Abstract
The low-density lipoprotein receptor-related protein (LRP) is a 600-kD scavenger receptor that binds a number of protein ligands with high affinity. Although some ligands do not compete with each other, binding of all is uniformly blocked by the 39-kD receptor-associated protein (RAP). RAP is normally found in the endoplasmic reticulum and seems to function as a chaperone for LRP. To identify the binding sites for RAP, lactoferrin, and plasminogen activator inhibitor-1 (PAI-1), a bacterial expression system has been developed to produce soluble LRP fragments spanning residues 783-1399. These residues overlap most of the CNBr fragment containing the second cluster of complement-type repeats (C). Solid phase binding assays show that 125I-RAP binds to fragments containing three successive complement-type repeats: C5-C7. PAI-1 and lactoferrin bind to the same fragments. A fragment containing C5-C7 also blocks uptake and degradation of 125I-RAP by fibroblasts in a concentration-dependent manner. Binding competition experiments show that RAP, PAI-1, and lactoferrin each inhibit the binding of the others, suggesting that at this site in LRP, RAP acts as a competitive, rather than an allosteric, inhibitor of PAI-1 and lactoferrin binding.
© 1998 by The American Society of Hematology.
THE LOW-DENSITY lipoprotein receptor-related protein (LRP) is a large (600 kD) scavenger receptor that is found on the surface of many types of cells, including fibroblasts, hepatocytes, macrophages, and neurons.1 The receptor contains four clusters of disulfide-bonded, complement-type, and epidermal growth factor (EGF)-like repeats that identify it as a member of the low-density lipoprotein (LDL) receptor family.2 LRP is an endocytic receptor with high selectivity; it is responsible for the uptake and degradation of more than a dozen proteins, most of which are evolutionarily unrelated.3 LRP ligands include α2-macroglobulin (α2M).proteinase and tissue plasminogen activator (t-PA).PA inhibitor-1 (PAI-1) complexes, lactoferrin, lipoprotein lipase, and apoE-containing lipoproteins.3 More recently, it was discovered that LRP internalizes β-amyloid precursor protein4 and thrombospondin.5
A 39-kD protein with high affinity for LRP, known as receptor-associated protein (RAP), was first encountered as a contaminant of α2M affinity-purified receptor.6,7 RAP is resident in the endoplasmic reticulum and functions as a chaperone for LRP folding and secretion.8-10 By binding to several regions of newly synthesized LRP, RAP prevents intracellular ligand-induced aggregation and degradation of the receptor. Although RAP is not normally found outside of the cell, RAP is a useful tool because it competes with all known ligands,1 although the reverse is not true. Estimates of the total number of RAP binding sites on LRP range from two to five.7,11 The number of binding sites for other ligands has not been determined. Examples of RAP competitors include lactoferrin, which binds to the chicken oocyte receptor,12 and lipoprotein lipase, which binds to LRP.13 Other ligands, such as t-PA and α2M, which bind to LRP on hepatoma cells,14 and very low-density lipoprotein (VLDL) and vitellogenin, which bind to the chicken oocyte receptor, do not block the binding of RAP to the same receptors.12 These observations have led to the proposal that RAP blocks binding of all LRP ligands by causing allosteric changes in the receptor.15-17
One or more RAP binding sites were identified in an approximately 600-amino acid, CNBr-generated fragment of LRP that corresponds to a cluster of EGF and complement-type repeats near the N-terminus of the molecule. Willnow et al16 18 showed that this fragment can be expressed as a recombinant minireceptor in mammalian cells and exhibits RAP-binding activity.
We have now expressed the same soluble minireceptor in the periplasm ofEscherichia coli and have shown its specific binding activity towards RAP. Further, by the use of bacterially expressed fragments of this minireceptor, we have localized the binding site for RAP to a small region in LRP consisting of complement-type repeats C5-C7. These residues also constitute the binding site for PAI-1 and lactoferrin. RAP competes with both ligands suggesting that, in this instance, RAP inhibition is due to direct competition rather than allosteric effects.
MATERIALS AND METHODS
Materials.
Human PAI-1 was expressed and purified using the pPAI-1.HIS vector.19 The GST 39-kD expression plasmid,6pLRPex192 containing the LRP cDNA, and mouse fibroblast cell lines20 were obtained from Dr Joachim Herz (University of Texas Southwestern Medical Center, Dallas). Plasmid pTMN was obtained from the American Type Culture Collection (Bethesda, MD), catalog #87197. All 125I-labeled proteins were radiolabeled with Iodobeads (Pierce Chemical Co, Rockford, IL) to a specific activity of 2,000 to 5,000 cpm/ng. Radionuclides were purchased from Amersham Life Sciences, Inc (Arlington Heights, IL). Protein concentrations were determined using the Bio-Rad Protein Assay (Hercules, CA).
Generation of LRP constructs.
cDNA encoding LRP fragments was inserted into a periplasmic expression vector using a polymerase chain reaction (PCR) cloning strategy. Fragment inserts were amplified from the LRP cDNA using the oligonucleotides shown in Table 1. The inserts were digested with BamHI and XhoI (Life Technologies, Gaithersburg, MD) and ligated into vector pSecTagB (Invitrogen) that had been digested with the same enzymes. Recombinant proteins expressed from this vector contain a myctag followed by a 6-His tag at the C-terminus. For insertion into the periplasmic expression vector pTMN,21 the mammalian constructs were amplified a second time with Expand taq polymerase (Boehringer Corp, Indianapolis, IN) to change the restriction enzyme sites. Forward primers were identical to those listed in Table 1, except that the BamHI sequence (GGATCC) was replaced with an NcoI site (CCATGGGA) in the same frame as the pTMN polylinker. The reverse primer was 5′-GGCGGATCCTCAATGATGATGATG-3′, which adds a stop codon followed by a BamHI site after the 6-His tag coding region of the inserts. On cleavage of the OmpA signal peptide, all LRP fragments had “Met-Gly-” as the amino terminus.
PCR Primers Used to Clone LRP Fragment Constructs
| Construct . | Amino Acids . | Oligos . | |
|---|---|---|---|
| E4 | 783-830 | Fwd: | GCCGGATCCGTTGGCACCAACAAATGCCGG |
| Rev: | CGGCTCGAGCACGTAGGATGGGTTCGC | ||
| E4-C3 | 783-875 | Rev: | GCGCCTCGAGGGTGTGCTGATGGCAGAG |
| E4-C5 | 783-956 | Rev: | CGCCTCGAGGGTGGGATAGGCACACGAAGC |
| E4-C7 | 783-1036 | Rev: | CGGCTCGAGGGCCTGGTTGGTGCA |
| E4-C8 | 783-1082 | Rev: | CGGCTCGAGGGTCACTCCCTCACA |
| E4-C10 | 783-1165 | Rev: | GCGCCTCGAGCTGGTCGCAGAGCTCGCC |
| E4-E6 | 783-1246 | Rev: | GCGCCTCGAGGTCCAGGCTGCGGCAGCT |
| C5-C10 | 913-1165 | Fwd: | GCGGGATCCTCAGCCCGCACCTGC |
| Rev: | GCGCCTCGAGCTGGTCGCAGAGCTCGCC | ||
| C5 | 913-956 | Rev: | CGCCTCGAGGGTGGGATAGGCACACGAAGC |
| C5-C7 | 913-1036 | Rev: | CGGCTCGAGGGCCTGGTTGGTGCA |
| C6-C7 | 953-1036 | Fwd: | GCGGGATCCGCCTATCCCACCTGC |
| C9-E6 | 1079-1246 | Fwd: | GCGGGATCCGAGGGAGTGACCCAC |
| Rev: | GCGCCTCGAGGTCCAGGCTGCGGCAGCT | ||
| E5-1399 | 1164-1399 | Fwd: | GCGGGATCCGACCAGTGCTCTCTG |
| Rev: | TGCCTCGAGCATGGAGGCTGCCTCAATGCG | ||
| Construct . | Amino Acids . | Oligos . | |
|---|---|---|---|
| E4 | 783-830 | Fwd: | GCCGGATCCGTTGGCACCAACAAATGCCGG |
| Rev: | CGGCTCGAGCACGTAGGATGGGTTCGC | ||
| E4-C3 | 783-875 | Rev: | GCGCCTCGAGGGTGTGCTGATGGCAGAG |
| E4-C5 | 783-956 | Rev: | CGCCTCGAGGGTGGGATAGGCACACGAAGC |
| E4-C7 | 783-1036 | Rev: | CGGCTCGAGGGCCTGGTTGGTGCA |
| E4-C8 | 783-1082 | Rev: | CGGCTCGAGGGTCACTCCCTCACA |
| E4-C10 | 783-1165 | Rev: | GCGCCTCGAGCTGGTCGCAGAGCTCGCC |
| E4-E6 | 783-1246 | Rev: | GCGCCTCGAGGTCCAGGCTGCGGCAGCT |
| C5-C10 | 913-1165 | Fwd: | GCGGGATCCTCAGCCCGCACCTGC |
| Rev: | GCGCCTCGAGCTGGTCGCAGAGCTCGCC | ||
| C5 | 913-956 | Rev: | CGCCTCGAGGGTGGGATAGGCACACGAAGC |
| C5-C7 | 913-1036 | Rev: | CGGCTCGAGGGCCTGGTTGGTGCA |
| C6-C7 | 953-1036 | Fwd: | GCGGGATCCGCCTATCCCACCTGC |
| C9-E6 | 1079-1246 | Fwd: | GCGGGATCCGAGGGAGTGACCCAC |
| Rev: | GCGCCTCGAGGTCCAGGCTGCGGCAGCT | ||
| E5-1399 | 1164-1399 | Fwd: | GCGGGATCCGACCAGTGCTCTCTG |
| Rev: | TGCCTCGAGCATGGAGGCTGCCTCAATGCG | ||
Abbreviations: Fwd, forward; Rev, reverse.
LRP repeats have been numbered according to Herz et al,2where “E” represents an epidermal growth factor-type repeat and “C” represents a complement-type repeat. Restriction sites (BamHI or Xho I) are underlined. Overhangs of several bases were included at the 5′ ends of the primers to facilitate restriction enzyme cleavage.
Expression and purification of LRP fragments.
Plasmids encoding the LRP fragments were transformed into strain BL21/DE3(pLysS) competent cells (Novagen, Madison, WI) for protein expression. Cultures were grown in Luria-Bertani medium broth containing 0.4% glucose, 15 μg/mL chloramphenicol, and 25 μg/mL ampicillin (LCA) at 37°C. Expressing clones were stored frozen as glycerol stocks and streaked out on LCA plates when required. For each liter of culture, 100 mL of LCA was inoculated with freshly streaked colonies and grown overnight. The bacteria were pelleted, resuspended in a liter of fresh media, and grown to an optical density of 0.5 to 0.6 at 600 nm. After induction with 0.5 mmol/L isopropylthio-β-D-galactoside (IPTG) and 5-hour growth, the cells were obtained by centrifugation at 4,000g for 20 minutes and periplasmic proteins were isolated by cold osmotic shock.22 Stock solutions were added to the osmotic shock fluid to give a final concentration of 50 mmol/L Tris-HCl, pH 8; 150 mmol/L NaCl; 0.1 mmol/L phenylmethylsulfonyl fluoride; 2 mmol/L CaCl2; and 10 mmol/L imidazole (buffer A). The osmotic shock fluid was stirred overnight with Ni-NTA agarose (Qiagen Inc, Santa Clarita, CA) at 4°C and pelleted by centrifugation at 500g. The pellet was then washed four times with fresh buffer A, followed by four washes with buffer A containing 500 mmol/L NaCl. LRP fragments were eluted from the Ni-NTA gel with buffer A containing 250 mmol/L imidazole and bound to a RAP affinity column (see below). RAP-Sepharose was washed with 50 mmol/L Tris-HCl, pH 7.5, containing 150 mmol/L NaCl, 2 mmol/L CaCl2, and 0.02% NaN3. LRP fragments were eluted with calcium-free Tris-HCl buffer containing 10 mmol/L EDTA. The correct amino acid sequence of the fragments was confirmed by double-stranded DNA sequencing using the Sequenase 7-Deaza-dGTP DNA Sequencing Kit (US Biochemical Corp, Cleveland, OH). N-terminal amino acid sequencing and electrospray mass spectrometry were performed on selected fragments by Dr Clive Slaughter (HHMI Biopolymer Facility, University of Texas Southwestern, Dallas, TX).
Preparation of RAP affinity column.
To obtain recombinant rat RAP in an unfused state, the GST-RAP expression plasmid was used as a PCR template with the following pair of primers: 5′-GCAGGATCCTACTCGCGGGAGAAG-3′ (forward) and 5′-ACGCTCGAGCTAGAGTTCGTTGTGCCGAGCCCT-3′ (reverse). The resulting insert was digested with BamHI and XhoI and ligated into the BamHI/XhoI-digested pET-28a vector (Novagen). RAP expressed from this plasmid contains a 6-His tag followed by a T7 epitope tag at the N-terminus. The pET28-RAP vector was transformed into strain BL21/DE3(pLysS) for protein expression. Cultures were induced and obtained as described for LRP fragments. Purification on Ni-NTA gel followed by a heparin column was essentially as described for active PAI-1.19 RAP was coupled to CNBr-activated Sepharose 4B (Pharmacia Biotech, Inc, Piscataway, NJ) according to the manufacturer’s instructions.
Direct binding assays.
Solid-phase binding assays using 125I-RAP were performed in 96-well microtiter plates (Immulon-4, Removawell; Dynatech, Chantilly, VA). Wells were coated with 5 pmol purified receptor in 100 μL Tris-HCl–buffered saline and allowed to bind overnight at 4°C. The plates were then blocked for 4 hours at room temperature with 300 μL binding buffer (50 mmol/L Tris-HCl, pH 7.5; 150 mmol/L NaCl; 0.02% NaN3; 0.05% Tween-20; 2 mmol/L CaCl2; 5% bovine serum albumin [BSA]) and incubated overnight at 4°C with increasing amounts of 125I-RAP diluted in binding buffer. The plates were washed three times with 200 μL binding buffer and the wells counted in a γ-counter. To assay nonspecific binding, an equal number of control wells were prepared with calcium-free binding buffer containing 40 mmol/L EDTA. Nonspecific binding was normally less than 5% of the total and was subtracted from the total cpm. Binding data were fit to a Langmuir isotherm by nonlinear regression using SigmaStat version 2.0 (Jandel Scientific, San Rafael, CA). In every case binding data were fit to a single-site model; multisite models failed to produce statistically significant (P < .05) improvements in χ2. Dissociation constants (Kd) were determined by nonlinear regression of the single-site Langmuir isotherm rather than using linear Scatchard analysis.23 Because iodinated lactoferrin and PAI-1 are insoluble at high concentrations, we were unable to determine their dissociation constants by direct binding.
Binding competition assays.
Binding competitions of 2 nmol/L 125I-RAP, 10 nmol/L125I-lactoferrin, and 15 nmol/L 125I-PAI-1 with unlabeled RAP, lactoferrin, and PAI-1 were performed using RAP-affinity-purified C5-C7 and E4-C10. To rule out nonspecific interactions, unlabeled RAP, lactoferrin, BSA, and PAI-1 were coated in wells and assayed to ensure that they did not bind to the iodinated ligands. The competition experiments were performed using 96-well microtiter plates in the same manner as were direct binding assays. The wells were counted in a γ-counter and the IC50 values were determined by nonlinear regression using a single-site model in the SigmaStat program.
Cell degradation assays.
Mouse fibroblasts (2 × 105 per well) were seeded in 12-well plates and grown for 24 hours in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum, 100 U/mL penicillin, and 100 mg/mL streptomycin. Cells were washed with DMEM (without glutamine or serum) containing 0.2% (wt/vol) BSA and incubated with 1 mL of the same media containing 1.5 μg125I-RAP.20 Competing ligands dissolved in 25 mmol/L HEPES, pH 7.4; 150 mmol/L NaCl; and 2 mmol/L CaCl2were added to the wells to give a final volume of 1.2 mL; controls received 200 μL HEPES buffer only. Cells were allowed to take up125I-RAP for 4 hours at 37°C. Degradation was assessed by measuring 125I-labeled trichloroacetic acid-soluble (noniodide) material released into the culture medium.24Amounts of degradation products were normalized to the values for wild-type mouse embryonic fibroblast (MEF 1) cells without competitors added. These values ranged from 520 to 866 ng/mg total cell protein/4 hours in different experiments. Background ligand degradation was measured in control wells lacking cells and was subtracted.
RESULTS
Periplasmic expression of LRP fragments.
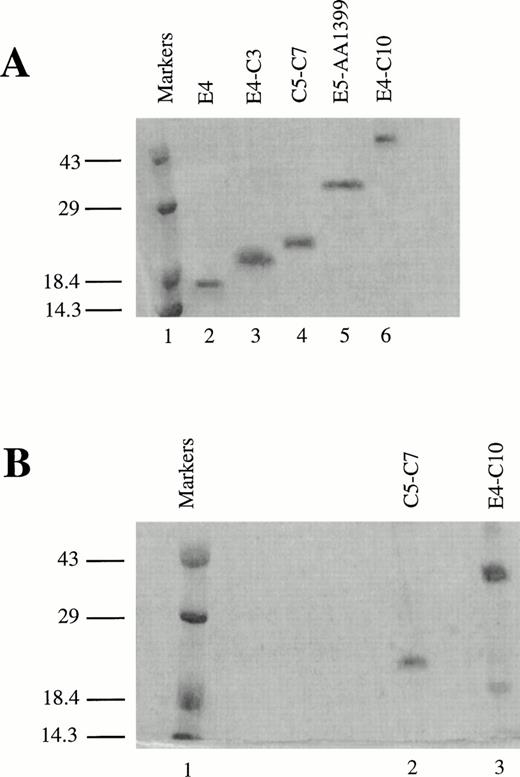
To identify the minimal binding sites for various LRP ligands, we cloned and expressed receptor fragments spanning most of the region that corresponds to the CNBr fragment (amino acids 776-1399), which contains the second cluster of complement-type repeats in LRP.16 The oligonucleotides used as PCR primers are listed in Table 1. All fragments comprised multiples of entire, contiguous repeats followed by a myc epitope tag and a 6-His tag at their C-termini. The fragments were secreted into the periplasm, isolated by cold osmotic shock, and subjected to further chromatographic purification. An approximately equal amount of inactive recombinant protein secreted into the media or the cytoplasm was discarded. LRP fragments separated on sodium dodecyl sulftate (SDS)-polyacrylamide gel electrophoresis and stained with Coomassie Brilliant blue (Bio-Rad) are shown in Fig 1. The mobility of the proteins under reducing conditions, particularly the smaller fragments, is lower than expected (Fig 1A). For example, the mass of E4-C3 measured by protein trap electrospray is 13,155 atomic mass units (amu) versus 13,160 amu computed from the amino acid sequence, although it seems to be greater than 18 kD according to the molecular weight markers. The anomalous mobility must be due to the presence of the 6-His tag. Figure 1B shows two of the RAP-binding fragments on a nonreducing SDS gel; both appear to have a major band consisting of the monomer. It is predicted that the fragments will migrate faster under nonreducing conditions because disulfide bonds prevent complete denaturation of the protein. The mobility of the smaller protein C5-C7 barely increases, presumably because the effect of the 6-His tag predominates over the presence of intact disulfides. E4-C10 migrates much faster on the nonreducing gel, indicating a more compact structure in the nonreduced state.
Recombinant LRP fragments from region II. (A) Electrophoresis of fragments on a 13.5% reducing SDS polyacrylamide gel. Lane 1 contains prestained low molecular weight markers (Life Technologies). The remaining lanes (2 through 6) contain the LRP fragments in the following order: E4 (8.2 kD), E4-C3 (13.2 kD), C5-C7 (16.8 kD), E5-AA1399 (29.5 kD), and E4-C10 (44.8 kD). The smaller proteins migrate abnormally slowly and appear to be larger than the actual molecular weights shown in parentheses due to the presence of the 6-His tag. (B) RAP affinity-purified fragments on a nonreducing 13.5% SDS polyacrylamide gel. Lane 1 contains low molecular weight markers, lane 2 contains C5-C7, and lane 3 contains E4-C10. Both gels were stained with Coomassie Brilliant blue.
Recombinant LRP fragments from region II. (A) Electrophoresis of fragments on a 13.5% reducing SDS polyacrylamide gel. Lane 1 contains prestained low molecular weight markers (Life Technologies). The remaining lanes (2 through 6) contain the LRP fragments in the following order: E4 (8.2 kD), E4-C3 (13.2 kD), C5-C7 (16.8 kD), E5-AA1399 (29.5 kD), and E4-C10 (44.8 kD). The smaller proteins migrate abnormally slowly and appear to be larger than the actual molecular weights shown in parentheses due to the presence of the 6-His tag. (B) RAP affinity-purified fragments on a nonreducing 13.5% SDS polyacrylamide gel. Lane 1 contains low molecular weight markers, lane 2 contains C5-C7, and lane 3 contains E4-C10. Both gels were stained with Coomassie Brilliant blue.
Analysis of RAP binding.
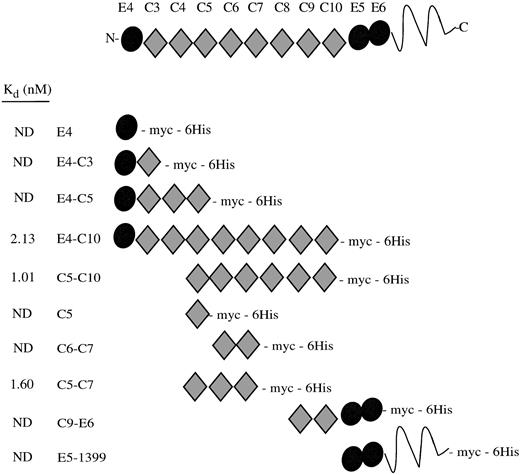
When we initiated our study, the 39-kD protein RAP was known to have a high affinity for cellular LRP and to bind to recombinant fragments corresponding to region II of the receptor, as shown by ligand blots and coimmunoprecipitation experiments.9,16 Therefore, we examined the ability of periplasmically expressed recombinant receptor fragments to bind 125I-RAP as an assay of native structure. A schematic of the repeat structure of the receptor fragments showing their Kd values are shown in Fig 2. EGF repeat E4 was not essential for RAP binding, in agreement with previous ligand blot experiments. The C-terminal EGF-precursor homologous domain, which contains E5 and E6, also did not seem to bind RAP. By comparison of the Kd values, the minimal RAP binding site can be localized to the three complement-type repeats, C5-C7 (Fig 3). Every protein containing these three repeats had a Kd of approximately 2 nmol/L, including protein fragments beginning with E4 and terminating at C7 or C8 (data not shown). The affinity of C5-C7 for RAP is similar to affinity constants reported for the binding of 125I-RAP to immobilized LRP: 14 nmol/L7 and 18 nmol/L.25Complement-type repeats contain conserved and essential disulfide bonds.6 As a control, we tested the RAP binding activity in the presence and absence of reducing agents. RAP has no cysteines and is unaffected by reducing agents. Five pmol of fragments C5-C7 and E4-C10 were coated on plates and assayed for binding to 2 nmol/L125I-RAP in the presence or absence of 10 mmol/L dithiothreitol (DTT). For C5-C7, counts per minute corrected for background were 7,959 ± 198 (no DTT) and 0 ± 101 (+ DTT); for E4-C10, 7,013 ± 231 (no DTT) and 0 ± 31 (+ DTT). DTT totally abolished the binding of 125I-RAP to the receptor fragments, suggesting that binding activity is dependent on the presence of intact disulfides in the receptor fragments.
RAP binding to LRP region II fragments. A schematic drawing of the recombinant LRP fragments shows their constituent EGF precursor type repeats (•) and complement-type repeats (⧫). The LRP repeats have been numbered according to Herz et al.2 The Kds determined by direct binding assays are shown at left; ND, no detectable binding.
RAP binding to LRP region II fragments. A schematic drawing of the recombinant LRP fragments shows their constituent EGF precursor type repeats (•) and complement-type repeats (⧫). The LRP repeats have been numbered according to Herz et al.2 The Kds determined by direct binding assays are shown at left; ND, no detectable binding.
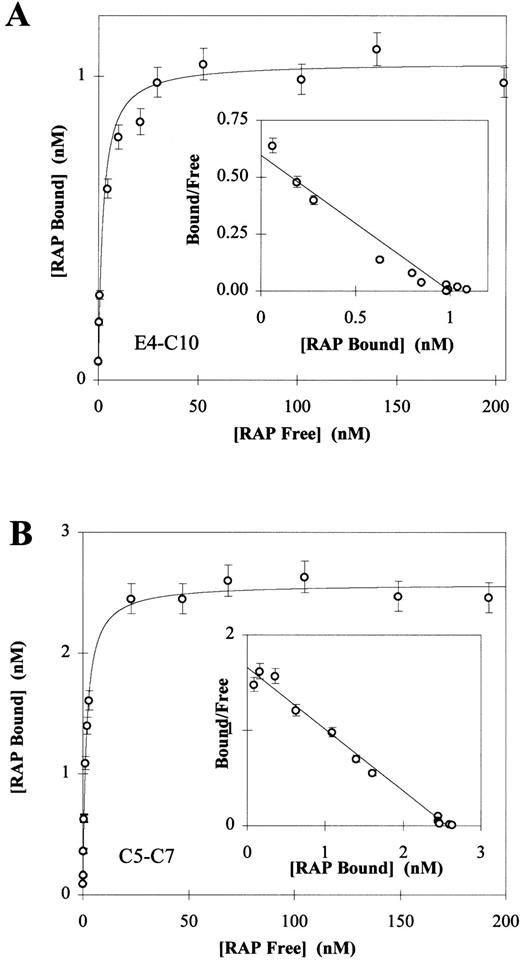
Direct binding of 125I-RAP to LRP fragments E4-C10 (A) and C5-C7 (B). 125I-RAP was added to receptor fragments (250 ng E3-C10 or 100 ng C5-C7) coated in 96-well microtiter plates as described in Materials and Methods. Triplicate experiments were used to calculate a mean and SD for each data point. In each experiment the total amount of 125I-RAP added ranged from 0.5 nmol/L to 200 nmol/L and the concentration of free125I-RAP was calculated by counting the well buffer after overnight binding. The amount of 125I-RAP bound was determined by counting the dry wells after washing. Nonspecific binding was determined by subtracting the value of wells in which125I-RAP was bound without calcium and in the presence of 40 mmol/L EDTA. The Kd values were determined to be 2.13 ± 0.67 for the fragment E3-C10 (A) and 1.60 ± 0.09 for C5-C7 (B). Inset to each panel is the Scatchard plot of the Langmuir isotherm calculated from the data.
Direct binding of 125I-RAP to LRP fragments E4-C10 (A) and C5-C7 (B). 125I-RAP was added to receptor fragments (250 ng E3-C10 or 100 ng C5-C7) coated in 96-well microtiter plates as described in Materials and Methods. Triplicate experiments were used to calculate a mean and SD for each data point. In each experiment the total amount of 125I-RAP added ranged from 0.5 nmol/L to 200 nmol/L and the concentration of free125I-RAP was calculated by counting the well buffer after overnight binding. The amount of 125I-RAP bound was determined by counting the dry wells after washing. Nonspecific binding was determined by subtracting the value of wells in which125I-RAP was bound without calcium and in the presence of 40 mmol/L EDTA. The Kd values were determined to be 2.13 ± 0.67 for the fragment E3-C10 (A) and 1.60 ± 0.09 for C5-C7 (B). Inset to each panel is the Scatchard plot of the Langmuir isotherm calculated from the data.
Effect on LRP fragments on uptake of 125I-RAP by fibroblasts.
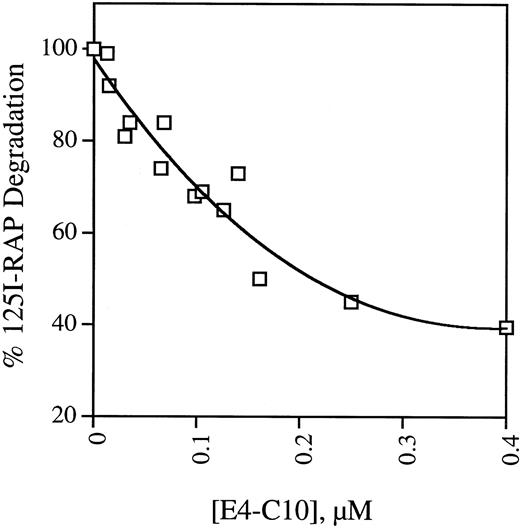
Most of the protein ligands internalized through LRP are degraded in a lysozomal pathway. To confirm the results of the plate assays, we examined the ability of soluble receptor fragments to bind125I-RAP and block its uptake and degradation by mouse fibroblast cells. The activity of five receptor fragments is shown in Table 2. MEF 1 express large amounts of LRP and the rate at which they take up and degrade 125I-RAP is considered 100% of full activity. The data shown in Table 2 mirror the findings of the binding assays in that only fragment E4-C10, which contains repeats C5-C7, is able to prevent 125I-RAP uptake and degradation. Unlabeled RAP is included as a control and is also effective at decreasing the amount of degradation. PEA 13 cells are homozygous LRP-deficient but take up and degrade 125I-RAP at one-seventh the rate observed in wild-type cells.20 This residual uptake activity is presumably LRP independent and thus cannot be blocked by ligands that bind exclusively to LRP, indicating that 400 nmol/L E4-C10 is able to block about 70% of the LRP-dependent degradation. The activity of E4-C10 to interfere with degradation is proportional to its concentration, as shown in Fig 4.
Inhibition of LRP-Mediated Degradation of125I-RAP by LRP Fragments
| Cell Line . | Unlabeled Competitor . | % Degradation of125I-RAP . | |
|---|---|---|---|
| MEF 1 | None | 100.0 ± 2.3 | |
| 400 nmol/L | E4 | 100.7 ± 2.9 | |
| E4-C3 | 99.8 ± 1.5 | ||
| E4-C10 | 39.9 ± 0.3 | ||
| C9-E6 | 103.6 ± 3.0 | ||
| E5-AA1399 | 101.8 ± 2.9 | ||
| 200 nmol/L | RAP | 24.9 ± 0.4 | |
| PEA 13 | None | 14.4 ± 0.3 | |
| Cell Line . | Unlabeled Competitor . | % Degradation of125I-RAP . | |
|---|---|---|---|
| MEF 1 | None | 100.0 ± 2.3 | |
| 400 nmol/L | E4 | 100.7 ± 2.9 | |
| E4-C3 | 99.8 ± 1.5 | ||
| E4-C10 | 39.9 ± 0.3 | ||
| C9-E6 | 103.6 ± 3.0 | ||
| E5-AA1399 | 101.8 ± 2.9 | ||
| 200 nmol/L | RAP | 24.9 ± 0.4 | |
| PEA 13 | None | 14.4 ± 0.3 | |
Replicate monolayers of wild-type (MEF 1) or LRP-deficient (PEA 13) mouse fibroblasts were incubated with serum-free media containing125I-RAP and recombinant LRP fragments. After incubation at 37°C for 4 hours, the total amount of 125I-RAP degradation products secreted into the media was measured. % Degradation is the mean of triplicates ± 1 SD and is normalized to the amount of degradation products released by MEF 1 cells with no competitor added.
Dependence of inhibition of 125I-RAP degradation on soluble E4-C10. Replicate monolayers of MEF 1 fibroblasts were incubated with serum-free DMEM containing125I-RAP and different concentrations of LRP fragment E4-C10. After incubation at 37°C for 4 hours, the total amount of125I-RAP degradation products secreted into the media was measured. The points are the mean of triplicates from one to three experiments and are normalized to the amount of degradation products released in the absence of E4-C10.
Dependence of inhibition of 125I-RAP degradation on soluble E4-C10. Replicate monolayers of MEF 1 fibroblasts were incubated with serum-free DMEM containing125I-RAP and different concentrations of LRP fragment E4-C10. After incubation at 37°C for 4 hours, the total amount of125I-RAP degradation products secreted into the media was measured. The points are the mean of triplicates from one to three experiments and are normalized to the amount of degradation products released in the absence of E4-C10.
Identification of ligand binding sites for RAP, PAI-1, and lactoferrin.
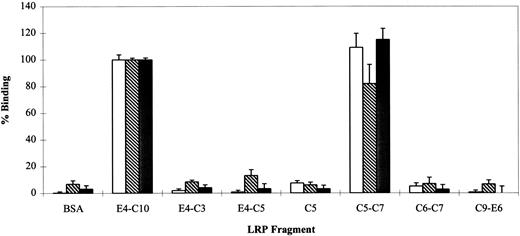
RAP is able to interfere with the binding of all known LRP ligands, including PAI-1 and lactoferrin, although it has been more problematic to show reciprocal competition between individual ligands.15 This has been interpreted as an indication of multiple independent or partially overlapping binding sites on LRP. To compare the binding sites of RAP, PAI-1, and lactoferrin, iodinated proteins (25 nmol/L) were incubated with various LRP fragments and assayed for binding activity (Fig 5). The data show that all three ligands interact predominately with C5-C7; a longer fragment (E4-C10) containing all eight complement-type repeats does not show appreciably better binding for any of the proteins. A cluster of three repeats seems to be the minimal binding site detectable by this assay, because C6-C7 and C5 alone have negligible binding. This is consistent with the observation that LRP ligand binding domains contain at least three consecutive complement-type repeats. The unusual cluster of two repeats found at the N-terminus of LRP (region I) is unable to bind RAP.9
Qualitative binding of lactoferrin (), PAI-1 (▩), and RAP (□) to LRP fragments. The ability of various LRP fragments to bind iodinated protein ligands at a single concentration (25 nmol/L) was investigated by direct binding assay. 125I-RAP,125I-lactoferrin, and 125I-PAI-1 were bound to 5 pmol of various LRP fragments in the presence of 2 mmol/L CaCl2. BSA was used as a nonreceptor control. Nonspecific binding was measured in parallel wells without calcium and in the presence of 40 mmol/L EDTA. Fragment E4-C10 was used as the 100% binding control and had the following cpm: 125I-RAP = 31,707 ± 1433; 125I-lactoferrin = 14,072 ± 255; and125I-PAI-1 = 4,098 ± 60. Triplicate points in duplicate experiments were used to calculate a mean and SD for each receptor.
Qualitative binding of lactoferrin (), PAI-1 (▩), and RAP (□) to LRP fragments. The ability of various LRP fragments to bind iodinated protein ligands at a single concentration (25 nmol/L) was investigated by direct binding assay. 125I-RAP,125I-lactoferrin, and 125I-PAI-1 were bound to 5 pmol of various LRP fragments in the presence of 2 mmol/L CaCl2. BSA was used as a nonreceptor control. Nonspecific binding was measured in parallel wells without calcium and in the presence of 40 mmol/L EDTA. Fragment E4-C10 was used as the 100% binding control and had the following cpm: 125I-RAP = 31,707 ± 1433; 125I-lactoferrin = 14,072 ± 255; and125I-PAI-1 = 4,098 ± 60. Triplicate points in duplicate experiments were used to calculate a mean and SD for each receptor.
Reciprocal competition of RAP, PAI-1 and lactoferrin on C5-C7 and E4-C10.
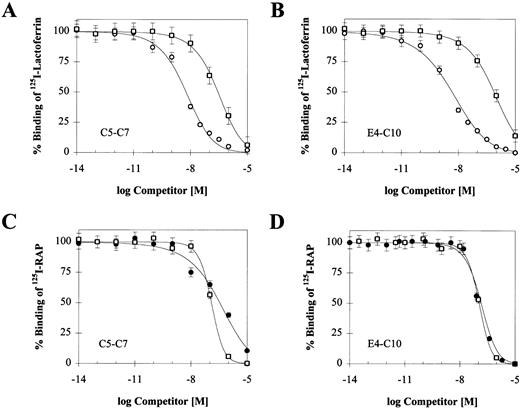
To confirm that RAP, PAI-1, and lactoferrin bind to a single region encompassed by C5-C7, we performed competition experiments between iodinated RAP, PAI-1, lactoferrin, and unlabeled ligands. The effect of increasing concentrations of the two unlabeled ligands on125I-RAP and 125I-lactoferrin binding is shown in Fig 6, and the corresponding IC50 values are summarized in Table 3 along with values from experiments with 125I-PAI-1. Each ligand is able to reduce binding of each of the others by nearly 100%. The IC50s for unlabeled RAP against the other ligands are similar to its Kd, consistent with its high affinity for the receptor. Cold PAI-1 competes equally well against 125I-RAP bound to either receptor, suggesting that C5-C7 contains the entire PAI-1 binding site. Differences in affinity for the two receptors are mainly observed with lactoferrin. Unlabeled lactoferrin is more efficient at competing with 125I-RAP bound to E4-C10, suggesting that its binding site may extend outside of C5-C7. Consistent with this, higher concentrations of PAI-1 are required to compete with 125I-lactoferrin bound to the larger receptor fragment. Further competition experiments are required to determine the exact boundaries of the lactoferrin binding site.
Reciprocal competition of LRP ligands on binding to C5-C7 or E4-C10. Binding of 10 nmol/L 125I-lactoferrin (A and B) was competed with RAP (○) and active PAI-1 (□); binding of 2 nmol/L125I-RAP (C and D) was competed with lactoferrin (•) and active PAI-1 (□). Triplicate experiments were used to calculate a mean and SD for each data point. In each experiment, the concentration of the competing unlabeled ligand ranged from 0.01 pmol/L to 10 μmol/L. Nonspecific binding was measured in parallel wells without calcium and in the presence of 40 mmol/L EDTA. The IC50value for each competing ligand was determined by using a single-site competition model and is shown in Table 3.
Reciprocal competition of LRP ligands on binding to C5-C7 or E4-C10. Binding of 10 nmol/L 125I-lactoferrin (A and B) was competed with RAP (○) and active PAI-1 (□); binding of 2 nmol/L125I-RAP (C and D) was competed with lactoferrin (•) and active PAI-1 (□). Triplicate experiments were used to calculate a mean and SD for each data point. In each experiment, the concentration of the competing unlabeled ligand ranged from 0.01 pmol/L to 10 μmol/L. Nonspecific binding was measured in parallel wells without calcium and in the presence of 40 mmol/L EDTA. The IC50value for each competing ligand was determined by using a single-site competition model and is shown in Table 3.
IC50 Values for Reciprocal Competition Experiments
| Receptor Fragment . | Radiolabeled Ligand . | IC50 of Unlabeled Competitor (nmol/L) . | ||
|---|---|---|---|---|
| Lactoferrin . | PAI-1 . | RAP . | ||
| E4-C10 | Lactoferrin | — | 766 ± 45 | 4.69 ± 1.33 |
| PAI-1 | 71 ± 3 | — | 2.03 ± 0.46 | |
| RAP | 143 ± 20 | 107 ± 21 | — | |
| C5-C7 | Lactoferrin | — | 280 ± 24 | 5.90 ± 1.63 |
| PAI-1 | 68 ± 10 | — | 1.69 ± 0.18 | |
| RAP | 352 ± 48 | 123 ± 11 | — | |
| Receptor Fragment . | Radiolabeled Ligand . | IC50 of Unlabeled Competitor (nmol/L) . | ||
|---|---|---|---|---|
| Lactoferrin . | PAI-1 . | RAP . | ||
| E4-C10 | Lactoferrin | — | 766 ± 45 | 4.69 ± 1.33 |
| PAI-1 | 71 ± 3 | — | 2.03 ± 0.46 | |
| RAP | 143 ± 20 | 107 ± 21 | — | |
| C5-C7 | Lactoferrin | — | 280 ± 24 | 5.90 ± 1.63 |
| PAI-1 | 68 ± 10 | — | 1.69 ± 0.18 | |
| RAP | 352 ± 48 | 123 ± 11 | — | |
IC50 values were calculated from the competitive binding curves shown in Fig 6 and from additional experiments performed with 125I-PAI-1. LRP fragment E4-C10 or C5-C7 was immobilized in microtiter wells and incubated with an iodinated ligand and increasing concentrations of the indicated unlabeled protein. Values represent the mean and SD of three separate experiments.
DISCUSSION
To obtain sufficient quantities of LRP receptor to perform structure-function studies, we have developed a prokaryotic secretion system that is capable of producing fragments of active LRP. Folding and disulfide formation occur in bacteria despite the absence of the mammalian molecular chaperone, RAP. This is not totally unexpected, because RAP-deficient mice have about 25% of the normal amount of LRP.26 Recombinant proteins representing portions of the LRP region II were purified from bacterial periplasm and tested for their ability to bind iodinated RAP, lactoferrin, and PAI-1 in a solid-phase assay. Binding activity of the fragments is dependent on the presence of their disulfide bonds and Ca2+, and can be reversibly blocked by EDTA. Comparison of different-sized fragments indicates that all three protein ligands bind to a group of complement-type repeats in the middle of region II, namely C5-C7. This is the first demonstration of the position of a lactoferrin binding site on LRP and that a single binding site on LRP is capable of binding three different proteins, including RAP. The fact that a three-complement repeat binds ligands as well as the eight-complement repeat cluster shows that the binding activity of the site is independent of its position or context within the cluster and is probably due to unique properties of the C5-C7 repeats.
LRP region II had previously been identified as a binding site for α1M light chain (and presumably α2M), uPA.PAI-1, and RAP on the basis of ligand blots.27 While this work was in progress, two reports further defining the RAP and PAI-1 binding sites of recombinant LRP region II were published. Obermoeller et al11 used ligand blots to examine binding of RAP and RAP subdomains to all four regions of LRP containing complement-type repeats, and Horn et al17analyzed RAP, PAI-1, t-PA.PAI-1, and Fab A8 binding by means of surface plasmon resonance. With respect to RAP, our results agree well with those of Horn et al because we both identify C5-C7 as the binding site, although Kds determined by the solid-phase binding assay are lower than those measured by surface plasmon resonance. However, Horn et al failed to observe PAI-1 binding to C5-C7 alone, and concluded that the PAI-1 binding site only partially overlaps the RAP binding site and extends towards the fourth EGF-like repeat at the N-terminus of region II. We did not measure the Kd values for125I-PAI-1 or 125I-lactoferrin by direct binding due to technical limitations, although we did examine their competition of equimolar amounts of 125I-RAP bound to different receptors that had the same Kd for RAP. In our experiments, cold PAI-1 gave a similar IC50 when competing against125I-RAP bound to either the eight complement-type repeat fragment (E4-C10) or C5-C7. This suggests that both fragments contain the entire PAI-1 binding site, and that RAP and PAI-1 may bind to a similar surface of the receptor. Possible reasons for discrepancies between Horn et al’s results and our solid-phase binding assays are that the ligands were immobilized for the surface plasmon resonance measurements, whereas we immobilized the receptor fragments,17 the presence of an epitope tag on the N-termini of their fragments versus on the C-termini of ours, and absence of glycosylation in the bacterial proteins.
In the case of lactoferrin, the approximately twofold higher IC50 against 125I-RAP bound to the small receptor fragment and its pattern of competition with cold PAI-1 could mean that its binding site extends beyond C5-C7. These results also suggest that, unlike RAP and PAI-1, lactoferrin interacts with a different subset of the receptor residues. Despite this, unlabeled RAP and PAI-1 are able to block binding of 125I-lactoferrin by 90% to 100%, indicating substantial overlap of the binding sites resulting in steric hindrance. Our results showing reciprocal-competition between RAP and lactoferrin seem to contradict the conclusions of cell-uptake studies. It was previously noted in uptake experiments on LDL-deficient fibroblasts that lactoferrin degradation was inhibited up to 60% by GST-RAP, but lactoferrin was unable to block uptake of125I-t-PA.PAI-1.15,28Nonreceptor-mediated binding of LRP ligands to heparin and the presence of additional lactoferrin receptors may complicate the results of cell culture experiments.12,29 In addition, the t-PA.PAI-1 complex seems to have a higher affinity for LRP and recombinant LRP fragments compared with PAI-1 alone, which may decrease its ability to be displaced by lactoferrin.17 30
In the study by Obermoeller et al,11 the eight complement-type repeats of region II were divided into two halves and assayed for RAP binding activity. Surprisingly, fragments representing C3-C6 and C7-C10 bound equally well to immobilized RAP. This suggested the possibility of two distinct RAP binding sites. However, our results and those of Horn et al17 argue against this. The C8-C10 fragment17 and C9-E6 fragment do not bind RAP, indicating that C7 must be crucial for the RAP binding observed on ligand blots. Because C7 is a part of C5-C7, region II may only contain one RAP binding site, as is consistent with our results on the direct binding assays. The number of binding sites presented by equimolar amounts of E4-C10 and C5-C7 for 125I-RAP, the Bmax, was actually less for E4-C10 than for C5-C7, lending support to the idea of a single binding site. Of course, the Bmax might be affected by the mode of binding of each fragment with the surface of the plate. Another question is whether two molecules of 39-kD RAP would be able to occupy adjacent binding sites on a series of four complement-type repeats, representing an 18- to 19-kD fragment.
Currently little information is available about the complement-repeat receptor:ligand interactions at the atomic level. The spatial relationship between the repeats constituting LRP is unknown, although structures of single repeats have been solved.31-33Mutagenesis and peptide competition studies have been used in attempts to identify the LRP binding sites on some of its ligands. The three binding sites on RAP have been most extensively studied.11,34,35 We and others showed that replacement of K1370 (human numbering) in the receptor binding fragment of α2M with an alanine effectively blocks receptor binding.36,37 Lysine residues have also been implicated inPseudomonas exotoxin A,38 lipoprotein lipase,39 and PAI-140 binding sites for LRP. The recent x-ray structure of the LDL ligand-binding repeat has ruled out the direct interaction of the lysines with the conserved aspartate residues, because the latter are involved in Ca2+ coordination.33 Details of the interactions will be best answered by solving the three-dimensional structure of an LRP ligand bound to a receptor fragment. The recombinant LRP fragments that we have produced in bacteria should prove useful in determining the stoichiometry of ligand binding and help advance our understanding of the protein:protein interactions between LRP and its ligands to the molecular level of detail.
ACKNOWLEDGMENT
We are indebted to our colleagues, Drs Elizabeth Goldsmith and Joachim Herz, at the University of Texas Southwestern Medical Center, Dallas, for providing plasmids and cDNA. We would also like to thank Dr Stephen Sprang of the Howard Hughes Medical Institute at the University of Texas Southwestern Medical Center, Dallas, for his comments on this manuscript.
From the Department of Pharmacology, University of Texas Southwestern Medical Center, Dallas, TX.
Submitted March 25, 1998; accepted June 25, 1998.
Supported by American Heart Association Texas Affiliate Grant-in-Aid 966-063.
Address reprint request to Dianne DeCamp, PhD, Department of Pharmacology, The University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd, Dallas, TX 75235-9041.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal