Abstract
The promoter region of the Bβ fibrinogen gene containing the polymorphic site (G−455-A) shows an increase in fibrinogen levels for individuals containing an adenine rather than a guanine. Two methods were used to explore the possible functional role of this region. Electrophoretic mobility shift assays (EMSAs) were performed using specific DNA probes containing base sequences pertinent to the allelic site. Specific DNA binding proteins were detected and their binding characteristics were determined. Secondly, we placed DNA fragments containing different −455 nucleotide substitutions of the Bβ promoter upstream of a luciferase reporter gene and transfected them into HepG2 cells to determine their effect on transactivation. An adenine at position −455 resulted in greater luciferase activity than when a guanine was present. UV cross-linking bound protein to the DNA demonstrated a 47-kD protein binding preferentially to the site when a guanine rather than an adenine was present at −455. We hypothesize that a transactivation protein complex associates with the site, but its association is stronger when guanine is present, thereby slowing downstream Bβ gene transcription. These data provide the first molecular evidence that accounts for the increase in fibrinogen in individuals carrying this allele.
© 1998 by The American Society of Hematology.
ELEVATED LEVELS OF fibrinogen are an independent risk factor for the development of cardiovascular diseases.1-9 Fibrinogen is one of the primary agents in blood clot formation at sites of vascular damage. In instances of physical trauma or infection, fibrinogen is transiently upregulated by the cytokine interleukin-6 (IL-6).10-13 However, elevated circulating fibrinogen levels are also associated with increased age, gender, race, smoking, obesity, stress, elevated cholesterol, and menopause.1 Structurally, fibrinogen is comprised of three pairs of nonidentical polypeptide chains: Aα, Bβ, and γ.10,12,13 The chains are encoded by three separate genes clustered on chromosome 4q23-32.13 These genes are under such stringent transcriptional control that if one chain is overexpressed, the other two will upregulate; however, the molecular basis of this coordinated signaling is not understood.14,15The identified regulatory elements further complicate the understanding of the coordinated transcriptional control, because the promoter regions of the three chains are not completely identical. All three genes have a type II IL-6 response element and TATA-like sequences; however, only the γ chain has a USF site and the Aα and Bβ chains have C/EBP sites immediately proximal to the IL-6 response sequence and, further downstream, HNF-1 sites.16-19 In addition to these identified regulatory elements, there may be a number of other, yet unidentified factors that are involved in regulating transcription of each gene.
All three fibrinogen chains are essential for function and secretion of the molecule; however, emphasis has been placed on the Bβ chain for two reasons. First, in studies performed to understand the order of fibrinogen assembly, it was suggested that the Bβ chain may be the more prominent chain in monomer assembly, thereby marking it as the nucleating chain.20-23 Second, additional experiments performed in human systems have shown that, when any one of the three fibrinogen chain genes is transcriptionally upregulated, the result is subsequent upregulation of the other two chains, with the largest increase in total chain and fibrinogen production occurring when the Bβ chain is overexpressed.14,15,24,25 These observations suggest that one of the determining factors of circulating fibrinogen levels could be the rate at which the Bβ chain gene is transcriptionally regulated. It has been suggested that polymorphisms found on the proximal promoter portion of the 5′ flanking region of the Bβ gene may influence the interaction of transcription factors that potentially bind to these polymorphic sites.26 To date, a total of 11 polymorphisms have been found on the Bβ gene.27 Presently, no evidence has shown that there is a direct correlation between Bβ chain alleles and cardiovascular disease development; however, there is a direct correlation between Bβ chain alleles and elevated circulating fibrinogen levels.26,28-34 One polymorphism of interest is a guanine-to-adenine (G → A) transition at the −455 position of the 5′ flanking region of the Bβ gene. Epidemiological evidence has shown that an adenine in the −455 position (the A allele) expresses increased levels of circulating fibrinogen.27-42 One previously reported study (abstract) using 20-bp oligonucleotide probes implicated a protein binding within this region, but no additional information has been reported.43
In this study, we have detected three complexes binding within the region −468 to −439 of the Bβ chain gene using electrophoretic mobility shift assays (EMSAs). All three complexes bind specifically to this site; however, one of the complexes preferentially binds when a guanine is present in the −455 position rather than an adenine. Functional analyses show that there is a 1.2- to 1.5-fold increase in Bβ chain gene transcription when an adenine is in the −455 position, instead of a guanine. Mutation of the Bβ chain gene sequence from −462 to −451 prevents binding of this approximately 47-kD complex. These observations show a protein complex that is allele specific and may potentially participate in Bβ chain regulation by partially repressing Bβ chain gene synthesis. Furthermore, binding of this complex may not only affect the amount of Bβ chain that is produced, but also total circulating fibrinogen levels, given the coordinated transcriptional control of all three chains.
MATERIALS AND METHODS
Cell culture.
The human hepatocarcinoma cell line, HepG2 (ATCC, Rockville, MD), was used and maintained in Dulbecco’s modified Eagle’s medium (DMEM; Cellgro, Herndon, VA, and Sigma, St Louis, MO), 1× nonessential amino acids (Cellgro), 1 mmol/L sodium pyruvate, 10% fetal bovine serum (Sigma), and 1× antibiotic/antimycotic (ciprofloxacin [Pentex, Kankakee, IL], and later penicillin/streptomycin [Cellgro] and nystatin [GIBCO-BRL, Grand Island, NY]). Cells were incubated at 37°C under 5% CO2.
Plasmid constructs and functional assays.
Constructs of the Bβ gene from −636 to +16 were derived by polymerase chain reaction (PCR) amplification. Primers for the constructs were designed to facilitate subcloning of the region from −623 to +9 into the pGL2 basic luciferase vector (Promega, Madison, WI), which uses the luciferase gene as its reporter. HepG2 DNA is heterozygous for both alleles; therefore, the amplified A allele construct was isolated by digesting G allele fragments with the restriction enzyme Hae III and then subcloned into the pGL2 vector, and the G allele was obtained by performing site-directed mutagenesis by the procedure provided by the manufacturer (Clontech, Palo Alto, CA) on the A allele construct in the pGL2 vector. The subcloned G and A allele constructs were sequenced (Amersham, Arlington Heights, IL) for verification of alleles and to ensure that no mutations were introduced during PCR amplification. The vectors plus construct were transfected into HepG2 cells using the calcium phosphate transfection method.44 Transfected cells were stimulated with 50 ng/mL of recombinant human IL-6 (rhIL-6; Promega) for 16 hours. The cells were harvested, lysates were prepared, and luciferase production was quantitated using a luciferase assay system kit (Promega) and a luminometer (Biorad, Hercules, CA).
Nuclear extract preparation.
Nuclear extracts were obtained from HepG2 cells by performing a modification of the published protocol.45 Cells were washed twice with cold phosphate-buffered saline (PBS), scraped from the plates, resuspended in PBS, and pelleted at 1,500 rpm for 5 minutes at 4°C. The pellets were resuspended in 5 mL of buffer A (10 mmol/L HEPES, pH 7.6, 15 mmol/L KCl, 0.15 mmol/L spermine, 0.5 mmol/L spermidine, 2 mmol/L EDTA, pH 8.0, 2.4 mol/L sucrose, 0.5 mmol/L dithiothreitol [DTT], 0.5 mmol/L phenylmethyl sulfonyl fluoride [PMSF], and 1% trasylol aprotinin [15 mg/mL]) and, using a tight pestle, homogenized with 20 to 25 strokes in a Dounce homogenizer. The cell homogenate volume was increased to 14 mL with buffer A, divided into four 3.5-mL volumes, and placed over 1.3 mL of buffer B (10 mmol/L HEPES, pH 7.6, 15 mmol/L KCl, 0.15 mmol/L spermine, 0.5 mmol/L spermidine, 2 mmol/L EDTA, pH 8.0, 2.0 mol/L sucrose, 10% glycerol, 0.5 mmol/L DTT, 0.5 mmol/L PMSF, and 1% trasylol aprotinin [15 mg/mL]) to form a sucrose gradient. The samples were centrifuged at 27,400 rpm for 2 hours at 2°C. The supernatants were removed and the pellets were pooled and resuspended in 200 mL of extraction buffer (0.4 mol/L KCl, 20 mmol/L HEPES, pH 7.0, 20% glycerol, 2 mmol/L DTT, 1 mmol/L benzamidine, 0.2 mmol/L PMSF, 0.2 mmol/L EDTA, pH 8.0, 1 mmol/L sodium fluoride, 1 mmol/L sodium diphosphate, and 0.1 mmol/L sodium orthovanadate) and centrifuged at 30,000 rpm for 1 hour at 0°C to pellet the chromatin. The supernatant was removed from the chromatin pellet, aliquoted, and stored at −80°C for use in EMSAs.
EMSAs.
Oligonucleotides of 34 bp were synthesized by Integrated DNA Technologies, Inc (Coralville, IA) and radiolabeled with [α-32P] TTP (0.4 ng/λ, ∼100,000 cpm/λ; Amersham) by the Klenow fragment of DNA Polymerase I (Boehringer Mannheim, Indianapolis, IN). Nuclear extracts were thawed on ice before being incorporated into the binding reaction. Protein concentrations of the nuclear extracts were determined by using a commercially available protein assay kit (Pierce, Rockford, IL). For the binding reactions, 20 μg of nuclear extract was added to binding buffer (10 mmol/L Tris, pH 7.5, 1 mmol/L DTT, 100 mmol/L KCl, 1 mmol/L EDTA, 0.2 mmol/L PMSF, 1 mg/mL bovine serum albumin [BSA], 5% glycerol), and 1 mmol/L DTT and preincubated at room temperature for 10 minutes with 1 μg of poly dI/dC · poly dI/dC in a reaction volume of 20 μL. Radiolabeled probe was then added to the binding reactions and incubation at room temperature continued for 20 minutes. The competition assays were performed using unlabeled probes combined with radiolabeled probes added to the binding reaction together to prevent proteins from binding to one probe before the other was introduced. The binding reactions were loaded onto 7% nondenaturing polyacrylamide gels and run in 0.5× TBE at 500 V for 2.5 to 3 hours. Gels were placed on Whatman 3 paper (Whatman, Maidstone, UK), dried, and exposed to autoradiograph film for 8 to 24 hours.
UV cross-linking assay.
UV cross-linking analysis was performed according to the published procedure46 and by using binding assays as described for the EMSAs; however, whole cell extract instead of nuclear extract was used in the binding reaction. The whole cell extract was obtained by using a modification of the published procedure.47 At 4°C, the cells were washed twice with cold PBS, scraped from the plates with 500 mL of PBS, and pelleted at 1,500 rpm for 1 minute, and the supernatants were discarded. Two volumes of extraction buffer (same used for nuclear extract), approximately 250 μL, was added, the pellets were vortexed, and the samples were put in an ethanol/dry-ice bath for 2 to 3 minutes. The contents were allowed to thaw on ice and vigorously mixed and vortexed, followed by centrifugation for 15 minutes at 13,000 rpm in the cold. The supernatants were removed and aliquoted, and the pellets were discarded. After the binding reactions were completed, the samples were covered with plastic wrap and exposed to UV light at 254 nm for 5 minutes at approximately 5 cm from the UV light source. The samples were then resolved on a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel.
For isolation of DNA-bound complex III, whole cell extract (as described) was used in the EMSA binding reaction and allowed to bind onto a 34-bp internally labeled oligonucleotide probe. The binding reactions were scaled up 5×, UV irradiated for 10 minutes, and treated with DNAse I (Promega; published procedure).46 The reactions were run on a 4% nondenaturing gel 2× thicker than the previous EMSAs; the gel was not dried and was exposed to autoradiograph film overnight. The band corresponding to DNA-bound complex III was cut from the gel, eluted (published procedure),48 and run on a 10% SDS-PAGE gel.
RESULTS
In vitro model of G/A allelic differences in Bβ fibrinogen expression.
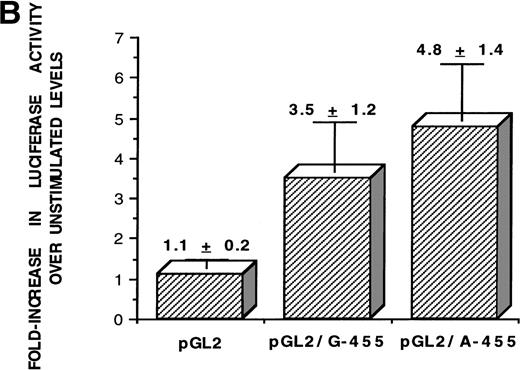
Epidemiological studies have indicated that individuals heterozygous or homozygous for an adenine in the −455 position of the Bβ chain gene have higher circulating fibrinogen levels than their homozygous G (guanine at −455) counterparts. To observe if the alleles affected transcriptional activity, the functionality of the promoter was assessed when the A allele (adenine in the −455 position) or the G allele (guanine in the −455 position) is present. Constructs of the region of the Bβ chain gene from −636 to +16, containing all of the known proximal promoter elements required for Bβ chain transcription (HNF-1 site, C/EBP consensus site, and IL-6 response element), were synthesized by PCR amplification from HepG2 DNA and subcloned into the pGL2 basic luciferase vector, containing an adenine or guanine in the −455 position (Fig 1A). After transfection into HepG2 cells and 16 hours of stimulation with rhIL-6, luciferase activity was quantitated (Fig 1B). A series of 10 independent luciferase experiments were performed and the increase in luciferase activity over basal level readings was averaged. Activity of the pGL2 vector only (control) was low, as expected, with an average luciferase activity of 1.1- ± 0.2-fold. There was approximately 1.4-fold greater luciferase activity from the pGL2/A-455 construct over the pGL2/G-455 construct, with average increases in luciferase activity after IL-6 stimulation of 3.2- ± 1.2-fold for pGL2/G-455 and 4.8- ± 1.4-fold for pGL2/A-455; the difference in luciferase activity between both alleles was statistically significant (P = .048). The pGL2/A-455 construct had higher luciferase activity than the pGL2/G-455 construct, between the range of 1.2- to 1.5-fold in the independent experiments. The results of these functional studies show that, in these in vitro conditions, the A allele is associated with increased Bβ chain transcription. These results coincide with the epidemiological evidence, correlating the A allele with higher levels of circulating fibrinogen.
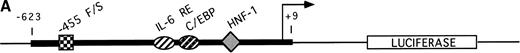
The A allele shows greater promoter functionality in response to IL-6. (A) The sequence of the Bβ chain gene from −623 to +9 was subcloned into the pGL2 basic luciferase vector, upstream from the luciferase gene. This portion of the Bβ chain gene contains all of the known proximal promoter elements required for induction of Bβ chain gene transcription: HNF-1 (hepatocyte nuclear factor-1), −79 to −91; C/EBP site (CAAT-enhancer binding protein site), −124 to −132; IL-6 RE (IL-6 response element), −137 to −142,16 and, in addition, the −455 F/S (−455 nucleotide and its proximal 5′ and 3′ flanking sites). (B) After stimulation of HepG2 cells transfected with either pGL2 vector only (control), pGL2/G-455 (guanine at −455 of the Bβ construct), and pGL2/A-455 (adenine at −455 of the Bβ construct) with IL-6 for 16 hours, the luciferase activity was quantitated and the fold increase in luciferase activity over basal level was statistically averaged for 10 independent experiments. The activity for the pGL2 vector only was 1.1- ± 0.2-fold, for pGL2/G-455 was 3.5- ± 1.2-fold, and for pGL2/A-455 was 4.8- ± 1.4-fold increase over basal level activity. Statistical analyses using a two-sample t-test for independent samples showed that luciferase activity under the control of the pGL2/A-455 construct was significantly different from pGL2/G-455 activity (P = .048).
The A allele shows greater promoter functionality in response to IL-6. (A) The sequence of the Bβ chain gene from −623 to +9 was subcloned into the pGL2 basic luciferase vector, upstream from the luciferase gene. This portion of the Bβ chain gene contains all of the known proximal promoter elements required for induction of Bβ chain gene transcription: HNF-1 (hepatocyte nuclear factor-1), −79 to −91; C/EBP site (CAAT-enhancer binding protein site), −124 to −132; IL-6 RE (IL-6 response element), −137 to −142,16 and, in addition, the −455 F/S (−455 nucleotide and its proximal 5′ and 3′ flanking sites). (B) After stimulation of HepG2 cells transfected with either pGL2 vector only (control), pGL2/G-455 (guanine at −455 of the Bβ construct), and pGL2/A-455 (adenine at −455 of the Bβ construct) with IL-6 for 16 hours, the luciferase activity was quantitated and the fold increase in luciferase activity over basal level was statistically averaged for 10 independent experiments. The activity for the pGL2 vector only was 1.1- ± 0.2-fold, for pGL2/G-455 was 3.5- ± 1.2-fold, and for pGL2/A-455 was 4.8- ± 1.4-fold increase over basal level activity. Statistical analyses using a two-sample t-test for independent samples showed that luciferase activity under the control of the pGL2/A-455 construct was significantly different from pGL2/G-455 activity (P = .048).
Detection of DNA binding proteins and assessment of binding specificity.
The increase in luciferase activity under control of the A allele implies that a transactivation factor(s) binds to this site, resulting in an increase in Bβ gene transcription. Furthermore, the presence of a particular nucleotide in the −455 position must affect the binding of the transactivating factor(s) by altering its binding affinity, which could affect Bβ transcription.
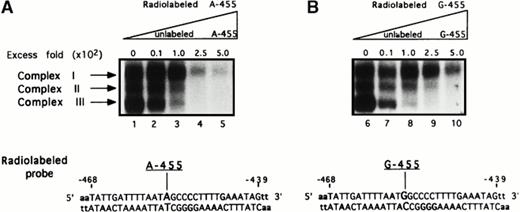
To detect the presence of DNA binding proteins, EMSAs were performed. The EMSAs were performed using 34-bp oligonucleotide probes, having the sequence from −468 to −439 of the Bβ chain gene with either a guanine (G-455 probe, representing the G-allele) or an adenine (A-455 probe, representing the A-allele) in the −455 position (Fig 2), that were allowed to bind with HepG2 nuclear extract. The results showed three constitutively bound complexes, designated complex I, complex II, and complex III on both A-455 and G-455 probes, present before and after IL-6 stimulation (data not shown), which is evident with the other Bβ chain gene response elements.18 19
Presence of complexes binding onto the −455 flanking region. To detect complexes that may potentially bind onto the −455 nucleotide and its proximal 5′ and 3′ flanking regions, two 34-bp oligonucleotide probes were used in EMSA analysis having the sequence of the Bβ chain from −468 to −439. The probes contained either a guanine (G-455) or an adenine (A-455) in the −455 position. EMSA results resolved on 7% nondenaturing gels show three complexes binding onto the radiolabeled probes when either an adenine (A-455), representing the A allele (A, lanes 1 through 5), or a guanine (G-455), representing the G allele (B, lanes 6 through 10), is present. To assess the specificity of the bound complexes, both probes were competed with increasing concentrations of unlabeled probe: 0-fold, 10-fold (4 ng), 100-fold (40 ng), 250-fold (100 ng), and 500-fold (200 ng) excess the radiolabeled probe concentration. Both competition assays, radiolabeled A-455 versus unlabeled A-455 (lanes 1 through 5) and radiolabeled G-455 versus unlabeled G-455 (lanes 6 through 10), show that all three complexes are binding onto both probes specifically; however, complexes II and III appear to bind more specifically.
Presence of complexes binding onto the −455 flanking region. To detect complexes that may potentially bind onto the −455 nucleotide and its proximal 5′ and 3′ flanking regions, two 34-bp oligonucleotide probes were used in EMSA analysis having the sequence of the Bβ chain from −468 to −439. The probes contained either a guanine (G-455) or an adenine (A-455) in the −455 position. EMSA results resolved on 7% nondenaturing gels show three complexes binding onto the radiolabeled probes when either an adenine (A-455), representing the A allele (A, lanes 1 through 5), or a guanine (G-455), representing the G allele (B, lanes 6 through 10), is present. To assess the specificity of the bound complexes, both probes were competed with increasing concentrations of unlabeled probe: 0-fold, 10-fold (4 ng), 100-fold (40 ng), 250-fold (100 ng), and 500-fold (200 ng) excess the radiolabeled probe concentration. Both competition assays, radiolabeled A-455 versus unlabeled A-455 (lanes 1 through 5) and radiolabeled G-455 versus unlabeled G-455 (lanes 6 through 10), show that all three complexes are binding onto both probes specifically; however, complexes II and III appear to bind more specifically.
To determine if these protein complexes were specific, a series of competition assays were performed in which the radiolabeled probes were competed against their unlabeled sequences, radiolabeled A-455 versus unlabeled A-455 (Fig 2, lanes 1 through 5) and radiolabeled G-455 versus unlabeled G-455 (Fig 2, lanes 6 through 10). The unlabeled competitors were competed against the labeled probes at concentrations of 10-fold (4 ng), 100-fold (40 ng), 250-fold (100 ng), and 500-fold (200 ng) the concentration of the radiolabeled probes. The results show that all three complexes are binding to both probes specifically; however, complexes II and III are competed away at lower concentrations of unlabeled competitor, implying a more specific binding affinity to this site. A 1,000-fold excess of cold competitor was required to compete away complex I (data not shown).
Detection of preferential allelic binding.
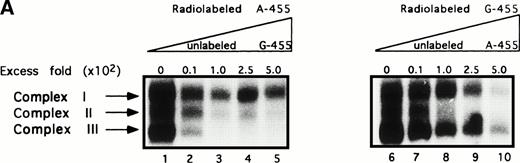
Three protein complexes have been shown to bind within the region −468 to −439. Furthermore, the three complexes are able to bind specifically to this site regardless of whether an adenine or a guanine is in the −455 position. It was of interest to determine whether these complexes may bind preferentially to either one of the two nucleotides. As a result, a set of cross-competition EMSAs were performed in which radiolabeled A-455 was competed against unlabeled G-455 (Fig 3A, lanes 1 through 5) and radiolabeled G-455 was competed against unlabeled A-455 (Fig 3A, lanes 6 through 10) at the same concentrations used in the previous competition assays. The competition assay involving radiolabeled A-455 versus unlabeled G-455 showed complexes II and III being competed away, as in the previous competition assays; however, complex I was not competed away to the same extent as the other two complexes, which is also comparable to the previous competition assays. In the radiolabeled G-455 versus unlabeled A-455 cross-competition assay, again as in the three previous competition assays, complex II was competed away and, to a lesser extent, complex I. Conversely, complex III was unable to be completely competed away from the radiolabeled G-455 probe by unlabeled A-455 competitor. In fact, at 1,000-fold unlabeled A-455 competitor, complex III is still present (data not shown). These results suggest that one of the three complexes, complex III, preferentially binds within the region of interest when a guanine rather than an adenine is present.
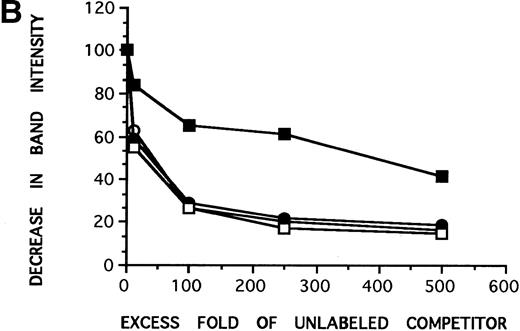
Complex III preferentially binds to the G allele. (A) In the cross-competition assays, performed to detect preferential allelic binding of the complexes, when radiolabeled A-455 probe is competed against increasing concentrations of unlabeled G-455 probe (lanes 1 through 5), the three complexes are able to be competed away from the radiolabeled A-455 probe, with competition patterns similar to the results in Fig 2. Complex III is not competed away from radiolabed G-455 by increasing concentrations of A-455 unlabeled competitor (lanes 6 through 10), even at 1,000-fold cold competitor (data not shown), as seen in the three previous competition assays, implying that complex III preferentially binds to the G allele or a guanine in the −455 position. (B) Decrease in band intensity of complex III at increasing concentrations of cold competitor was analyzed by scanning densitometer analysis to observe the strength with which the unlabeled competitiors competed against the radiolabeled probes. Results for all of the competition assays were comparable with the exception of the labeled G-455 probe versus unlabled A-455 probe competition assay. The unlabeled A-455 competitor was unable to competitively bind with complex III in the presence of labeled G-455 at the same rate as the other competition assays, also showing that complex III preferentally binds to the G allele, rather than to the A allele. The assays were resolved on 7% nondenaturing gels. (○), Labeled A-455 probe versus unlabeled A-455 probe; (•), labeled A-455 probe versus unlabeled G-455 probe; (□), labeled G-455 probe versus unlabeled G-455 probe; (▪), labeled G-455 probe versus unlabeled A-455 probe.
Complex III preferentially binds to the G allele. (A) In the cross-competition assays, performed to detect preferential allelic binding of the complexes, when radiolabeled A-455 probe is competed against increasing concentrations of unlabeled G-455 probe (lanes 1 through 5), the three complexes are able to be competed away from the radiolabeled A-455 probe, with competition patterns similar to the results in Fig 2. Complex III is not competed away from radiolabed G-455 by increasing concentrations of A-455 unlabeled competitor (lanes 6 through 10), even at 1,000-fold cold competitor (data not shown), as seen in the three previous competition assays, implying that complex III preferentially binds to the G allele or a guanine in the −455 position. (B) Decrease in band intensity of complex III at increasing concentrations of cold competitor was analyzed by scanning densitometer analysis to observe the strength with which the unlabeled competitiors competed against the radiolabeled probes. Results for all of the competition assays were comparable with the exception of the labeled G-455 probe versus unlabled A-455 probe competition assay. The unlabeled A-455 competitor was unable to competitively bind with complex III in the presence of labeled G-455 at the same rate as the other competition assays, also showing that complex III preferentally binds to the G allele, rather than to the A allele. The assays were resolved on 7% nondenaturing gels. (○), Labeled A-455 probe versus unlabeled A-455 probe; (•), labeled A-455 probe versus unlabeled G-455 probe; (□), labeled G-455 probe versus unlabeled G-455 probe; (▪), labeled G-455 probe versus unlabeled A-455 probe.
To estimate the binding kinetics occurring in the competition assays, the decrease in band intensity for complex III with increasing concentrations of unlabeled competitor was established by using densitometer scanning analysis. The results were plotted with decrease in band intensity of complex III against increasing concentrations of unlabeled competitor (Fig 3B). The plots show similar patterns of competition for radiolabeled A-455 versus unlabeled A-455, radiolabeled G-455 versus unlabeled G-455, and radiolabeled A-455 versus unlabeled G-455. However, the plot of radiolabeled G-455 versus unlabeled A-455 shows that the unlabeled A-455 competitor is unable to compete complex III away from the radiolabeled G-455 probe to the extent that it is competed away in the other three competition assays.
Importance of the Bβ fibrinogen −455 nucleotide.
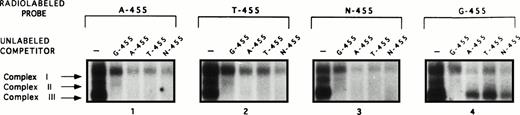
The EMSAs performed used probes that were representative of the G and A alleles, both of which are purines. It was of interest to observe how a pyrimidine, or even the absence of a nucleotide, in the −455 position would affect binding of the complexes, especially complex III. Another series of competition assays involving EMSAs were performed using two additional probes again containing the sequence from −439 to −468, denoted as T-455 (thymine in the −455 position) and N-455 (absence of a nucleotide in the −455 position; Fig 4). Binding of all three complexes was evident on the T-455 and N-455 probes, just as on the G-455 and A-455 probes. To assess the binding specificity of these complexes on the two additional probes, the radiolabeled A-455, T-455, N-455, and G-455 probes were competed against 500-fold unlabeled G-455, A-455, T-455, and N-455 competitors (Fig 4). In the competition assays involving radiolabeled A-455, T-455, and N-455, all three complexes were competed away, with complexes II and III being competed away to a greater extent than complex I. However, the only probe that was able to compete complex III away from radiolabeled G-455 probe (window no. 4) was itself, unlabeled G-455.
A guanine in the −455 position is important for preferential binding of complex III. In addition to G-455 and A-455 oligonucleotide probes being used in the EMSA analyses, two additional probes with the sequence from −468 to −439 with the following changes, T-455 (thymine in −455 position) and N-455 (no nucleotide in −455 position), were used to determine the effect that the nucleotide in the −455 position has on the binding patterns of the complexes. The radiolabeled probes (A-455, T-455, N-455, and G-455) were competed against all four unlabeled probes at a concentration of 500-fold excess labeled probe. Evident on the 7% nondenaturing gel, complex III was completely competed away in all of these competition assays, except the radiolabeled G-455 competition assay (window no. 4), thus providing more evidence supporting the allelic specficity of complex III to the G-allele.
A guanine in the −455 position is important for preferential binding of complex III. In addition to G-455 and A-455 oligonucleotide probes being used in the EMSA analyses, two additional probes with the sequence from −468 to −439 with the following changes, T-455 (thymine in −455 position) and N-455 (no nucleotide in −455 position), were used to determine the effect that the nucleotide in the −455 position has on the binding patterns of the complexes. The radiolabeled probes (A-455, T-455, N-455, and G-455) were competed against all four unlabeled probes at a concentration of 500-fold excess labeled probe. Evident on the 7% nondenaturing gel, complex III was completely competed away in all of these competition assays, except the radiolabeled G-455 competition assay (window no. 4), thus providing more evidence supporting the allelic specficity of complex III to the G-allele.
The results of these assays show that all three complexes are able to bind within the region of interest when an adenine, a guanine, or even a pyrimidine, such as thymine, or no nucleotide is present in the −455 position. These data further indicate that complex III appears to preferentially bind within the region of interest when a guanine is in the −455 position.
Analysis of complex III.
To explore in more detail the importance of the nucleotide sequence surrounding the −455 nucleotide, we constructed a series of mutations in the 5′ and 3′ flanking regions. EMSAs were performed using probes with the sequence of the Bβ chain gene from −468 to −439 having the G allele, but hexanucleotide sequences from −462 to −457 (GM-2), −456 to −451 (GM-1), and −450 to −445 (GM-3) were mutated by changing the purines to pyrimidines and the pyrimidines to purines (Fig 5A). Probes GM1 and GM2 show marked alteration of binding of all three complexes; however, GM3 shows no hindrance in complex binding. The results of the EMSAs showed that mutation of the sequence from −462 to −451 affected the binding of all three complexes (Fig 5B). These findings indicate that this sequence, which immediately flanks the −455 nucleotide, is important for recognition and binding of all three complexes, especially complex III.
Analysis of complex formation. (A) To determine the effect of sequence disruption on formation of the complexes, EMSA analyses were performed using three new oligonucleotide probes in which three hexanucleotide sequences, −462 to −457 (probe GM-2), −456 to −451 (probe GM-1), and −450 to −445 (probe GM-3), were mutated by changing the purines to pyrimidines and the pyrimidines to purines. (B) The results show that all three complexes, especially complex III, were unable to sufficiently bind when the sequence from −462 to −451 was mutated. (C) UV cross-linking analysis of complex III was performed to determine the specificity of its binding onto the G-455 probe and to approximate its molecular weight. All samples were binding reactions containing whole cell extract, were irradiated for 5 minutes (unless indicated otherwise), and were resolved on a 10% SDS-PAGE gel. Lane 1, no protein extract; lane 2, no irradiation; lane 3, protein extract (nonreduced); lane 4, protein extract (reduced); lane 5, addition of 100-fold unlabeled G-455 probe; lane 6, no extract, instead BSA (50 μg); lane 7, a probe with the same sequence as G-455 (Fig2A), but internally labeled on the bottom strand with [-32P] dCTP, was used to verify the molecular weight of complex III, obtained from previous UV cross-linking analyses. After UV irradiation, DNAse I treatment, and isolation of the DNA-bound complex III band from a 4% nondenaturing gel, resolution on a 10% SDS-PAGE gel still shows that the approximate molecular weight of complex III is 47 kD. The results show that complex III is a protein of approximately 47 kD that specifically binds to the G-455 probe, containing the sequence from −468 to −439 of the Bβ chain gene, flanking the −455 nucleotide.
Analysis of complex formation. (A) To determine the effect of sequence disruption on formation of the complexes, EMSA analyses were performed using three new oligonucleotide probes in which three hexanucleotide sequences, −462 to −457 (probe GM-2), −456 to −451 (probe GM-1), and −450 to −445 (probe GM-3), were mutated by changing the purines to pyrimidines and the pyrimidines to purines. (B) The results show that all three complexes, especially complex III, were unable to sufficiently bind when the sequence from −462 to −451 was mutated. (C) UV cross-linking analysis of complex III was performed to determine the specificity of its binding onto the G-455 probe and to approximate its molecular weight. All samples were binding reactions containing whole cell extract, were irradiated for 5 minutes (unless indicated otherwise), and were resolved on a 10% SDS-PAGE gel. Lane 1, no protein extract; lane 2, no irradiation; lane 3, protein extract (nonreduced); lane 4, protein extract (reduced); lane 5, addition of 100-fold unlabeled G-455 probe; lane 6, no extract, instead BSA (50 μg); lane 7, a probe with the same sequence as G-455 (Fig2A), but internally labeled on the bottom strand with [-32P] dCTP, was used to verify the molecular weight of complex III, obtained from previous UV cross-linking analyses. After UV irradiation, DNAse I treatment, and isolation of the DNA-bound complex III band from a 4% nondenaturing gel, resolution on a 10% SDS-PAGE gel still shows that the approximate molecular weight of complex III is 47 kD. The results show that complex III is a protein of approximately 47 kD that specifically binds to the G-455 probe, containing the sequence from −468 to −439 of the Bβ chain gene, flanking the −455 nucleotide.
UV cross-linking assays were performed to approximate the molecular mass of complex III (Fig 5C). For this assessment, whole cell extract was used in the binding assays along with the G-455 probe, because complexes I and II are not as enriched as they are in nuclear extract and complex III becomes the most prominent band (data not shown). This was advantageous in avoiding a partial purification step. After 5 minutes of irradiation (or indicated otherwise), results show that, in lane 1, in which no protein extract has been added to the binding reaction, there is no band present, indicating that there are no extraneous proteins in the binding reaction that would bind to the G-455 probe. Lane 2 shows the absence of band formation before irradiation. Both lanes 3 and 4, representing nonreduced and reduced protein, respectively, show a prominent band representing complex III, migrating at approximately 47 kD. Lane 5 shows a decrease in band intensity upon the addition of 100-fold unlabeled G-455 probe, indicating that the complex binds specifically to the G-455 probe and is able to be competed away from the radiolabeled probe by its unlabeled sequence as a competitor. In lanes 3 through 5, in addition to complex III, there are higher molecular weight complexes present that have been observed in all of the EMSAs; the importance of these complexes is presently being investigated. Lane 6, consisting of the binding reaction with 50 μg of BSA instead of protein extract, shows no band formation, confirming that the DNA-protein interaction between complex III and the G-455 probe is specific. To exclude the possibility that the oligonuceotide probe may alter the molecular weight, the DNA-bound complex III was UV irradiated, treated with DNAse I, run, and cut out of the nondenaturing gel. The eluted DNA-protein complex was subsequently run on a 10% SDS-PAGE gel. The complex continued to migrate at approximately 47 kD (lane 7). These results suggest that the protein comprising complex III is approximately 47 kD and specifically recognizes and binds to the DNA sequence of the G-455 probe.
DISCUSSION
The G-to-A transition at the −455 position of the 5′ flanking portion of the Bβ gene is an allele that has been associated with varying levels of circulating fibrinogen. Individuals that are homozygous for the G allele (guanine at −455) have the lowest circulating levels in comparison to heterozygous and homozygous A allele (adenine at −455) individuals. The epidemiological evidence for these findings provides results that are generally statistically significant; however, the increase in circulating fibrinogen levels with regard to the presence of A alleles is not dramatic.27-42 Presently, it is not known how such a slight increase in circulating fibrinogen levels can potentially have injurious physiological consequences. To better understand the possible connection between increased fibrinogen levels and regulation of the Bβ gene, we performed molecular analyses to observe transcriptional activities occurring at the −455 site and proximal flanking regions.
We detected three complexes that bind onto the region of the Bβ gene from −468 to −439. The three complexes are able to specifically bind to this region whether an adenine (the A allele), a guanine (the G allele), a pyrimidine, or no nucleotide is present in the −455 position. One of those three complexes, complex III, preferentially binds to the G allele, which is associated with the lower circulating fibrinogen levels. These data imply that the peculiar DNA-protein interaction that is occurring, with regard to complex III, is of a quantitative rather than of a qualitative nature, meaning that the presence of a particular nucleotide does not completely abolish binding, but the strength of the interaction is affected.
Information from functional analyses showed that, under in vitro conditions, the A allele is correlated with greater luciferase activity, implying that the A allele is somehow associated with increased Bβ chain transcription. The results of the functional assays coincide with the evidence provided in the epidemiological studies that connect the A allele with higher circulating fibrinogen levels. From these observations, it can be inferred that the detected complex, complex III, may have potential repressor-like functions in the transcriptional regulation of the Bβ chain gene. Furthermore, its partial repressive activities may be altered if it is not able to sufficiently bind to its recognition sequence if not conserved, as in the case of the A allele being present. If this occurs, Bβ chain transcription may be elevated, even slightly, resulting in increased levels of circulating fibrinogen. The existence of a negative regulatory element associated with this chain is not without precedent. Other studies have shown repressive elements associated with the human Aα, γ, and a more proximal portion of the Bβ chain.16,17 19
Complex III was the only complex identified that showed preferential allelic binding. Analysis of this protein complex suggested that it has a molecular weight of approximately 47 kD. Furthermore, its recognition sequence appears to reside within the region of −462 to −451.
The data shown in this study provide the initial steps in identifying a differential element in Bβ chain regulation that may be one of the factors causing increased amounts of circulating fibrinogen. Furthermore, an increase in fibrinogen, in conjunction with other genetic or external factors, may be one of the many predisposing components in cardiovascular disease development.
Supported in part by the GAANN Fellowship (to E.T.B.).
Address reprint requests to Gerald M. Fuller, PhD, University of Alabama at Birmingham, Department of Cell Biology, 1918 University Blvd, MCLM 680, Birmingham, AL 35294-0005.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

![Fig. 5. Analysis of complex formation. (A) To determine the effect of sequence disruption on formation of the complexes, EMSA analyses were performed using three new oligonucleotide probes in which three hexanucleotide sequences, −462 to −457 (probe GM-2), −456 to −451 (probe GM-1), and −450 to −445 (probe GM-3), were mutated by changing the purines to pyrimidines and the pyrimidines to purines. (B) The results show that all three complexes, especially complex III, were unable to sufficiently bind when the sequence from −462 to −451 was mutated. (C) UV cross-linking analysis of complex III was performed to determine the specificity of its binding onto the G-455 probe and to approximate its molecular weight. All samples were binding reactions containing whole cell extract, were irradiated for 5 minutes (unless indicated otherwise), and were resolved on a 10% SDS-PAGE gel. Lane 1, no protein extract; lane 2, no irradiation; lane 3, protein extract (nonreduced); lane 4, protein extract (reduced); lane 5, addition of 100-fold unlabeled G-455 probe; lane 6, no extract, instead BSA (50 μg); lane 7, a probe with the same sequence as G-455 (Fig2A), but internally labeled on the bottom strand with [-32P] dCTP, was used to verify the molecular weight of complex III, obtained from previous UV cross-linking analyses. After UV irradiation, DNAse I treatment, and isolation of the DNA-bound complex III band from a 4% nondenaturing gel, resolution on a 10% SDS-PAGE gel still shows that the approximate molecular weight of complex III is 47 kD. The results show that complex III is a protein of approximately 47 kD that specifically binds to the G-455 probe, containing the sequence from −468 to −439 of the Bβ chain gene, flanking the −455 nucleotide.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/9/10.1182_blood.v92.9.3286/5/m_blod42120005aw.jpeg?Expires=1766037944&Signature=HF7~G9F12T1BVDV75EIdn~T4SmZltRdEhPSZIhWIh3gLtA86GUVCPwRVGr6i-fvZ1hQo25eDT8CVqznhLmu66b8jZClJ0npbnxBwqxzbPyjbCl5TZJ68hh55Ud15lpBGP87CcfIbZkySZJ2nFPvNAduxLM0kswBHOX4hWfMiW4MqSX0V08O9Rv6sekr3ZQbbCr8eUZNDwsks4DXXO~KgH9xtELSTh95XD~taGtg6NNsc1hdTwm8mXqMiVlXUlwW6uzJRlvzNr4VSrPoAfRRj1s4Q0aym3AGvWM8khyj9Map~Ws5uKvQ2j4QhiV5rDQM2iUTsTxnxxYMCukdqa~s-MA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. Analysis of complex formation. (A) To determine the effect of sequence disruption on formation of the complexes, EMSA analyses were performed using three new oligonucleotide probes in which three hexanucleotide sequences, −462 to −457 (probe GM-2), −456 to −451 (probe GM-1), and −450 to −445 (probe GM-3), were mutated by changing the purines to pyrimidines and the pyrimidines to purines. (B) The results show that all three complexes, especially complex III, were unable to sufficiently bind when the sequence from −462 to −451 was mutated. (C) UV cross-linking analysis of complex III was performed to determine the specificity of its binding onto the G-455 probe and to approximate its molecular weight. All samples were binding reactions containing whole cell extract, were irradiated for 5 minutes (unless indicated otherwise), and were resolved on a 10% SDS-PAGE gel. Lane 1, no protein extract; lane 2, no irradiation; lane 3, protein extract (nonreduced); lane 4, protein extract (reduced); lane 5, addition of 100-fold unlabeled G-455 probe; lane 6, no extract, instead BSA (50 μg); lane 7, a probe with the same sequence as G-455 (Fig2A), but internally labeled on the bottom strand with [-32P] dCTP, was used to verify the molecular weight of complex III, obtained from previous UV cross-linking analyses. After UV irradiation, DNAse I treatment, and isolation of the DNA-bound complex III band from a 4% nondenaturing gel, resolution on a 10% SDS-PAGE gel still shows that the approximate molecular weight of complex III is 47 kD. The results show that complex III is a protein of approximately 47 kD that specifically binds to the G-455 probe, containing the sequence from −468 to −439 of the Bβ chain gene, flanking the −455 nucleotide.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/9/10.1182_blood.v92.9.3286/5/m_blod42120005cw.jpeg?Expires=1766037944&Signature=reFlo0744K34-VCtZm6ZI5uKmGbb0BDhv4ZSVTXQnCtFrFmI~jzqRyC~r1mDIff8S2klr0FszHSbHzr3244ikfWbxZI~STQu4gjeRDSvKH6CV4mkhh8gHeu0cCZA-ftkLbW6joLO9If5IUzTGCuPrIaim~HdhutPG2NSu8oWF9mtVs2BFeVL3cFAjHzKHgGmIGggCrmFUjafMA9j5h4Qe5~99UP-WgkqLQvlr-q2rLq5YQuldEZ6pof4X2uQnx6276NL1ORJ99lAyW5jRYclyuvHd-LG99bcnZ5CMFBhSimmtnye7VS~kkVV8U~8kJUeJcZ7uI6deE9lBGQ0JEeBiA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal