Abstract
Earlier, we described a stromal cell-free two-step clonal culture system in which murine primitive lymphohematopoietic progenitors produce myeloid and B-lymphoid lineage cells. In the same culture T-cell potential of the progenitors was maintained. We now report that, in addition to myeloid and B-lymphoid cells, putative T-cell progenitors are also produced in culture. Lineage-negative (Lin−) Ly-6A/E+ c-kit+ bone marrow cells from 5-fluorouracil–treated mice were cultured in methylcellulose in the presence of SF (Steel factor), interleukin (IL)-11, and IL-7, and the resulting primary colonies were picked and pooled. When injected into severe combined immune deficiency (scid) mice, the pooled cells reconstituted the T-cell compartment of the scid mice earlier than freshly prepared primitive marrow cells. This reconstitution activity of the pooled primary colony cells was enriched in the Ly-6A/E+ and FcγRII/III−/low cell fractions. Reverse transcriptase-polymerase chain reaction (RT-PCR) and DNA-PCR analyses showed that some of the primary colony cells are differentiated sufficiently to express messenger RNA (mRNA) of T-cell receptor (TCR) β-chain and pre-TCR alpha (pT) and, although not frequently, to perform Dβ-Jβ rearrangement of the TCR gene. Micromanipulation studies confirmed the clonal origin of myeloid lineage cells and the cells positive for the T-cell–specific transcripts and D-J rearrangement of TCR β-chain. These results suggested that, in the presence of SF, IL-11, and IL-7, primitive lymphohematopoietic progenitors differentiate toward T-cell lineage in addition to myeloid and B-cell lineages.
ALTHOUGH T CELLS ARE derived from the pluripotent hematopoietic stem cells, their development is different from that of other blood cells. T cells differentiate primarily in the thymus after prethymic T-cell progenitors migrate into the thymus, whereas the entire development of other blood cells takes place in the fetal liver or bone marrow. In the thymus, T-cell progenitors begin rearrangement of the T-cell–receptor (TCR) genes at the CD4− CD8− double-negative (DN) cell stage, and pass through the CD4−CD8low stage into the CD4+ CD8+double-positive (DP) TCR−/low stage. In the DP cell stage, the thymocytes whose TCR react strongly to self-major histocompatability complex (MHC) antigens are deleted (negative selection). The thymocytes whose TCR bind moderately to self-MHC antigens will survive and acquire self-MHC-restriction (positive selection). DP thymocytes then differentiate into TCRhighsingle positive (SP) CD4+ or CD8+ mature thymocytes.1-3 A number of cytokines, such as interleukin (IL)-1∼4, IL-6, IL-7, IL-9, IL-10, IL-12, steel factor (SF), flt3 ligand (FL), tumor necrosis factor (TNFα), transforming growth factor (TGF) β, and granulocyte-macrophage colony-stimulating factor (GM-CSF), have been reported to support the proliferation of immature thymocytes.4,5 Among them, crucial roles of IL-7,6-14 SF,15,16 and FL15 in the development of thymocytes have been shown by the in vivo injection of neutralizing antibodies and by the use of spontaneous mutant mice or gene-targeted mutant mice.
Compared with the intrathymic development, less is known about the early stages of T-cell development. One example is the nature of prethymic T-cell progenitors. Wu et al17 and Matsuzaki et al18 enriched small-cell populations from murine thymocytes that have T-, B-, and natural killer (NK)-cell potentials but not myeloid potentials. Their studies suggested the existence of common lymphoid progenitors and support the model of early diversion of myeloid from lymphoid lineages. In agreement with this, Akashi et al19 recently identified T/B common lymphoid progenitors in murine bone marrow. These results suggested that the diversion of lymphoid from myeloid lineages takes place before seeding into the thymus and that the common lymphoid progenitors commit to T-cell lineage in the thymus. In contrast to these observations, however, Rodewald et al20 identified in murine fetal blood committed T-cell progenitors that have neither B-cell nor myeloid potentials. This indicated that the commitment to T-cell lineage can occur before progenitor seeding into the thymus. Their observations did not exclude the possibility that common lymphoid progenitors or multipotential progenitors migrate into the thymus. Indeed, in addition to the committed T-cell progenitors, they also identified multipotential progenitors in the fetal blood.
One of the reasons for our poor understanding of the early stages of T-cell development is the lack of a culture assay that allows clonal analysis of T-cell development from primitive hematopoietic progenitors. Such a culture would significantly facilitate our study of the early stages of T-cell development. Earlier, we developed a two-step clonal culture system that supports the differentiation along the myeloid and B-cell lineages of murine bone marrow progenitors.21 Subsequently we documented that the cells in the primary colonies in this assay possess T-cell potential.22 In the current study, we showed that, in the presence of SF, IL-11, and IL-7, the lymphohematopoietic progenitors differentiate along T-cell lineage into progenitors that express messenger RNA (mRNA) of pre-TCR alpha (pTα) and TCR β-chain, as well as along myeloid and B-cell lineages.
MATERIALS AND METHODS
Growth factors.
Murine SF was provided by Immunex Corporation, Seattle, WA and Kirin Brewery Co, Ltd, Tokyo, Japan. Human IL-7 was a gift from C. Faltynek of Sterling Drug, Malvern, PA. Human IL-11 and erythropoietin (Epo) were provided by the Genetics Institute, Cambridge, MA. Murine FL was a gift from S. Lyman of Immunex Corp. All of the cytokines we used were recombinant and purified, and their concentrations were as follows: SF, 100 ng/mL; IL-7, 5 ng/mL; IL-11, 50 ng/mL; FL, 1000 ng/mL; Epo, 2 units/mL.
Progenitor purification.
Bone marrow cells were harvested from 10- to 20-week old BDF1 mice (Charles River, Raleigh, NC and Clea Japan, Inc, Tokyo, Japan) that had been injected intravenously with 5-fluorouracil (5-FU, Adria Laboratories, Columbus, OH) at 150 mg/kg (body weight) 2 days before. The bone marrow cells were enriched for primitive progenitor cells by using the technique we described previously.23 Briefly, cells at densities ranging from 1.0631 to 1.0770 g/mL were harvested with Nycodenz (Accurate Chemical & Scientific, Westbury, NY) density centrifugation. The samples were then depleted of cells expressing lineage-specific antigens by negative immunomagnetic bead selection with Dynabeads M-450 sheep-antirat IgG (Dynal, Great Neck, NY). The monoclonal antibodies (MoAbs) used were anti-B220 (14.8),24anti-Mac-1 (M1/70),25 anti-Gr-1 (RB6-8C5),26anti-CD4 (GK1.5),27 anti-CD8 (53-6.72),28 and TER-119.29 The density-separated, lineage-negative (Lin−) cells were then stained with fluorescein isothiocyanate (FITC)-conjugated anti–Ly-6A/E Ab (D7),30and biotin-conjugated anti–c-kit Ab (ACK4, a gift from S-I Nishikawa, Kyoto, Japan31). The cells were then stained with streptavidin-conjugated phycoerythrin (PE) (Jackson ImmunoResearch Laboratories, West Grove, PA). Ly-6A/E+ c-kit+cells were collected by sorting on a FACStarplus, FACS Vantage (Becton Dickinson, San Jose, CA), or Epics Elite (Coulter KK, Tokyo, Japan) cell sorter.
Clonal methylcellulose culture.
The methylcellulose culture was performed by using 35-mm suspension culture dishes (Becton Dickinson Labware, Lincoln Park, NJ). Forty enriched marrow cells were plated in the methylcellulose media containing SF and IL-11 with or without IL-7. In some experiments, FL was also added additionally. The culture media consisted of α-MEM (Flow Laboratories, Rockville, MD), 1.2% 1500-centipoise methylcellulose (Shinetsu Chemical, Tokyo, Japan), 25% fetal calf serum (FCS, Intergen, Purchase, NY and Hyclone, Logan, UT), 1% deionized fraction V bovine serum albumin (BSA, Sigma, St. Louis, MO), and 0.1 mmol/L 2-ME (Sigma). Dishes were incubated at 37°C in a humidified atmosphere flushed with 5% CO2 for 9 to 12 days.
In vivo transfer of enriched primitive progenitors and primary colony cells.
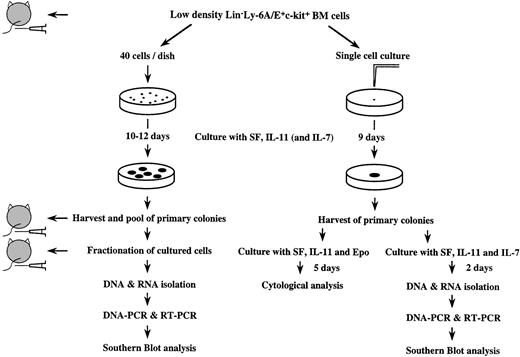
Figure 1 gives a diagrammatic outline of our experimental protocol. Four hundred resulting primary colonies were individually picked, pooled, and washed. Ten percent of the pooled cells (equivalent to 40 primary colonies) and 5,000 freshly prepared enriched marrow cells were injected intravenously into C.B.-17 scid mice (Ly-1.2, Taconic Farms, Germantown, NY and Clea Japan, Inc). Fifteen, 20, and 60 days later, recipient mice were sacrificed and thymocytes were stained with FITC-conjugated anti-Ly-1.1 Ab (clone: H11-86.1, PharMingen, San Diego, CA,32), PE-conjugated anti-CD8 antibody and biotin-conjugated anti-CD4 Ab followed by streptavidin-Tri-Color (Caltag Laboratories, South San Francisco, CA). The stained cells were analyzed for Ly-1.1+ donor-derived cells using a FACStarplus, FACS Vantage, or Epics Elite. Based on isotype control and control-noninjected mice, we defined recipient mice as being reconstituted when donor-derived cells composed more than 1% of thymocytes.
Schematic presentation of experimental protocol (for detail refer to text).
Schematic presentation of experimental protocol (for detail refer to text).
Enrichment of the pooled primary colony cells for T-cell progenitors.
Approximately 100 resulting primary colonies were individually picked and pooled during days 10 to 12 of culture. The cells were stained with FITC-conjugated anti–Ly-6A/E Ab and PE-conjugated anti-FcγRII/III Ab and sorted on the basis of the expression of Ly-6A/E and/or FcγRII/III. Crude and sorted cells were then injected intravenously into C.B.-17 scid mice based on the percentages of the cells in the sorting windows. Three weeks after the cell transfer, thymocytes of each recipient mouse were analyzed by flow cytometry for the expression of Ly-1.1. In some experiments, pooled primary colony cells were subjected to DNA and RNA extraction for DNA-polymerase chain reaction (PCR) and reverse transcriptase (RT)-PCR analysis, respectively.
Cultured pre-B cells.
Cultured pre-B cells were obtained as we described previously.21 Briefly, 40 enriched marrow cells were cultured in the methylcellulose media containing SF, IL-11, and IL-7. On day 10 of culture, resulting primary colonies were individually picked, pooled, and recultured in the methylcellulose media containing SF and IL-7. Large compact unicentric colonies consisting of small round cells were harvested, and the cells were used as pre-B cells.
DNA isolation and PCR.
Genomic DNA was extracted by using an isolation kit, Micro-TurboGen (Invitrogen, San Diego, CA) or QIAamp Blood Kit (Qiagen, Chatsworth, CA) following the manufacturers’ instructions. PCR was performed by using 100 ng DNA, 360 ng/reaction of primers, and 1U Taq DNA polymerase (GIBCO-BRL, Gaithersburg, MD) or 1U AmpliTaq Gold DNA polymerase (Perkin Elmer, Foster City, CA). The sequences of primers used to detect the Dβ2-Jβ2 rearrangement of the TCRβ gene were sequences 5′ of the Dβ2.1 (5′-GTAGGCACCTGTGGGGAAGAAACT-3′) and 3′ of the Jβ2.6 (5′-TGAGAGCTGTCTCCTACTATCGATT-3′).33 To examine the Vβ-DJβ TCR gene rearrangement, the Dβ2.1 primer was replaced with a mixture of Vβ6 (5′-GAAGGCTATGATGCGTCTCG-3′) and Vβ8 (5′-TCCCTGATGGGTACAAGGCC-3′) primers.34
RNA isolation and RT-PCR.
Total RNA was extracted with TRIzol reagent (GIBCO-BRL, Grand Island, NY) according to the manufacturer’s instructions. Complementary DNA (cDNA) was prepared from total RNA (0.3 to 0.5 μg/reaction), by using 50 ng of random primers (GIBCO-BRL), 200 μmol/L of each deoxynucleotide 5′-triphosphate (Epicentre Technologies, Madison, WI and Takara Biomedicals, Tokyo, Japan), and M-MLV RT (GIBCO-BRL). The oligonucleotide primers used to detect mRNA of constant region of TCR β-chain and pTα were as follows: sense primer (TCR β-chain): GTTTGAGCCATCAAAAGCAGA, antisense primer (TCR β-chain): AGGATCTCATAGAGGATGGT,35 sense primer (pTα): CATGCTTCTCCACGAGTG, antisense primer (pTα): CTATGTCCAAATTCTGTGGGTG.36 In some experiments, the RT-PCR products for pTα were further amplified by nested PCR by using a second set of primers; these primers were as follows: sense primer: TGGTGGTTTGCCTGGTCCTCGATG, antisense primer: GGTCAGGAGCACATCGAGCAGAAG.
Southern blot analysis.
Both DNA-PCR and RT-PCR products were analyzed by Southern blot analysis. Aliquots from each product were size-fractionated on a 1.5% agarose gel, denatured in 0.5 mol/L NaOH, neutralized in 0.5 mol/L Tris-HCl (pH 7.5), and blotted onto Hybond-N+ membranes (Amersham, Arlington Heights, IL). Membranes were hybridized with digoxigenin (DIG)-conjugated oligonucleotide probes. The probes used were as follows: Dβ2-Jβ2 and Vβ-DJβ rearrangement: TTTCCCTCCCGGAGATTCCCTAA,37 TCR β-chain transcripts: GCCTGAGCAGCCGCCTGA,35 and pTα transcripts: CAGGTACTGTGGCTGAGCCTACTG.36 The conjugation of oligonucleotide probes with DIG was performed using a DIG oligonucleotide tailing kit (Boehringer Mannheim, Indianapolis IN). The hybridization was visualized using a DIG luminescent detection kit (Boehringer Mannheim).
T-cell potential of individual progenitors.
Enriched marrow cells were individually plated by micromanipulation into methylcellulose media containing SF, IL-11, and IL-7. On day 9 of the culture, the primary colonies were individually harvested. One-tenth of each primary colony was replated in suspension culture containing SF, IL-11, and Epo for examination of myeloid differentiation. After 5 days of incubation, the cells were centrifuged onto a slide for cytological examination with May-Grunwald Giemsa staining. The remainder of the cells were plated in suspension culture containing SF, IL-11, and IL-7. On day 11 of culture, DNA and RNA were extracted from the cells for study of rearrangement of TCR β-chain gene and mRNA expression of TCR β-chain and pTα.
RESULTS
Differences in the time course of T-cell reconstitution between freshly prepared primitive progenitors and their progenies developing in culture.
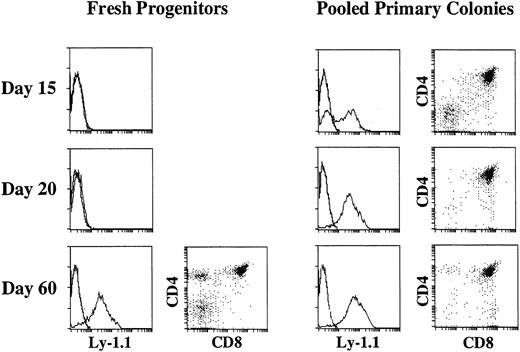
We previously showed that primary colonies developing in the presence of SF and IL-11 from murine bone marrow progenitors have the potential to reconstitute the T-cell compartments of scid mice on intravenous injection.22 This observation suggested that the differentiation of primitive hematopoietic progenitors along T-cell lineage took place in culture. However, considering that stem cells can be maintained in culture under similar conditions for 2 to 3 weeks,38 39 it is also possible that the transplanted stem cells were responsible for the T-cell reconstitution. Therefore, to determine whether or not the differentiation toward T-cell lineage takes place in culture, we first compared the time course of T-cell reconstitution of scid mice by 40 pooled primary colonies (1.5 × 106 cells on average) growing in the presence of SF and IL-11 with the time course of T-cell reconstitution by freshly prepared 5,000 primitive progenitors. Because the plating efficiency of the enriched marrow cells observed in cultures supported by SF and IL-11 is approximately 50%, 5,000 enriched marrow cells should give rise to 2,500 primary colonies, which is 65 times the number of the primary colonies injected into the first group of mice. Representative analyses are presented in Fig 2. It is evident that both primary colonies and fresh primitive progenitors had potential to reconstitute the thymus of scid mice. When primary colonies were injected, donor-derived Ly-1.1+ cells appeared in the thymi (1.9 × 106 Ly-1.1+ cells on average) of all of the mice as early as on day 15 of the cell injection. Although the majority of the donor-derived cells were CD4+CD8+ DP cells, significant numbers of CD4−CD8− DN cells were also observed. On day 20 the thymi became approximately 10-times larger (1.6 ×107 Ly-1.1+ cells on average). Even on day 60 donor-derived thymocytes (1.0 × 107Ly-1.1+ cells on average) were still found. In contrast, Ly-1.1+ donor-derived cells were absent on day 15 and 20, but appeared by day 60 in the thymi of the scid mice injected with 5,000 freshly prepared primitive progenitors. The fact that the primary colony cells reconstituted the thymus earlier than the freshly prepared primitive progenitors suggested that differentiation toward T-cell lineage took place in culture in the presence of SF and IL-11. The primary colonies growing in culture with IL-7 in addition to SF and IL-11 also reconstituted the thymus earlier than freshly prepared primitive progenitors (data not shown).
Three-color flow cytometric analysis of thymocytes of C.B.-17 scid mice injected with freshly prepared primitive marrow progenitors or pooled primary colonies. Five thousand enriched marrow cells (left) or the equivalent of 40 pooled primary colonies (right) were injected intravenously into scid mice. On days 15, 20, 25, and 60 after cell transfer, thymocytes were analyzed for Ly-1.1, CD4, and CD8. The expression of Ly-1.1 on whole thymocytes and that of CD4 and CD8 on Ly-1.1+ cells at each time is shown.
Three-color flow cytometric analysis of thymocytes of C.B.-17 scid mice injected with freshly prepared primitive marrow progenitors or pooled primary colonies. Five thousand enriched marrow cells (left) or the equivalent of 40 pooled primary colonies (right) were injected intravenously into scid mice. On days 15, 20, 25, and 60 after cell transfer, thymocytes were analyzed for Ly-1.1, CD4, and CD8. The expression of Ly-1.1 on whole thymocytes and that of CD4 and CD8 on Ly-1.1+ cells at each time is shown.
Enrichment for thymic reconstitution activity.
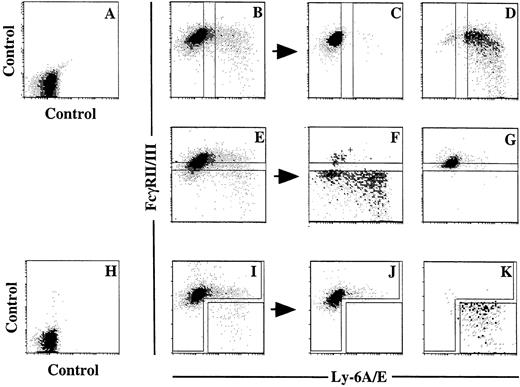
We next attempted to enrich the progenitors that reconstituted the thymus of scid mice from the pooled primary colony cells. After testing several cell surface markers including CD25, CD44, c-kit, Thy-1, and IL-7 receptor, we found Ly-6A/E and FcγRII/III to be useful. A summary of the results of 4 representative experiments is presented in Table 1. Primary colonies supported by the combination of SF and IL-11 with (experiments 3 and 4) or without (experiments 1 and 2) the addition of IL-7 were sorted on the basis of the expression of Ly-6A/E and FcγRII/III. FL was also added in addition to IL-7 in experiment 4. Crude and sorted cells that were in proportion to the percentages of the cells in the sorting windows were injected into C.B.-17 scid mice. Representative dot-plots of pooled primary colony cells, sorting windows, and dot-plots of sorted cells are presented in Fig 3. In experiment 1, in which culture was performed without IL-7, the percentages of the cells in the Ly-6A/E−, Ly-6A/E+, FcγRII/III−/low, and FcγRII/IIIhighwindows were 80%, 12%, 8.4%, and 75%, respectively. Therefore, 6.7 × 105 crude, 5.3 × 105Ly-6A/E−, 8.4 × 104Ly-6A/E+, 5.6 × 104FcγRII/III−/low, and 5.0 × 105FcγRII/IIIhigh cells were injected into each of three scid mice. In all of the mice injected with crude cells, Ly-6A/E+ cells and FcγRII/III−/lowcells, the thymi were reconstituted with Ly-1.1+thymocytes, whereas no donor-derived cells were detected in the mice receiving Ly-6A/E−cells and FcγRII/IIIhigh cells. The same results were obtained in experiments 3 and 4 in which IL-7 was added. Approximately half of the cultured cells were positive for Ly-6A/E in experiment 4 in which FL was also added. Finally, experiment 2 showed that the reconstituting cells were enriched in the Ly-6A/E+FcγRII/III−/low cell population.
Enrichment of Thymic Reconstitution Activity
| Exp . | Cells Injected . | Thymus Reconstitution* . | Mean No. (Mean %) of DonorDerived Thymocytes . | ||
|---|---|---|---|---|---|
| Cytokines Used . | Phenotype . | Cell No (%) . | |||
| 1 | SF IL-11 | Crude | 6.7 × 105 (100) | 3/3 | 4.6 × 105 (37) |
| Ly-6A/E− | 5.3 × 105 (80) | 0/3 | |||
| Ly-6A/E+ | 8.4 × 104 (12) | 3/3 | 1.1 × 106 (54) | ||
| FcγRII/III−/low | 5.6 × 104 (8.4) | 3/3 | 8.1 × 106 (83) | ||
| FcγRII/IIIhigh | 5.0 × 105 (75) | 0/3 | |||
| 2 | SF IL-11 | Ly-6A/E+ FcγRII/III−/low | 2.8 × 104 (2.3) | 3/3 | 1.8 × 107 (73) |
| Rest of the cells | 1.2 × 106 (95) | 0/3 | |||
| 3 | SF IL-11 IL-7 | Ly-6A/E− | 1.1 × 106 (70) | 0/2 | |
| Ly-6A/E+ | 1.7 × 105 (11) | 1/2 | 5.9 × 104 (3.1) | ||
| FcγRII/III−/low | 1.0 × 105 (6.5) | 2/2 | 1.1 × 105 (8.6) | ||
| FcγRII/IIIhigh | 1.1 × 106 (72) | 0/2 | |||
| 4 | SF IL-11 IL-7 | Ly-6A/E− | 6.0 × 105 (43) | 0/2 | |
| FL | Ly-6A/E+ | 6.0 × 105 (43) | 2/2 | 4.6 × 107 (82) | |
| FcγRII/III−/low | 1.2 × 105 (8.8) | 2/2 | 9.2 × 106 (60) | ||
| FcγRII/IIIhigh | 7.0 × 105 (63) | 0/2 | |||
| Exp . | Cells Injected . | Thymus Reconstitution* . | Mean No. (Mean %) of DonorDerived Thymocytes . | ||
|---|---|---|---|---|---|
| Cytokines Used . | Phenotype . | Cell No (%) . | |||
| 1 | SF IL-11 | Crude | 6.7 × 105 (100) | 3/3 | 4.6 × 105 (37) |
| Ly-6A/E− | 5.3 × 105 (80) | 0/3 | |||
| Ly-6A/E+ | 8.4 × 104 (12) | 3/3 | 1.1 × 106 (54) | ||
| FcγRII/III−/low | 5.6 × 104 (8.4) | 3/3 | 8.1 × 106 (83) | ||
| FcγRII/IIIhigh | 5.0 × 105 (75) | 0/3 | |||
| 2 | SF IL-11 | Ly-6A/E+ FcγRII/III−/low | 2.8 × 104 (2.3) | 3/3 | 1.8 × 107 (73) |
| Rest of the cells | 1.2 × 106 (95) | 0/3 | |||
| 3 | SF IL-11 IL-7 | Ly-6A/E− | 1.1 × 106 (70) | 0/2 | |
| Ly-6A/E+ | 1.7 × 105 (11) | 1/2 | 5.9 × 104 (3.1) | ||
| FcγRII/III−/low | 1.0 × 105 (6.5) | 2/2 | 1.1 × 105 (8.6) | ||
| FcγRII/IIIhigh | 1.1 × 106 (72) | 0/2 | |||
| 4 | SF IL-11 IL-7 | Ly-6A/E− | 6.0 × 105 (43) | 0/2 | |
| FL | Ly-6A/E+ | 6.0 × 105 (43) | 2/2 | 4.6 × 107 (82) | |
| FcγRII/III−/low | 1.2 × 105 (8.8) | 2/2 | 9.2 × 106 (60) | ||
| FcγRII/IIIhigh | 7.0 × 105 (63) | 0/2 | |||
Enriched marrow cells were cultured (40 cells/dish) in methylcellulose with designated cytokines for 10 to 12 days. Approximately 100 primary colonies were pooled and sorted for Ly-6A/E−, Ly-6A/E+, FcγRII/III−/low, and FcγRII/IIIhighcells. In experiment 2, Ly-6A/E+FcγRII/III−/low cells and the rest of the cells were sorted. Crude and sorted cells in proportion to the percentage of the cells in the sorting windows were then injected intravenously into C.B.-17 scid mice. Three weeks after the cell transfer, thymocytes were analyzed for Ly-1.1 expression. Expression of Ly-6A/E and FcγRII/III by pooled primary colony cells and the sorting windows are presented in Fig 3. Reanalysis of the sorted cells are also shown in Fig 3.
Number of reconstituted mice per number of scid mice injected.
Flow cytometric analyses of Ly-6A/E and FcγRII/III expression by pooled primary colony cells. (A and H) Isotype control. (B) Expression of Ly-6A/E and FcγRII/III by pooled primary colony cells and sorting windows for Ly-6A/E− and Ly-6A/E+ cells. (C) Sorted Ly-6A/E− cells. (D) Sorted Ly-6A/E + cells. (E) Expression of Ly-6A/E and FcγRII/III by pooled primary colony cells and sorting windows for FcγRII/ III−/low and FcγRII/IIIhigh cells. (F) Sorted FcγRII/III−/low cells. (G) Sorted FcγRII/IIIhigh cells. (I) Expression of Ly-6A/E and FcγRII/III by pooled primary colony cells and sorting windows for Ly-6A/E+ FcγRII/III−/low cells and the rest of the cells. (J and K) Sorted cells using the windows presented in (I).
Flow cytometric analyses of Ly-6A/E and FcγRII/III expression by pooled primary colony cells. (A and H) Isotype control. (B) Expression of Ly-6A/E and FcγRII/III by pooled primary colony cells and sorting windows for Ly-6A/E− and Ly-6A/E+ cells. (C) Sorted Ly-6A/E− cells. (D) Sorted Ly-6A/E + cells. (E) Expression of Ly-6A/E and FcγRII/III by pooled primary colony cells and sorting windows for FcγRII/ III−/low and FcγRII/IIIhigh cells. (F) Sorted FcγRII/III−/low cells. (G) Sorted FcγRII/IIIhigh cells. (I) Expression of Ly-6A/E and FcγRII/III by pooled primary colony cells and sorting windows for Ly-6A/E+ FcγRII/III−/low cells and the rest of the cells. (J and K) Sorted cells using the windows presented in (I).
Rearrangement status of TCR β-chain gene.
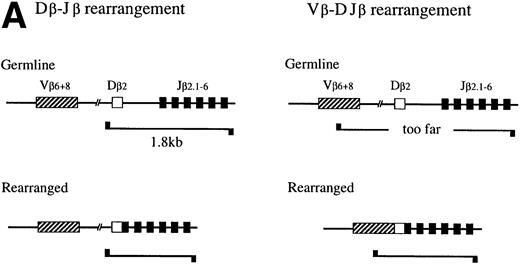
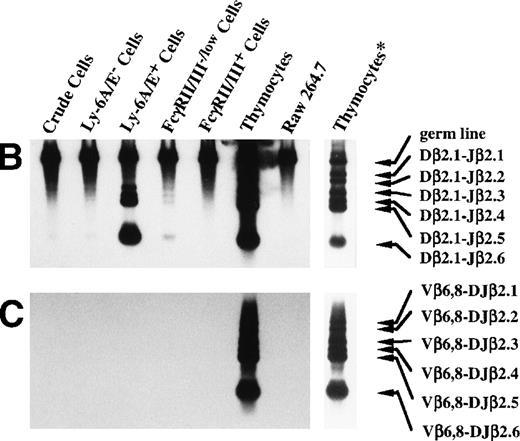
Using the enrichment method described above, we next examined the degree to which the primitive hematopoietic progenitors can differentiate toward T-cell lineage in culture. Because it has been reported that IL-7 enhances the TCR β-chain gene rearrangement40,41 and block apoptosis of pro-T cells,13,14 we used the cells cultured in the presence of IL-7. Although TCR β-chain gene rearrangement is generally believed to start in DN stage in the thymus,42,43 recent reports suggested that TCR β-chain gene rearrangement could take place in prothymic T-cell progenitors. For example, it has been reported that the prethymic T-cell progenitors isolated from fetal murine blood20 and Sca-1+ Lin−marrow cells from euthymic and athymic adult mice35 exhibit the D-J rearrangement of the TCR β-chain gene. Therefore, we first focused on the TCR β-chain gene rearrangement. DNA was extracted from Ly-6A/E−, Ly-6A/E+, Fc γRII/III−/low, and FcγRII/IIIhighprimary colony cells for the PCR analysis of the D-J and the V-DJ rearrangement status of the TCR β-chain gene as outlined in Fig 4A. Dβ2-Jβ2 rearrangement was observed in 2 of 10 analyses. The results of one of the two Southern blot analyses are shown in Fig 4B and 4C. In the analysis of the Dβ2-Jβ2 rearrangement, the genomic DNA from a control monocyte/macrophage cell line (Raw 264.7)44 gave only a 1.8-kb germ line band, whereas thymocytes showed six additional Dβ2-Jβ2 rearranged bands, ie, Dβ2-Jβ2.1∼Dβ2-Jβ2.6, corresponding to rearranged Dβ2-Jβ2 genes using six Jβ2 segments. The crude cultured cells, Ly-6A/E− cells, and FcγRII/IIIhigh cells barely showed the Dβ2-Jβ2.6 band. In contrast, the Ly-6A/E+ cells gave rise to all six D-J rearranged bands, Dβ2-Jβ2.1∼Dβ2-Jβ2.6. The FcγRII/III−/low cells showed four bands, Dβ2-Jβ2.3∼Dβ2-Jβ2.6 (Fig 4B). In the remaining eight analyses, Dβ2-Jβ2 rearrangement was not observed. We also examined rearrangement of Vβ6 and Vβ8 segments to study V-DJ rearrangement. However, none of the fractions of cultured cells showed the Vβ-DJβ2 rearrangement in any analyses even after a long exposure (Fig 4B). These results suggested that, although not frequently, primitive hematopoietic progenitors can differentiate along T-cell lineage in culture to the stage in which the D-J rearrangement, but not the the V-DJ rearrangement of the TCR β-chain gene is initiated. Although there was a marked difference in signal intensity between the Ly-6A/E+ and FcγRII/III−/low cell populations in the experiment presented in Fig 4B, such difference was not observed in the other analysis. Infrequency of the cells possessing Dβ2-Jβ2 rearrangement may have caused uneven distribution of the cells to the two cell fractions in that experiment.
PCR analysis of the Dβ-Jβ and Vβ-DJβ rearrangements of the TCR gene. (A) Schematic presentations of the PCR-based analyses of β-chain D-J (left) and V-DJ (right) gene rearrangements. (B) and (C) Enriched primitive marrow progenitors were cultured with SF, IL-11, and IL-7 in methylcellulose. On day 11 of culture, resulting primary colonies were individually harvested, pooled, and sorted on the basis of the expression of Ly-6A/E and FcγRII/III. DNA was extracted from crude and sorted cells, control monocyte/macrophage cell line (Raw 264.7) and thymocytes. One hundred micrograms of DNA was PCR amplified for 25 cycles for (Dβ-Jβ, B) or for 35 cycles (Vβ-DβJβ, C). Signals were visualized by using a DIG-conjugated probe and a DIG luminescent detection kit. * Signals were obtained after a short exposure.
PCR analysis of the Dβ-Jβ and Vβ-DJβ rearrangements of the TCR gene. (A) Schematic presentations of the PCR-based analyses of β-chain D-J (left) and V-DJ (right) gene rearrangements. (B) and (C) Enriched primitive marrow progenitors were cultured with SF, IL-11, and IL-7 in methylcellulose. On day 11 of culture, resulting primary colonies were individually harvested, pooled, and sorted on the basis of the expression of Ly-6A/E and FcγRII/III. DNA was extracted from crude and sorted cells, control monocyte/macrophage cell line (Raw 264.7) and thymocytes. One hundred micrograms of DNA was PCR amplified for 25 cycles for (Dβ-Jβ, B) or for 35 cycles (Vβ-DβJβ, C). Signals were visualized by using a DIG-conjugated probe and a DIG luminescent detection kit. * Signals were obtained after a short exposure.
mRNA transcription of TCR β-chain and pTα.
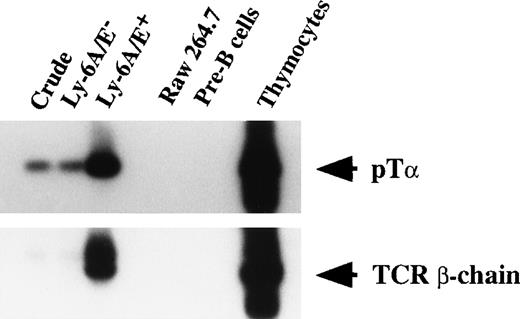
Since TCR gene gives rise to unrearranged or sterile transcript before the gene rearrangement starts,45 we next examined transcription of TCR β-chain. We also studied the mRNA expression of pTα. pTα expression has been reported to be exquisitely T-cell–lineage specific and found in the precursors of αβ T cells outside the thymus as well as in intrathymic sites in both murine20,34,46 and human.47-49 Primary colonies supported by SF, IL-11, and IL-7 were used. In all the three experiments performed, primarily Ly-6A/E+ and FcγRII/III−/low cells expressed TCR β-chain and pTα transcripts. A representative analysis is shown in Fig 5. These results indicated that primitive hematopoietic progenitors differentiate to progenitors in which TCR β-chain and pTα are actively transcribed. Because V-DJ rearrangement of the TCR β-chain gene was not observed, as stated earlier, it is likely that transcripts of TCR β-chain are derived from sterile transcription of unrearranged or partially rearranged TCR β-chain loci.
mRNA expression of pT and TCR β-chain by fractionated pooled primary colony cells. Total RNA was purified from a monocyte/macrophage cell line (Raw 264.7), thymocytes, and crude and fractionated pooled primary colony cells. Next, 0.3 μg to 0.5 μg of RNA was subjected to reverse transcription. One-tenth of the cDNA was then PCR-amplified for 45 cycles. Signals were visualized by using a DIG-conjugated probe and a DIG luminescent detection kit.
mRNA expression of pT and TCR β-chain by fractionated pooled primary colony cells. Total RNA was purified from a monocyte/macrophage cell line (Raw 264.7), thymocytes, and crude and fractionated pooled primary colony cells. Next, 0.3 μg to 0.5 μg of RNA was subjected to reverse transcription. One-tenth of the cDNA was then PCR-amplified for 45 cycles. Signals were visualized by using a DIG-conjugated probe and a DIG luminescent detection kit.
T-cell transcripts of pre-B cells.
Considerable TCR gene rearrangement within B-cells have been noted in transformed as well as nontransformed cells.50-53 We previously reported that, under similar culture conditions, murine lymphohematopoietic progenitors can give rise to B-cell progenitors, which further differentiate into pre-B cells on reculture in secondary culture containing SF and IL-7.21,53 55 Therefore, one may consider that the B-cell progenitors developed in culture may have contributed the transcripts of TCR β-chain and pTα. To test this possibility, we examined pre-B cells developed in culture in mRNA expression of TCR β-chain and pTα. As shown in Fig 6, pre-B cells did not show mRNA of TCR β-chain and pTα, whereas primary colony cells, especially Ly-6A/E+ cells did. This result suggested that the mRNA expression of TCR β-chain and pTα are derived from putative T-cell progenitors.
mRNA expression of pT and TCR β-chain by cultured pre-B cells. Fractionated pooled primary colony cells, Raw 264.7, pre-B cells, and thymocytes were analyzed as to mRNA expression of pT and TCR β-chain.
mRNA expression of pT and TCR β-chain by cultured pre-B cells. Fractionated pooled primary colony cells, Raw 264.7, pre-B cells, and thymocytes were analyzed as to mRNA expression of pT and TCR β-chain.
Study of individual primitive progenitors.
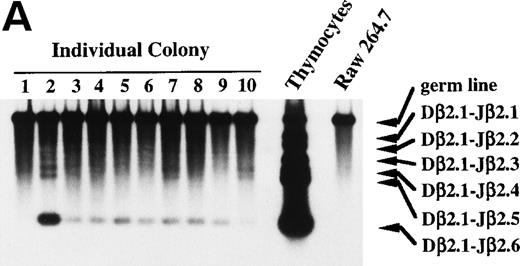
To show unequivocally that primitive hematopoietic progenitors, but not contaminating committed T-cell progenitors differentiate in culture to the stage in which the Dβ-Jβ rearrangement and the expression of T-cell transcripts start, we performed micromanipulation of the progenitors. Eighty-six primary colonies developed from micromanipulated single progenitors in the presence of SF, IL-11, and IL-7 yielded sufficient DNA for the subsequent PCR analysis. All primary colonies showed differentiation into at least granulocyte and macrophage lineages in the myeloid secondary culture. Although most of the primary colonies showed Dβ2-Jβ2.6 rearrangement bands, we assumed that only the primary colonies that showed Dβ2-Jβ2.1∼Dβ2-Jβ2.5 rearrangement bands are positive for DJ rearrangement, because non–T-lineage control cells sometimes gave rise to the Dβ2-Jβ2.6 band. According to this criterion, 4 (4.7%) of the 86 primary colonies proved positive for Dβ-Jβ rearrangement. The results of the Southern blot analysis of 10 individual primary colonies are presented in Fig 7A. Sample 2 is an example showing Dβ2-Jβ2.4 and Dβ2-Jβ2.5 rearrangement bands in addition to a distinct Dβ2-Jβ2.6 band. None of the colonies showed Vβ-DJβ rearrangement (data not shown).
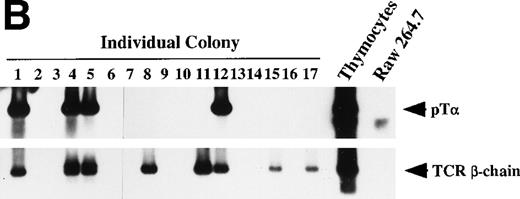
Dβ-Jβ TCR gene rearrangement and mRNA expression of pT and TCR β-chain by individual primary colonies. Genomic DNA and total RNA were extracted from aliquots of each primary colony derived from micromanipulated individual primitive marrow progenitors. (A) The DNA of each primary colony was used for the PCR analysis of the Dβ-Jβ rearrangement of the TCR gene. A Southern blot analysis of 10 representative primary colonies. (B) The RNA of each primary colony was examined for the presence of pT and TCR β-chain mRNA by RT-PCR. Southern blot analyses of 17 primary colonies.
Dβ-Jβ TCR gene rearrangement and mRNA expression of pT and TCR β-chain by individual primary colonies. Genomic DNA and total RNA were extracted from aliquots of each primary colony derived from micromanipulated individual primitive marrow progenitors. (A) The DNA of each primary colony was used for the PCR analysis of the Dβ-Jβ rearrangement of the TCR gene. A Southern blot analysis of 10 representative primary colonies. (B) The RNA of each primary colony was examined for the presence of pT and TCR β-chain mRNA by RT-PCR. Southern blot analyses of 17 primary colonies.
We also examined mRNA expression of TCR β-chain and pTα in a total of 17 primary colonies that yielded sufficient RNA to perform RT-PCR. The results of the Southern blot analysis of these primary colonies are shown in Fig 7B. Of the 17 colonies, 4 (24%) showed transcript of TCR β-chain, but not that of pTα, and 4 (24%) produced both. There was no primary colony that showed only pTα transcript. All 17 colonies showed differentiation potentials in at least 2 myeloid lineages on reculture in SF, IL-11, and Epo. These results confirmed that in the presence of SF, IL-11 and IL-7 significant number of primitive progenitors can differentiate along T-cell lineage in addition to myeloid lineages. None of these 17 primary colonies showed Dβ-Jβ rearrangement of TCR β-chain (data not shown). It suggested that most of transcripts of TCR β-chain were sterile transcripts of unrearranged TCR loci.
DISCUSSION
In this study, we showed that, in the presence of SF, IL-11, and IL-7, murine primitive hematopoietic progenitors can differentiate to express mRNA of TCR β-chain and pTα. These putative T-cell progenitors were enriched in Ly-6A/E+ and FcγRII/III−/low cell populations, and in the same cell fractions the accelerated thymic reconstitution activity was found as well. Although not frequently, the differentiation progressed to involve the Dβ-Jβ rearrangement but not the Vβ-DJβ rearrangement of the TCR gene. Dβ-Jβ rearrangement suggested activation of the rearrangement machinery. Indeed, RAG-1 mRNA was also expressed by primary colony cells, particularly by the Ly-6A/E+ and FcγRII/III−/low cell populations (data not shown). B-cell progenitors generated in culture were also enriched in the same cell fractions and possibly express RAG-1 (data not shown), however, it is not clear whether the putative T-cell progenitors expressed RAG-1. We have used micromanipulation of single progenitor cells to show unequivocally that the putative T-cell progenitors are derived from lymphohematopoietic progenitors. These results suggested the differentiation of lymphohematopoietic progenitors to T-cell progenitors in culture.
However, sorting experiments showed that myeloid and B-lymphoid progenitors were always coenriched with the putative T-cell progenitors in the same cell fractions (data not shown). Therefore, we have no direct evidence at this time for the differentiation of committed T-cell progenitors that have T-cell, but not myeloid nor B-lymphoid potential. In addition, It is also unknown whether the cells expressing the T-cell transcripts reconstituted the thymus of scid mice. The accelerated thymic reconstitution by cultured cells suggested, but did not necessarily indicate that the reconstituting cells were already committed to T-cell lineage. One possibility is that primitive pluripotent progenitors like stem cells were responsible for the thymic reconstitution. As we reported in the previous study,22donor-derived thymocytes were observed even 4 months after the injection of cultured cells. This long-term reconstitution suggests the maintenance of stem cells in culture and supports this possibility. However, they may not account for the accelerated thymic reconstitution. Another possibility is that the reconstituting cells had already differentiated toward T-cell lineage to some extent but not committed to T-cell lineage yet. Further characterization of the putative T-cell progenitors developed in culture remains to be performed in future study.
Recently, investigators in a number of laboratories documented the presence of committed T-cell progenitors outside the thymus. Rodewald et al20 isolated Thy-1+ c-kitlowCD3− committed T-cell progenitors from murine fetal blood, some of which have expressed pTα mRNA and have performed the Dβ-Jβ but not the Vβ-DJβ rearrangement. Soloff et al35 also observed the presence of germline transcripts and partial D-J rearrangement of TCR β-chain gene in adult murine bone marrow. The T-cell progenitors presented in this paper may be analogous to these committed T-cell progenitors. Bruno et al49identified, in adult human peripheral blood, CD4+CD3− CD14− committed T-cell progenitor populations that express T-cell transcripts including pTα, RAG-1, CD3γ, CD3δ, and CD3ε and have initiated D-J rearrangement of TCR β-chain. Ktorza et al56 recently found a similar population in human cord blood. These results and ours suggest that the commitment to T-cell lineage can occur prethymically. However, it does not exclude the possibility that stem cells or common lymphoid progenitors migrate into the thymus and then perform final commitment to the T-cell lineage.
We used SF, IL-11, and IL-7 to induce differentiation toward T-cell lineage. It has been shown that IL-7 is an essential cytokine for the proliferation and survival of early lymphoid progenitors including immature thymocytes.6-12,57-63 Akashi et al13and Maraskovsky et al14 recently reported that IL-7 blocks apoptosis of pro-T cells. In agreement with this, recent preliminary single-cell culture experiments showed that the removal of IL-7 from the culture reduced the expression of mRNA of TCR β-chain and pTα. It has also been reported that IL-7 could support the Dβ-Jβ,41 Vβ-DJβ,43 and Vγ-Jγ64 rearrangements of the TCR genes. We do not know at this time whether or not IL-7 is required for the Dβ-Jβ rearrangement because of the low efficiency of Dβ-Jβ rearrangement in culture. We have reported earlier that the two cytokines SF and IL-11 synergistically support the early proliferation of primitive hematopoietic progenitors and their differentiation toward myeloid lineages in both human65 and murine66 systems as well as toward B-cell lineage in a murine system.21 We also reported that SF, IL-11, and IL-7 can support the development of natural killer (NK) cell progenitors from murine lymphohematopoietic progenitors.67 Thus, primitive lymphohematopoietic progenitors can differentiate into all lymphohematopoietic lineages, including myeloid, B-lymphoid, and NK cell lineages in the presence of SF, IL-11, and IL-7, and in this report we have shown that differentiation toward T-cell lineage also takes place in culture. This culture system will be useful in delineating the mechanisms regulating commitment of common lymphoid progenitors and characterization of the cytokines specific for individual lymphoid lineages.
Supported by National Institutes of Health Grants No. DK32294 and DK48714, Office of Research and Development, Medical Research Service, Department of Veterans Affairs, and contributions from Kirin Brewery Co, Ltd, Japan.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Fumiya Hirayama, MD, Hokkaido Red Cross Blood Center, Yamanote 2-2, Nishi-ku, Sapporo 063-0002, Japan.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal