show that the regulation of NF-YA subunit expression is not mediated by transcriptional mechanisms; indeed, NF-YA mRNA levels do not change, and no differential expression of the two isoforms is observed during monocytic maturation. Conversely, a posttranscriptional or a translational mechanism seems to be operative in primary monocytes. A translational mechanism of NF-Y expression regulation is supported by the observation that the 5′ untranslated region of NF-YA mRNA from chicken, mouse, and human shares an extremely high homology (95%) and thus may be involved in a mechanism of regulation leading to rapid availability of active NF-Y heteromeric complex during macrophage activation.
A crucial role in controlling gene expression during developmental processes is also played by transcriptional repressors. In particular, it has been shown that in immature phagocytic cells, in which the endogenous gp91-phox gene is transcriptionally inactive, NF-Y binding to the CCAAT boxes present in the gp91-phox gene promoter is prevented by the transcriptional repressor cut/CDP.23 No cut/CDP protein or binding activity was detected in freshly isolated monocytes, but both were present in U937 promonocytic cell line. The lack of cut/CDP protein in monocytes with respect to the monocytic cell line may be explained either by the more advanced stage of monocyte differentiation or by the differences between a transformed cell line and primary cells. Our results suggest that, in monocytes, in which the amount of the gp91-phox is still quite low,35 one of the limiting factors for gp91-phox gene expression, as well as for the other genes examined, may be the lack of the NF-YA subunit that prevents the formation of the active NF-Y heteromeric complex.
Altogether, our results show a pivotal role of the NF-YA subunit in controlling the overall activity of the NF-Y transcription factor during monocyte maturation. This conclusion is in line with structural studies showing that the conserved sequences of the NF-YB and NF-YC subunits show sequence similarity with the histone fold motifs of the histone proteins H2A and H2B47,48 and that the YB/YC dimer can be found associated with other proteins in high molecular weight complexes, even in the absence of NF-YA.49 Because all three subunits are necessary for DNA binding, we speculate that NF-YB and NF-YC play some basic role in gene activation, whereas NF-YA represents the regulatory subunit of the heteromeric complex.
ACKNOWLEDGMENT
The authors gratefully acknowledge A. Nepveu for providing antibodies against cut/CDP. The authors also thank S. Mochi for oligonucleotide preparation, S. Tocchio for editorial assistance, and R. Gilardi for graphics.
Supported by grants from Istituto Superiore di Sanitè (special project on AIDS) and from Italy-USA program on “Therapy of Tumors” (A.B.). The financial support of Telethon Italy (Grant No. 582 to R.M.) is gratefully acknowledged.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Angela Battistini, PhD, Department of Virology, Istituto Superiore di Sanità, Viale Regina Elena, 299, 00161, Rome, Italy.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal