Abstract
Previously we reported that a karyotypically silent t(4;14)(p16.3;q32.3) translocation is present in about 25% of multiple myeloma (MM) tumors, and causes overexpression of FGFR3, which is 50 to 100 kb telomeric to the 4p16 breakpoints. Frequent FGFR3 kinase activating mutations in MM with t(4;14) translocations substantiate an oncogenic role for FGFR3. We now report that the 4p16 breakpoints occur telomeric to and within the 5′ introns of a novel gene,MMSET (Multiple Myeloma SETdomain). In normal tissues, MMSET has a complex pattern of expression with a short form (647 amino acids [aa]) containing an HMG box andhath region, and an alternatively spliced long form (1365 aa) containing the HMG box and hath region plus 4 PHD fingers and a SET domain. Although t(4;14) translocation results in IgH/MMSET hybrid transcripts, overexpression of MMSET also occurs from endogenous promoters on 4p16. Given the homology to HRX/MLL1/ALL1at 11q23 that is dysregulated by translocations in acute leukemia, we hypothesize that dysregulation of MMSET contributes to neoplastic transformation in MM with t(4;14) translocation. This is the first example of an IgH translocation that simultaneously dysregulates two genes with oncogenic potential: FGFR3 on der(14) andMMSET on der(4).
© 1998 by The American Society of Hematology.
DYSREGULATION of an oncogene by translocation to an Ig locus is a seminal event in the pathogenesis of most B-cell tumors.1 Recently, we have determined that multiple myeloma (MM) is characterized by frequent translocations into an Ig locus, including all members of our panel of 21 MM cell lines. Moreover, at least three of these lines have two independent translocations involving two IgH loci or an IgH locus plus and IgL locus.2-5 Others have also reported a high incidence of Ig translocations in MM, including occasional examples of coincidence of two IgH translocations or an IgH translocation and an IgL translocation in the same tumor.6-9 We have found that translocations to the IgH locus at 14q32.3 primarily involve IgH switch regions, and involve a large array of translocation partners.2 Three loci are frequently involved (20% to 25% each): cyclin D1 on 11q13,FGFR3 on 4p16, and c-maf on 16q23.3-5 In each of these cases we have cloned the breakpoints and shown that they are between 50 and 500 kb centromeric to the ectopically expressed oncogene on the der(14) chromosome. The expression of these genes is thought to be dysregulated by juxtaposition of endogenous promoters to powerful regulatory regions of the IgH locus (eg, 3′ enhancers downstream of Cα), and not through the formation of hybrid transcripts or fusion proteins. For the t(4;14) translocation we have shown in addition that in three of six cell lines and in one of three patient samples there are activating mutations of FGFR3, indicating that it plays a critical role in the tumor development. We now report that the translocation breakpoints on 4p16 are telomeric to and within a novel gene, MMSET (Multiple Myeloma SET domain protein), that is also dysregulated as a result of this translocation.
MATERIALS AND METHODS
Cell Culture
Cosmid Clones
The sequences of L75b9, L184d6, L190b4, L19h1, and L96a2 were obtained from GenBank. In addition, L75b9 cosmid, used to subclone a 5.3-kbSst I restriction fragment containing MMSET exon 1, was kindly provided to us by M.R. Altherr (Los Alamos National Laboratory, Los Alamos, NM).
Library
The human testis 5′-Stretch Plus cDNA library was purchased from Clontech (Palo Alto, CA). It contains both random-primed and oligo-dT primed cDNA phage clones.
Primers and Probes
The sequence of the primers used in a polymerase chain reaction (PCR) assay to amplify probes for the Northern blot analysis and to screen the cDNA library, in rapid amplification of cDNA ends (RACE) experiments, for sequencing analysis, and for amplification of hybrid transcripts, are listed in Table 1. All the following MMSET PCR reactions have been conducted on UTMC2 cDNA, unless specified. The exon 3 probe, used in the Northern blot analysis and to screen the cDNA library, was a 475-bp fragment PCR amplified using the primers pair 5541 and 5540. The exon 6-10 probe is a 900-bp fragment PCR amplified from the phage clone no. 714, using o58 primer and the T7 primer at the 3′ end of the clone. The exon 19-23 probe is an 822-bp fragment amplified with primers o76 and o77. The 3′ exon 24 probe is a 402-bp fragment amplified with primers o83 and o82. Using o93 and o58 primer pair, a 1.7-kb fragment was amplified to cover the coding sequence in exon 11. A 3340-bp fragment crossing the stop codon in exon 24 has been amplified using o80-o58 primer pairs. The Iμ probe has been generated by PCR amplification using the primer pair 5518 and o52. In addition to o99 and o48, the IgH primers used for PCR amplification of hybrid transcripts are 5536 (Iμ), 5590 (JH1), 5592 (JH3), o64 (Cγ), o65 (Cα), and o132 (Cμ).
PCR and RACE Primers
| Exon . | Sense . | Antisense . | Sequence . |
|---|---|---|---|
| 1a | o134 | CTTGCCTGGCTATCACCAG | |
| 1 | o99 | CCGAGGATGCGACGCACCGCAG | |
| 3 | 5541 | CTGACTGAGACACATCAGCAGCAC | |
| 3 | 5760 | GGGGACTCTGCTTGATGCTAA | |
| 3 | 5540 | CATCTGGGCTGGATGGAATTTAG | |
| 4 | 5761 | CTCTTCCAGTGTTTGGACAAG | |
| 6 | o58 | AAGCCAAGTTCACCTTTCTCTATGTG | |
| 6 | o48 | CCTCAATTTCCCTGAAATTGGTT | |
| 7 | o103 | AAGAGCTGCTCAGGTCACAGTGG | |
| 10 | o143 | CTCCTTCTGCATCCTTAACTG | |
| 11 | o93 | CCTTACCATGCAAAAATGCGGAC | |
| 19 | o76 | GTGCAACTGCAAGCCCACAGATG | |
| 24 | o77 | GCCACACACGTCACAATGATGCC | |
| 24 | o80 | ATATCTAGAGTGGCGGTAACGCTGAGGAGTGA | |
| 24 | o83 | TCCTTGGCATCCGAAACCAG | |
| 24 | o82 | CCGTGGGACACAGGCATTTTAC | |
| Iμ | 5518 | CAGATCTGAAAGTGCTCTACTG | |
| Iμ | o52 | AGGATCCGGCAGCAGAAGCCACGCATCCCAGCTCTG | |
| Iμ | 5536 | AGCCCTTGTTAATGGACTTGGAGG | |
| JH1 | 5590 | CCCTGGTCACCGTCTCCTCA | |
| JH3 | 5592 | CAATGGTCACCGTCTCTTCA | |
| Cγ | o64 | TGTCCTTGGGTTTTGGGGGGAA | |
| Cα | o65 | TCTCTCAGGCCGGTCAGTGTGC | |
| Cμ | o132 | GAAGACGCTCACTTTGGGAGG | |
| RACE | 5051 | TAGGAATTCGATCTCGAGGCTTTTTTTTTTTTTTTTT(ACG) | |
| RACE | 5052 | TAGGAATTCGATCTCGAGGC |
| Exon . | Sense . | Antisense . | Sequence . |
|---|---|---|---|
| 1a | o134 | CTTGCCTGGCTATCACCAG | |
| 1 | o99 | CCGAGGATGCGACGCACCGCAG | |
| 3 | 5541 | CTGACTGAGACACATCAGCAGCAC | |
| 3 | 5760 | GGGGACTCTGCTTGATGCTAA | |
| 3 | 5540 | CATCTGGGCTGGATGGAATTTAG | |
| 4 | 5761 | CTCTTCCAGTGTTTGGACAAG | |
| 6 | o58 | AAGCCAAGTTCACCTTTCTCTATGTG | |
| 6 | o48 | CCTCAATTTCCCTGAAATTGGTT | |
| 7 | o103 | AAGAGCTGCTCAGGTCACAGTGG | |
| 10 | o143 | CTCCTTCTGCATCCTTAACTG | |
| 11 | o93 | CCTTACCATGCAAAAATGCGGAC | |
| 19 | o76 | GTGCAACTGCAAGCCCACAGATG | |
| 24 | o77 | GCCACACACGTCACAATGATGCC | |
| 24 | o80 | ATATCTAGAGTGGCGGTAACGCTGAGGAGTGA | |
| 24 | o83 | TCCTTGGCATCCGAAACCAG | |
| 24 | o82 | CCGTGGGACACAGGCATTTTAC | |
| Iμ | 5518 | CAGATCTGAAAGTGCTCTACTG | |
| Iμ | o52 | AGGATCCGGCAGCAGAAGCCACGCATCCCAGCTCTG | |
| Iμ | 5536 | AGCCCTTGTTAATGGACTTGGAGG | |
| JH1 | 5590 | CCCTGGTCACCGTCTCCTCA | |
| JH3 | 5592 | CAATGGTCACCGTCTCTTCA | |
| Cγ | o64 | TGTCCTTGGGTTTTGGGGGGAA | |
| Cα | o65 | TCTCTCAGGCCGGTCAGTGTGC | |
| Cμ | o132 | GAAGACGCTCACTTTGGGAGG | |
| RACE | 5051 | TAGGAATTCGATCTCGAGGCTTTTTTTTTTTTTTTTT(ACG) | |
| RACE | 5052 | TAGGAATTCGATCTCGAGGC |
The sequence of the primers used in a PCR assay to amplify probes for the Northern blot analysis and to screen the cDNA library, in RACE experiments, for sequencing analysis, and for amplification of hybrid transcripts are listed below in a 5′-3′ orientation.
RACE
5′ RACE experiments were performed on 100 ng of poly(A)+ cDNA from testis and KMM1, UTMC2, JIM3, and LP1 MM cell lines using the 5′ RACE System for Rapid Amplification of cDNA Ends, Version 2.0 (GIBCO-BRL, Gaithersburg, MD). Specifically, the first-strand cDNA was synthesized from 5761, the first PCR reaction was performed using the 5540 primer, and the reaction product was nested using 5760. The final product was fractionated on an agarose gel, blotted, and hybridized with 5541-labeled oligonucleotide to confirm the specificity of the PCR products. The DNA was then subcloned into the Original TA Cloning Kit (Invitrogen, San Diego, CA). To obviate the extremely high GC content of the MMSET exon 1, the cDNA synthesis and the PCR reactions have been performed in the additional presence of 5% dimethyl sulfoxide (DMSO) and 1 mol/L Betaine (Sigma Chemical Co, St Louis, MO).
For the 3′ RACE experiments, 5 μg of total RNA from UTMC2 MM cell lines have been primed with 5051 for the first-strand cDNA synthesis; for the second-strand cDNA synthesis and first amplification the primer o103 has been used with 5052; the product of the first PCR reaction has been nested using the primer pairs 5052 and o143. The 250-bp final product (no. 1112) has been subcloned as described above, and has also been used as a probe in a Northern blot.
Other Procedures
Northern blot analysis, cDNA synthesis, PCR, and sequencing are described elsewhere.3 To sequence the GC-rich region upstream of exon 1, 1 mol/L Betaine was added to the sequencing reactions.
GenBank accession numbers for MMSET.
The GenBank accession number for the 7,418-bp MMSET mRNA (type II), encoding for a 1365 amino acid (aa) protein is AF071593; the accession number for the 8389-bp mRNA (type I), encoding for a 647-aa protein isAF071594; the accession number for exon 1, and 5′ and 3′ flanking regions on L75b9 cosmid is AF071595.
RESULTS
The MMSET Gene Is Identified by Sequence Analysis
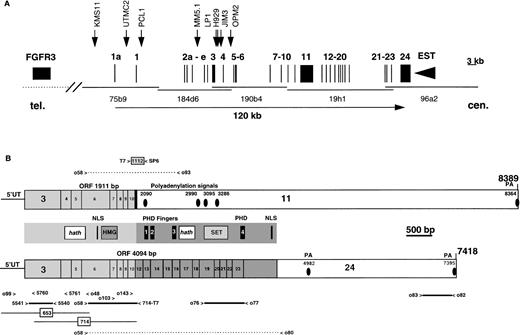
The translocation breakpoints on 4p16 are at the telomeric end of a 2-Mb cosmid contig that was fully sequenced during the search for the Huntington’s disease gene4,10(Fig 1). The sequence from cosmid 184d6 was analyzed for potential coding exons using the Gene Recognition and Analysis Internet Link (GRAIL),11 and the region corresponding to exon 3 identified. PCR primers (5540-5541) were used to generate a probe to screen a Northern blot, confirming that this region was expressed, and subsequently to screen a testis cDNA library. Ten independent phage clones were isolated. There was considerable heterogeneity in the 5′ end, with one clone (no. 653) containing 25 bp in exon 1 and splicing to exon 3, one starting in exon 2a and splicing to exon 3, and others starting at bp 39, 72 (no. 714), 83, 115, 254, and 435 of exon 3. All three of the clones extending beyond exon 10 spliced to exon 12.
Identification and mapping of MMSETgene on 4p16.3. (A) The diagram, drawn to scale, represents the distal 200 kb of the 2-Mb cosmid contig spanning the Huntington’s disease region, with the telomeric side of the contig, containing theFGFR3 gene, to the left. Vertical lines represent the exons distribution of the MMSET gene within about 120 kb of the cosmid contig. The MMSET gene includes at least 24 exons that are transcribed from the telomeric to the centromeric end. The solid arrowhead indicates the 3′ end of an EST that is localized 530 bp centromeric to the end of the last MMSET exon, but is transcribed in the opposite orientation. The solid arrows indicate the position of the previously cloned translocation breakpoints for KMS11, UTMC2, H929, JIM3, OPM2 MM cell lines, and for the tumor sample PCL1; The LP-1 breakpoint has been mapped by sequence analysis of a hybrid transcript splicing to exon 4, and MM5.1 between exons 2b and 3. The KMS11 t(4;14) translocation breakpoint is localized at the 5′ end of MMSET exon 1a, about 15 kb from exon 1; the UTMC2 breakpoint is localized between exon 1a and 1, about 2.5 kb from exon 1; the PCL1 breakpoint is in the intron between exons 1 and 2a; the LP-1, H929, and the JIM3 breakpoints are between exons 3 and 4; and the OPM2 breakpoint between exons 4 and 5. (B) The MMSET transcription units. MMSET is expressed as transcripts that polyadenylate either in exon 11 (type I), or exon 24 (type II) as a result of alternative splicing occurring from exon 10 to 11 (top), or exon 10 to 12 (bottom). The position of the polyadenylation signals (PA) is indicated by black circles. Because of the heterogeneity of the 5′ untranslated region (5′UT), we chose to start the numbering of MMSET from the first nucleotide in exon 3. The type I transcript contains an ORF of 1911 bp, encoding a 647-aa protein with the first methionine at nt 30, in exon 3, and the stop codon at nt 1971 after the first 20 aa of exon 11. The type II transcript contains a longer ORF of 4094 bp, encoding a 1365-aa protein, that is identical to the shorter protein up to the splicing site in exon 10, as indicated by the light gray shading. The two proteins differ after exon 10, with the unique portion of the short protein shaded in black, and the portion unique to the long protein shaded in dark gray. In the middle panel are shown characteristic domains of MMSET. A putative nuclear localization signal (NLS, indicated by a thick vertical line) is common to the two proteins, as well as the HMG domain (dark gray rectangle), and the hathdomain (white rectangle). The long protein is characterized by four PHD fingers (black rectangles), a SET domain (light gray rectangle), an additional hath domain, and another putative NLS. The thick horizontal lines represent the PCR amplified probes used in the Northern blot assay, with the primers number indicated. Numbers within boxes indicate two of the bacteriophage clones isolated from a testis cDNA library. To obtain a complete sequence of the ORFs of MMSET, cDNA fragments have been amplified by PCR as indicated by the dashed horizontal lines. 1112 (gray rectangle) is a clone obtained by 3′ RACE and sequenced with SP6 and T7 primers. (C) Amino acid sequence of MMSET long and short proteins. The numbers above the aa indicate the first residue of the corresponding exon. The first methionine in exons 3, 4, and 6 are in bold. In a white box are shown the two hathdomains; two putative nuclear localization signals (NLS) are underlined; in a dark grey box is indicated the HMG domain; the four black boxes show the PHD fingers, with the consensus histidine residues in bold; in a dark grey box is shown the SET domain.
Identification and mapping of MMSETgene on 4p16.3. (A) The diagram, drawn to scale, represents the distal 200 kb of the 2-Mb cosmid contig spanning the Huntington’s disease region, with the telomeric side of the contig, containing theFGFR3 gene, to the left. Vertical lines represent the exons distribution of the MMSET gene within about 120 kb of the cosmid contig. The MMSET gene includes at least 24 exons that are transcribed from the telomeric to the centromeric end. The solid arrowhead indicates the 3′ end of an EST that is localized 530 bp centromeric to the end of the last MMSET exon, but is transcribed in the opposite orientation. The solid arrows indicate the position of the previously cloned translocation breakpoints for KMS11, UTMC2, H929, JIM3, OPM2 MM cell lines, and for the tumor sample PCL1; The LP-1 breakpoint has been mapped by sequence analysis of a hybrid transcript splicing to exon 4, and MM5.1 between exons 2b and 3. The KMS11 t(4;14) translocation breakpoint is localized at the 5′ end of MMSET exon 1a, about 15 kb from exon 1; the UTMC2 breakpoint is localized between exon 1a and 1, about 2.5 kb from exon 1; the PCL1 breakpoint is in the intron between exons 1 and 2a; the LP-1, H929, and the JIM3 breakpoints are between exons 3 and 4; and the OPM2 breakpoint between exons 4 and 5. (B) The MMSET transcription units. MMSET is expressed as transcripts that polyadenylate either in exon 11 (type I), or exon 24 (type II) as a result of alternative splicing occurring from exon 10 to 11 (top), or exon 10 to 12 (bottom). The position of the polyadenylation signals (PA) is indicated by black circles. Because of the heterogeneity of the 5′ untranslated region (5′UT), we chose to start the numbering of MMSET from the first nucleotide in exon 3. The type I transcript contains an ORF of 1911 bp, encoding a 647-aa protein with the first methionine at nt 30, in exon 3, and the stop codon at nt 1971 after the first 20 aa of exon 11. The type II transcript contains a longer ORF of 4094 bp, encoding a 1365-aa protein, that is identical to the shorter protein up to the splicing site in exon 10, as indicated by the light gray shading. The two proteins differ after exon 10, with the unique portion of the short protein shaded in black, and the portion unique to the long protein shaded in dark gray. In the middle panel are shown characteristic domains of MMSET. A putative nuclear localization signal (NLS, indicated by a thick vertical line) is common to the two proteins, as well as the HMG domain (dark gray rectangle), and the hathdomain (white rectangle). The long protein is characterized by four PHD fingers (black rectangles), a SET domain (light gray rectangle), an additional hath domain, and another putative NLS. The thick horizontal lines represent the PCR amplified probes used in the Northern blot assay, with the primers number indicated. Numbers within boxes indicate two of the bacteriophage clones isolated from a testis cDNA library. To obtain a complete sequence of the ORFs of MMSET, cDNA fragments have been amplified by PCR as indicated by the dashed horizontal lines. 1112 (gray rectangle) is a clone obtained by 3′ RACE and sequenced with SP6 and T7 primers. (C) Amino acid sequence of MMSET long and short proteins. The numbers above the aa indicate the first residue of the corresponding exon. The first methionine in exons 3, 4, and 6 are in bold. In a white box are shown the two hathdomains; two putative nuclear localization signals (NLS) are underlined; in a dark grey box is indicated the HMG domain; the four black boxes show the PHD fingers, with the consensus histidine residues in bold; in a dark grey box is shown the SET domain.
MMSET mRNA Transcripts Primarily Initiate in Exon 1
To analyze the 5′ end, several 5′ RACE experiments were performed on poly-A enriched RNA from testis and the MM cell line KMM1 [that does not have a t(4;14) translocation]. The results were the same for both samples, and confirmed the heterogeneous use of different exons upstream of exon 3, that we have called 2a, 2b, 2c, and 2d, all of which contain Alu repetitive elements. Additionally in the 5′ RACE, exon 1 was identified spliced to various of the exon 2 and also directly to exon 3. A primer from exon 1 (o99) was used in RT-PCR with a primer from exon 3 to confirm the results of the 5′ RACE, with amplification of a heterogeneous PCR product, consistent with variable exon usage between exons 1 and 3. Furthermore, amplification using o99 with primers in exon 11 (o93) or exon 24 (o80) generated products of the expected size, with no evidence of downstream splicing (data not shown). The sequence of the genomic segment that includes exon 1 and the 5′ flanking region has an extremely high GC nucleotide (85%) content. This may explain the reason why exon 1 falls within an approximately 1.1-kb gap in the published sequence of cosmid 75b9, between the two sequences hsl75b9a and hsl75b9b. We sequenced this region, and by computer analysis identified a potential promoter with TATA box 3037 bp upstream the first Sst I site (nt 2501) of 75b9b and by 5′RACE identified transcripts that initiated 149 bp from this TATA box. We analyzed the published sequence of cosmid 75b9 with the programs TSSG, TSSW, and ProScan12 13 and identified a putative promoter approximately 8 kb telomeric to exon 1. We designed a PCR primer (o134) in the exon 1a immediately downstream from this promoter and demonstrated by hemi-nested reverse transcription (RT)-PCR that this exon was expressed at a very low level in KMM1, and spliced appropriately to exon 3, and to no other exons. Furthermore, no amplification was obtained using a pair of primers in exon 1a and exon 1. Hence, we believe exon 1a represents the use of an alternative promoter and first exon that is not very active in MM. This suggests that there may be multiple promoters for this gene that may be active in different cell types or at different stages of differentiation. Based on our library screening and 5′ RACE experiments in testis and KMM1, it appears that the promoter upstream of exon 1 is used most commonly.
MMSET mRNA Undergoes Complex Alternative Splicing and Differential Polyadenylation
The sequence from the telomeric five cosmids (tel-75b9-184d6-190b4-19h1-96a2-cen) was stripped of repeats using RepeatMasker (Smit AFA, Green P: RepeatMasker athttp://ftp.genome.washington.edu/RM/RepeatMasker.html), and used to probe the dbEST database.14 Expressed regions were identified and used to design oligonucleotide in the 3′ untranslated exons 11 (o93) and 24 (o80) (Fig 1B). These primers were used to amplify the intervening region from exon 6 (o58) by RT-PCR on UTMC2 mRNA. Polyadenylation sites were identified by clustered initiation of 3′ EST sequences downstream from consensus polyadenylation signals (in exon 11 at bp 2990, 3095, 3286 and 8364; in exon 24 at bp 4982 and 7395). By 3′ RACE (no. 1112) we confirmed that the additional polyadenylation signal in exon 11 at bp 2090 was also used in MM. A series of Northern blots identified transcripts of a size consistent with the use of each of these polyadenylation signals (see below). Sequence analysis identified another gene transcribed in the opposite orientation with polyadenylation at 29270 of cosmid 96a2, only 529 bp from the polyadenylation in exon 24 of MMSET, serving to delimit the 3′ end of the MMSET gene. The complete gene organization is summarized in Table 2. The sequence of the phage clones and PCR products agreed with the published genomic sequence, and the whole open reading frame (ORF) was sequenced in both orientations.
MMSET Exon Structure
| Exon . | Size bp . | First nt . | First Codon . | 4p16 Cosmid . | First nt . | Last nt . |
|---|---|---|---|---|---|---|
| 1a | 218 | 75b9b | 8204 | 7986 | ||
| 1 | 244 | gap a-b | −3008 | −2743 | ||
| 2a | 100 | 184d6 | 14925 | 14803 | ||
| 2b | 139 | 184d6 | 14339 | 14200 | ||
| 2c | 200 | 184d6 | 13376 | 13199 | ||
| 2d | 174 | 184d6 | 7888 | 7714 | ||
| 2e | 41? | 184d6 | 7536 | 7496? | ||
| 3 | 626 | 1 | 1 | 184d6 | 6483 | 5858 |
| 4 | 163 | 627 | 200 | 184d6 | 2893 | 2731 |
| 5 | 167 | 790 | 254 | 190b4 | 19885 | 19719 |
| 6 | 483 | 957 | 310 | 190b4 | 18615 | 18133 |
| 7 | 145 | 1440 | 471 | 190b4 | 6129 | 5985 |
| 8 | 119 | 1585 | 519 | 190b4 | 1610 | 1492 |
| 9 | 82 | 1704 | 559 | 19h1 | 38351 | 38270 |
| 10 | 125 | 1786 | 586 | 19h1 | 37148 | 37024 |
| 11 | 6454 | 1911 | 628 | 19h1 | 34463 | 27985 |
| 12 | 132 | 1911 | 628 | 19h1 | 25731 | 25601 |
| 13 | 124 | 2043 | 672 | 19h1 | 24719 | 24596 |
| 14 | 201 | 2167 | 713 | 19h1 | 23503 | 23303 |
| 15 | 180 | 2368 | 780 | 19h1 | 21666 | 21487 |
| 16 | 157 | 2548 | 840 | 19h1 | 21134 | 20978 |
| 17 | 206 | 2705 | 893 | 19h1 | 20844 | 20639 |
| 18 | 104 | 2911 | 961 | 19h1 | 18894 | 18791 |
| 19 | 270 | 3015 | 996 | 19h1 | 17356 | 17087 |
| 20 | 118 | 3285 | 1086 | 19h1 | 15792 | 15676 |
| 21 | 142 | 3402 | 1125 | 19h1 | 1956 | 1815 |
| 22 | 107 | 3544 | 1172 | 19h1 | 1525 | 1419 |
| 23 | 205 | 3651 | 1208 | 19h1 | 344 | 140 |
| 24 | 3563 | 3856 | 1276 | 96a2 | 33362 | 29800 |
| Exon . | Size bp . | First nt . | First Codon . | 4p16 Cosmid . | First nt . | Last nt . |
|---|---|---|---|---|---|---|
| 1a | 218 | 75b9b | 8204 | 7986 | ||
| 1 | 244 | gap a-b | −3008 | −2743 | ||
| 2a | 100 | 184d6 | 14925 | 14803 | ||
| 2b | 139 | 184d6 | 14339 | 14200 | ||
| 2c | 200 | 184d6 | 13376 | 13199 | ||
| 2d | 174 | 184d6 | 7888 | 7714 | ||
| 2e | 41? | 184d6 | 7536 | 7496? | ||
| 3 | 626 | 1 | 1 | 184d6 | 6483 | 5858 |
| 4 | 163 | 627 | 200 | 184d6 | 2893 | 2731 |
| 5 | 167 | 790 | 254 | 190b4 | 19885 | 19719 |
| 6 | 483 | 957 | 310 | 190b4 | 18615 | 18133 |
| 7 | 145 | 1440 | 471 | 190b4 | 6129 | 5985 |
| 8 | 119 | 1585 | 519 | 190b4 | 1610 | 1492 |
| 9 | 82 | 1704 | 559 | 19h1 | 38351 | 38270 |
| 10 | 125 | 1786 | 586 | 19h1 | 37148 | 37024 |
| 11 | 6454 | 1911 | 628 | 19h1 | 34463 | 27985 |
| 12 | 132 | 1911 | 628 | 19h1 | 25731 | 25601 |
| 13 | 124 | 2043 | 672 | 19h1 | 24719 | 24596 |
| 14 | 201 | 2167 | 713 | 19h1 | 23503 | 23303 |
| 15 | 180 | 2368 | 780 | 19h1 | 21666 | 21487 |
| 16 | 157 | 2548 | 840 | 19h1 | 21134 | 20978 |
| 17 | 206 | 2705 | 893 | 19h1 | 20844 | 20639 |
| 18 | 104 | 2911 | 961 | 19h1 | 18894 | 18791 |
| 19 | 270 | 3015 | 996 | 19h1 | 17356 | 17087 |
| 20 | 118 | 3285 | 1086 | 19h1 | 15792 | 15676 |
| 21 | 142 | 3402 | 1125 | 19h1 | 1956 | 1815 |
| 22 | 107 | 3544 | 1172 | 19h1 | 1525 | 1419 |
| 23 | 205 | 3651 | 1208 | 19h1 | 344 | 140 |
| 24 | 3563 | 3856 | 1276 | 96a2 | 33362 | 29800 |
In the first column are listed the MMSET exons; a double line separates exon 11 and 12, indicating that they are contained in two alternative MMSET transcripts (type I and II, respectively). In the second column is reported the size of each exon. In the third column is indicated the first nucleotide of the exon, numbered starting from exon 3. The fourth column shows the first codon of the exon; the first Methionine in exon 3 corresponds to nucleotide 30. The fifth column lists in which 4p16 cosmid the exon is contained, with the first and last nucleotide indicated in columns six and seven. The position of exon 1 is arbitrarily given as distance in nt from the firstSst I site (at position 2501) of 75b9b.
MMSET Encodes 647- and 1365-aa Proteins With Domains Homologous to Those Found in the Trithorax Group
There is at least one AUG codon in exon 1, and in each of the alternatively spliced exons 2a, 2b, 2c, and 2d, but none in exon 1a. In exon 3 the first AUG is immediately downstream from an in-frame stop codon, and is followed by a long ORF that covers many exons. Following exon 10 there is an alternative splice either to exon 11 (type I transcripts) or exon 12 (type II transcripts). In exon 11 there is a termination codon 60 nucleotides (nt) after the splice site, and four polyadenylation signals. Following exon 12 the ORF continues to exon 24 where there is a stop codon, and two polyadenylation signals. These two alternatively spliced mRNAs are predicted to encode proteins of 647 aa and 1365 aa that share a common amino terminal (Fig 1C). This shared region contains a putative nuclear localization signal (NLS), an HMG (high mobilitygroup) box that is characteristic of a large number proteins that bind DNA including SRY, the SOX family of transcription factors, the Hrx fusion partner AF17, and the RNA polymerase I transcription factor UBF,15 and a hath (homologous to the amino terminus of hepatoma derived growth factor [HDGF]) region that is also found in HRP-1 and HRP-2, identified by homology screening.16 HDGF has an NLS and an HMG box, and HRP-1 has an NLS, suggesting that these proteins may function in the nucleus. Not reported previously, the human and mouse G/T mismatch repair proteins MSH6 also contain the hath domain, although it is not conserved in yeast MSH6. The long MMSET protein also contains another copy of the hath motif, as well as another putative nuclear localization signal. In addition, it has four PHD (plant homeodomain) zinc fingers17 characterized by the C4-H-C3 motif, and an SET (Suvar3-9, Enhancer-of-zeste, Trithorax) domain,18,19 domains characteristic of the trithorax group proteins that during Drosophila development are required to maintain stable expression of the clustered homeotic genes.20 The entire carboxy terminal half is most homologous (81% over 666 aa) to the carboxy terminal of the recently described murine protein NSD1,21 a much larger protein of 2588 aa whose amino terminal portion interacts with several nuclear receptors (retinoic acid receptor, thyroid hormone receptor, and estrogen receptor).
MMSET Is Highly Expressed in Testis and Thymus
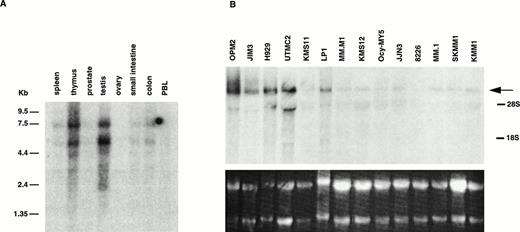
Using a probe from exon 3, a high level of expression of 5.2- and 7.7-kb mRNAs was detected in oligo-dT selected RNA from testis and thymus, and a much lower level in the other tissues examined (Fig 2A). Based on multiple probes on MM cell line RNA (see below), these appear to represent type II transcripts that use the two polyadenylation sites in exon 24. In addition, several fainter bands at 2.4, 3.1, and 4.0 kb are seen, consistent with type I transcripts that use the polyadenylation sites in exon 11 (see below).
Expression of MMSET mRNA in myeloma cell lines and normal tissues. (A) A Northern blot containing 2 μg of poly(A)+ RNA from each of several normal tissues was assessed for MMSET expression hybridizing with an exon 3 probe. The two major bands of 7.7 and 5.2 kb reflect the two polyadenylation signals in exon 24; the lower bands (4.0, 3.1, and 2.8 kb) correspond to the alternative spliced form of MMSET type I containing exon 11, with polyadenylation at different sites. (B) Northern blot analysis of 15 μg of total RNA from 14 myeloma cell lines hybridized with a 3′ exon 24 MMSET probe. The arrow shows the 7.7-kb band, corresponding to the type II MMSET mRNA, that is polyadenylated at nt 7395 of exon 24. The position of 5.0- and 2.0-kb ribosomal RNAs is indicated. The lower panel shows the ethidium bromide staining of the blotted RNAs. (C) A Northern blot containing 2 μg of oligo-dT selected total RNA from MM cell lines with (JIM3 and UTMC2) and without (KMM1) a t(4;14) translocation was repeatedly hybridized with MMSET probes covering different exons. The size of the bands referred in the text is indicated. All three MMSET probes detected the 7.7-kb band (MMSET type II) that, in UTMC2 but not KMM1, cohybridize to the Iμ probe, consistent with the presence of a t(4;14) translocation. Exon 6-10 and exon 19-23, but not 3′ exon 24 probes, also hybridize to the 5.2-kb type II transcript. In addition, exon 6-10 probes detected an 8.8-kb and lower 4.0- and 3.1-kb bands, corresponding to MMSET type I transcript. In JIM3, the size of the bands detected by the exon 6-10 probes is about 600 bp smaller than in the other lines, because the translocation breakpoint is between exon 3 and 4. To obtain comparable signals, the KMM1 blot has been exposed approximately three times longer.
Expression of MMSET mRNA in myeloma cell lines and normal tissues. (A) A Northern blot containing 2 μg of poly(A)+ RNA from each of several normal tissues was assessed for MMSET expression hybridizing with an exon 3 probe. The two major bands of 7.7 and 5.2 kb reflect the two polyadenylation signals in exon 24; the lower bands (4.0, 3.1, and 2.8 kb) correspond to the alternative spliced form of MMSET type I containing exon 11, with polyadenylation at different sites. (B) Northern blot analysis of 15 μg of total RNA from 14 myeloma cell lines hybridized with a 3′ exon 24 MMSET probe. The arrow shows the 7.7-kb band, corresponding to the type II MMSET mRNA, that is polyadenylated at nt 7395 of exon 24. The position of 5.0- and 2.0-kb ribosomal RNAs is indicated. The lower panel shows the ethidium bromide staining of the blotted RNAs. (C) A Northern blot containing 2 μg of oligo-dT selected total RNA from MM cell lines with (JIM3 and UTMC2) and without (KMM1) a t(4;14) translocation was repeatedly hybridized with MMSET probes covering different exons. The size of the bands referred in the text is indicated. All three MMSET probes detected the 7.7-kb band (MMSET type II) that, in UTMC2 but not KMM1, cohybridize to the Iμ probe, consistent with the presence of a t(4;14) translocation. Exon 6-10 and exon 19-23, but not 3′ exon 24 probes, also hybridize to the 5.2-kb type II transcript. In addition, exon 6-10 probes detected an 8.8-kb and lower 4.0- and 3.1-kb bands, corresponding to MMSET type I transcript. In JIM3, the size of the bands detected by the exon 6-10 probes is about 600 bp smaller than in the other lines, because the translocation breakpoint is between exon 3 and 4. To obtain comparable signals, the KMM1 blot has been exposed approximately three times longer.
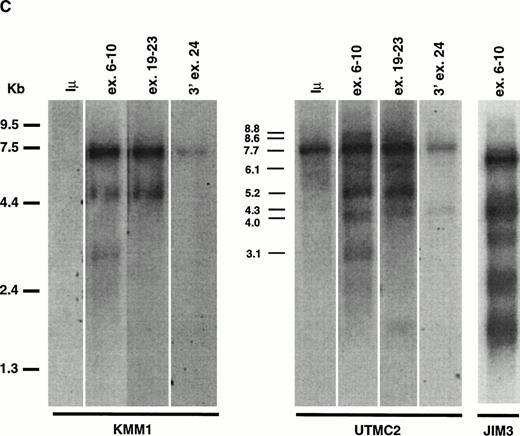
MMSET Is Over-Expressed in the MM Cell Lines With t(4;14)
We probed a Northern blot of total RNA from 14 MM cell lines using a probe from the 3′ end of exon 24 (Fig 2B). A 7.7-kb mRNA was detected in all 14 MM cell lines, with a higher level of expression in 5 of 6 cell lines with t(4;14) translocations: OPM2, JIM3, H929, UTMC2, and LP-1, but not in KMS11. To study the effects of the translocation on the expression of the different splice and polyadenylation forms of MMSET, we used different regions of the gene to repeatedly probe a Northern blot of oligo-dT selected RNA from MM cell lines with (JIM3 and UTMC2) and without (KMM1) the t(4;14) translocation. In Fig 2C a much longer exposure of KMM1 is shown to demonstrate that there are three bands (3.1, 5.2, and 7.7 kb) detected with an exon 6-10 probe, consistent with 5.2- and 7.7-kb type II transcripts and, to a lesser extent, a 3.1-kb type I transcript. In contrast, the expression in UTMC-2 is not only more abundant, but the pattern is more complex. Using the same exon 6-10 probe, in addition to the 5.2- and 7.7-kb type II transcripts, 3.1-, 4.0-, and 8.8-kb type I transcripts are more prominent than in KMM1. These were confirmed to be type I transcripts by using a probe from the 5′ end of exon 11 (no. 1112) that hybridized only to bands of approximately 3.1, 4.0, and 8.8 kb (data not shown). With the exon 19-23 probe, in addition to the major 5.2- and 7.7-kb type II transcripts, there are fainter bands at 4.3, 6.1, and 8.6 kb. We have not determined what mRNAs these minor bands represent, although the 4.3- and 8.6-, but not the 6.1-kb, bands are also detected with a probe from 3′ exon 24, suggesting that they use the distal polyadenylation signal in exon 24. These mRNAs may be formed by alternative splicing that we have been unable to detect, or by the use of alternative promoters, or by specific mRNA degradation. In JIM3, the translocation breakpoint is between exon 3 and 4 and, as predicted, the mRNAs detected with an exon 6-10 probe are each about 600 bp smaller then the corresponding bands in UTMC2. This confirms that it is the translocated MMSET allele that is over-expressed. To determine if the translocation significantly altered the ratio of the two transcripts, we performed a competitive RT-PCR using a 5′ primer in exon 10 and 3′ primers in exon 11 and exon 16 that indicated that the type II transcripts are relatively more abundant in both the lines with and without the translocation (data not shown).
The Dysregulated Expression of MMSET Initiates Both From Promoters on 4p16 and the IgH Locus
Hypothesizing that the dysregulated MMSET expression may be the result of hybrid mRNA transcripts initiating in the IgH locus, we hybridized the same blot mentioned above with a probe from the Iμ exon (we did not probe with a JH probe because the JH exons are too short to hybridize efficiently). In UTMC2, the Iμ probe uniquely detected a 7.7-kb band that exactly cohybridized with the 7.7-kb type II MMSET transcript. Similarly, in JIM3 the same probe detected a band of 7.0 kb that exactly co-hybridized with the 7.0-kb band detected by the MMSET probes (data not shown). There was no expression detected in KMM1 that lacks a t(4;14) translocation. Although this result implied the existence of hybrid type II transcripts, it did not explain the over-expression of all of the different splice and polyadenylated forms of MMSET. To determine the transcription initiation of these forms, a 5′ RACE was performed from exon 3 in UTMC2, and from exon 4 in JIM3. In UTMC2 the results we obtained were similar to those in KMM1 and testis, with evidence of exon 2d upstream of exon 3, but primarily exon 1 spliced directly to exon 3. In JIM3 the 5′ RACE indicated that transcription started in the intron upstream of exon 4 (at 2293 of cosmid 184d6) and in switch gamma with splicing to exon 4. This apparent use of cryptic promoters may explain the very broad appearance of the MMSET bands detected on the JIM3 Northern blot, as though the mRNAs initiate over an ill-defined area. Although in neither JIM3 or UTMC2 did the 5′ RACE identify the JH or Iμ hybrid transcripts, a competitive RT-PCR in KMS11 and UTMC2 using 5′ primers from exon 1, JH, Iμ, and a 3′ primer from exon 3 suggested equal or greater amplification with Iμ and JH then exon 1 (data not shown). This discrepancy suggests some artifactual skewing during the PCR so that further analysis will be required to determine the relative contribution of the different promoters.
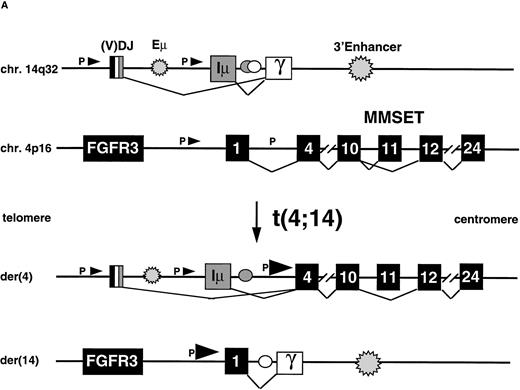
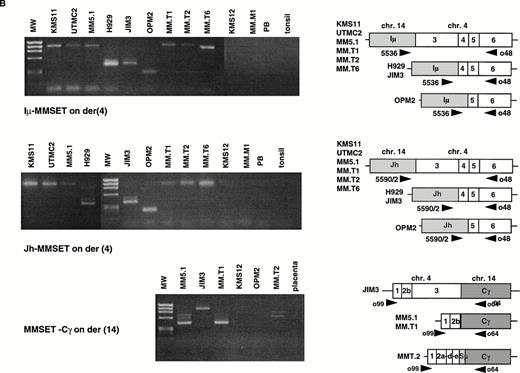
The t(4;14) Translocation Results in Hybrid JH-MMSET and Iμ-MMSET mRNA Transcripts
The t(4;14) translocation is predicted to result in IgH-MMSET and MMSET-IgH hybrid transcripts (Fig 3A). To confirm the existence of IgH-MMSET hybrid transcripts we performed RT-PCR using consensus JH primers (5590/2) with an exon 6 primer (o48), and an Iμ primer (5536) with the same exon 6 primer (Fig 3B). We detected PCR products of the predicted size only in the cell lines with the translocation, and not in other cell lines, peripheral blood, or tonsil. The cell lines with breakpoints telomeric to exon 3 (KMS11, UTMC2, and MM5.1) all had mRNAs that spliced appropriately from JH or Iμ to exon 3. The cell lines with breakpoints between exon 3 and 4 (JIM3 and H929) had hybrid mRNAs that spliced to exon 4, and OPM2, with a breakpoint between exon 4 and 5 had hybrid mRNAs that spliced to exon 5. By Northern blot we showed that Iμ-MMSET transcripts are primarily type II transcripts, with use of the distal polyadenylation signal in exon 24 (see above). RT-PCR results suggest that the JH-MMSET transcripts are also primarily type II: in UTMC2 mRNA, very strong amplification of a 2.7-kb PCR product was obtained with a JH-exon 17 primer pair, and only very weak amplification of a similar sized product with a JH-exon 11 primer pair (data not shown).
The t(4;14) translocation results in IgH-MMSET hybrid transcripts. (A) The top diagram shows the genomic organization of the IgH locus on 14q32 in an IgG isotype switched plasma cell. The structural elements include the recombined VDJ exons, 5′ and 3′ enhancers (Eμ and 3′E), the noncoding Ιμ (gray rectangle) and coding Cγ (white rectangle) exons, and the switch regions (circles). The transcription may initiate either from promoters (black arrowhead) upstream of JH or Iμ, in both cases with splicing to the Cγ exon. The second diagram shows the 4p16 locus, with FGFR3 on the telomeric side, and MMSET on the centromeric side. The MMSET exons are depicted as black rectangles. The main promoter upstream of exon 1 and a cryptic promoter upstream of exon 4 are depicted. The alternative splicing occurring between exon 10-11 or exon 10-12 is shown. The lower two diagrams represent the der(4) and der(14) as result of the t(4;14) translocation in JIM3 MM cell line. On der(4), the transcription may initiate at a lower level either from (VD)J or Iμ, and in both cases it splices to exon 4 of MMSET, proceeds up to exon 10 and then it splices to exon 12, but not exon 11. However, the transcription preferentially initiates from the upregulated MMSET promoter upstream of exon 4 and generates both types of alternatively spliced forms of MMSET. On the der(14), the reciprocal hybrid transcript is formed when the transcription starts from the MMSET promoter upstream of exon 1, and it splices to the Cγ exon. (B) An RT-PCR assay detects hybrid transcripts Iμ-MMSET (top panel) and JH-MMSET (middle panel) on der(4), and MMSET-Cγ (bottom panel) on der(14) in myeloma cell lines and primary tumor samples with t(4;14) translocation. The IgH exons are shaded in gray, and the MMSET exons are depicted as white rectangles. The exon structure of each hybrid transcripts is depicted and has been obtained by sequencing of the PCR product (Iμ-MMSET and MMSET-Cγ), or deduced by the size of the PCR products, consistent with the breakpoint position (JH-MMSET).
The t(4;14) translocation results in IgH-MMSET hybrid transcripts. (A) The top diagram shows the genomic organization of the IgH locus on 14q32 in an IgG isotype switched plasma cell. The structural elements include the recombined VDJ exons, 5′ and 3′ enhancers (Eμ and 3′E), the noncoding Ιμ (gray rectangle) and coding Cγ (white rectangle) exons, and the switch regions (circles). The transcription may initiate either from promoters (black arrowhead) upstream of JH or Iμ, in both cases with splicing to the Cγ exon. The second diagram shows the 4p16 locus, with FGFR3 on the telomeric side, and MMSET on the centromeric side. The MMSET exons are depicted as black rectangles. The main promoter upstream of exon 1 and a cryptic promoter upstream of exon 4 are depicted. The alternative splicing occurring between exon 10-11 or exon 10-12 is shown. The lower two diagrams represent the der(4) and der(14) as result of the t(4;14) translocation in JIM3 MM cell line. On der(4), the transcription may initiate at a lower level either from (VD)J or Iμ, and in both cases it splices to exon 4 of MMSET, proceeds up to exon 10 and then it splices to exon 12, but not exon 11. However, the transcription preferentially initiates from the upregulated MMSET promoter upstream of exon 4 and generates both types of alternatively spliced forms of MMSET. On the der(14), the reciprocal hybrid transcript is formed when the transcription starts from the MMSET promoter upstream of exon 1, and it splices to the Cγ exon. (B) An RT-PCR assay detects hybrid transcripts Iμ-MMSET (top panel) and JH-MMSET (middle panel) on der(4), and MMSET-Cγ (bottom panel) on der(14) in myeloma cell lines and primary tumor samples with t(4;14) translocation. The IgH exons are shaded in gray, and the MMSET exons are depicted as white rectangles. The exon structure of each hybrid transcripts is depicted and has been obtained by sequencing of the PCR product (Iμ-MMSET and MMSET-Cγ), or deduced by the size of the PCR products, consistent with the breakpoint position (JH-MMSET).
RT-PCR Amplification of JH-MMSET and Iμ-MMSET mRNA Transcripts Identifies MM Patient Samples With t(4;14) Translocation
Previously we had identified three patients with MM in whom we could detect FGFR3 by RT-PCR of the BM RNA. In these three samples, but not in others, we detected hybrid mRNAs that spliced to exon 3, confirming the presence of the t(4;14) translocation. In LP1 we failed to detect a JH-, Iμ-, Iγ-, or Iα-MMSET hybrid transcripts, although we have demonstrated a t(4;14) translocation by fluorescence in situ hybridization (FISH) analysis (data not shown). To analyze this further, we performed a 5′ RACE from exon 4 and identified transcripts that appeared to initiate in switch gamma and spliced to exon 4. This shows that although frequent, JH- and Iμ-MMSET transcripts are not always seen with t(4;14) translocation. We expect that the Iμ-MMSET hybrid transcripts initiate from the Iμ promoter. The JH-MMSET hybrid transcripts may theoretically initiate from a V region promoter if there is a VDJ rearrangement, or from a promoter upstream of the D or J exons. Similar Iμ-Bcl-6 and JH-Bcl-6 hybrid transcripts have been described that are associated with the t(3;14) translocation.22 Using a consensus V region framework 3 (FR3) oligo we did not detect any FR3-MMSET hybrid transcripts, suggesting that there is no sense V region transcribed upstream of JH-MMSET mRNAs. A 5′ RACE in UTMC2 from exon 3, nested to JH, identified transcripts that appeared to initiate in the intron upstream of J5. From our analysis it appears that the t(4;14) translocation is frequently associated with JH and Iμ hybrid transcripts, and this RT-PCR assay represents a convenient and sensitive method to identify this translocation in patients samples.
The Reciprocal Hybrid MMSET-Cγ mRNA Is Also Expressed
This translocation is also predicted to result in hybrid transcripts initiating in MMSET and splicing to the IgH locus. To identify these transcripts we performed RT-PCR with an MMSET exon 1 primer (o99) and a pool of primers for Cμ, Cα, and Cγ (o132, o64, o65) in the samples with t(4;14) translocation. We detected MMSET-Cγ transcripts in LP1, JIM3, MM5.1, MM.T1, MM.T2, but not in OPM2 that also has a translocation to switch gamma (Sγ); however, we did not detect MMSET-IgH in UTMC2 or H929 in which the translocations are into Sμ and Sα, respectively (Fig 3B). In comparison with the nearly universal expression of IgH-MMSET hybrid transcripts, the reciprocal MMSET-IgH hybrid transcripts are less frequently detected by our assay.
Most of the Hybrid Transcripts Would Not Be Predicted to Result in Fusion Proteins
To determine whether the hybrid transcripts resulting from the t(4;14) translocation may potentially encode fusion proteins, we sequenced the PCR products and looked for ORFs. Because exon 3 has an in-frame stop codon upstream of the AUG, none of the transcripts that splice to exon 3 can result in a fusion protein, and they would be predicted to encode the full-length type II MMSET protein. Similarly, no fusion protein would be predicted for hybrid transcripts between JH-exon 4, exon 2b-Cγ, exon 2e-Cγ, or exon 3-Cγ, because they are all out of frame. Iμ transcripts are said to be sterile, but the Iμ exon has several AUGs, and a translatable polypeptide chain has been described.23 Iμ-exon 4 could result in a fusion protein containing 17 aa from Iμ; however, the Iμ-exon 5 is out of frame. In sterile transcripts that splice to exon 4 (or initiate upstream of exon 4), the first AUG is in frame at nt 744, and in transcripts that splice to exon 5, the first AUG at nt 908 is out of frame, but the second one, at nt 999, is in frame. The nucleotide context of AUG999 (it has a G−3 and G+4) and AUG744 (only G+4) suggest that they may potentially serve to initiate translation according to the Kozak consensus sequence.24 Among the other hybrid transcripts that might possibly result in a fusion protein is the JH-exon 5, if the translocation occurred on the productive allele, and finally, the reciprocal MMSET exon 4-Cγ mRNA. In summary, with the exception of the possible fusion proteins mentioned above, the translocation may be predicted to result in the full-length MMSET protein if the breakpoint is upstream of exon 3, or truncated proteins lacking either the amino terminal 238, or 323 aa, if the breakpoints are upstream of exon 4 or 5, respectively.
DISCUSSION
We have shown previously that the t(4;14) translocation bringsFGFR3 within 50 to 100 kb of the strong 3′ IgH enhancers.4 Normally FGFR3 expression is undetectable by RT-PCR in MM cell lines lacking the t(4;14) translocation, in peripheral blood or in bone marrow. We have shown that in the presence of the translocation FGFR3 is ectopically expressed, with selective expression of only one allele in all the informative cases. In addition, we have shown that the same kinase-activating mutations that cause thanatophoric dysplasia when inherited in the germline are present in three of six MM cell lines and in one of three primary tumor samples with t(4;14) translocation. In one patient sample we were able to show that the translocation occurred first, causing upregulation of the translocated FGFR3 allele. The development of an activating mutation was a secondary event that may have been associated with tumor progression (unpublished results, May 1997). We now show that this same translocation that dysregulates FGFR3 brings the weaker intronic enhancer adjacent to a novel gene, MMSET, resulting in its dysregulated expression. In the cases we examined, this dysregulation appears to arise both from endogenous promoters on chromosome 4, and from IgH promoters resulting in JH-MMSET and Iμ-MMSET mRNA. Similar JH-BCL6 and Iμ-BCL6 hybrid transcripts have been described associated with t(3;14) translocations in about 5% of diffuse large cell lymphoma.22 In addition, BCL2-JH hybrid transcripts occur in follicular lymphoma with t(14;18) translocation. Otherwise hybrid transcripts involving the IgH locus have not been described in recurrent 14q32 translocations (eg, 8q24 c-myc, 11q13 cyclin D1, 16q23 c-maf). These hybrid transcripts have important clinical implications because they provide a convenient way to identify the t(4;14) translocation in patients’ samples, and to monitor patients for minimal residual disease. In general, the hybrid transcripts do not appear to undergo alternative splicing or differential polyadenylation, and are not predicted to encode fusion proteins. They appear to encode only the full-length type II MMSET transcript, or 5′ truncated type II full-length transcripts that lack the 238 or 323 amino terminal amino acids if the breakpoints are upstream of exon 4 or 5, respectively. Additionally, the t(4;14) translocation is associated with over-expression of both type I and type II alternatively spliced and differentially polyadenylated MMSET transcripts.
Sequence analysis of the predicted MMSET protein suggests that it may also play an important role in the neoplastic transformation. MMSET has a number of domains found in nuclear proteins involved in chromatin remodeling, and in the epigenetic regulatory machinery: most notably the PHD fingers and SET domain characteristic of the trithorax group proteins in Drosophila (eg, trx and Ash1). Other mammalian proteins with these features include Hrx (also called All-1, Htrx, or Mll), the mammalian homologue of trx,20 and the recently described NSD1.21 The SET domain was shown to interact with dual specificity phosphatases, and recently with Sbf1 (SET binding factor 1) that contains a SET binding domain but lacks the catalytic phosphatase domain. When either the isolated SET domain, or Sbf1, were over-expressed they were shown to be transforming in fibroblasts.25 This interaction provides a model linking proteins involved with chromatin remodeling to signaling factors controlling cellular growth and differentiation. As a consequence of 11q23 chromosomal translocation with a promiscuous array of partner chromosomes, fusion proteins are formed that replace the PHD fingers and SET domain of Hrx with a variety of protein domains, some of which appear to have roles in transcriptional activation.26 It is unresolved whether these fusion proteins act through a gain of function mechanism, loss of function mechanism, or a combination of both. We have shown here that the t(4;14) translocation results in a marked dysregulation of MMSET expression. MMSET encodes a short protein lacking the PHD and SET domains that may potentially function as a dominant negative regulator of the more abundantly expressed long protein. In the presence of the translocation, the expression of both of these forms is increased, although the type II remains predominant.
Because Hrx+/− mice have a severe phenotype (including hematologic abnormalities), it is evident that gene dosage is critical for normal development.27 Interestingly, the loss of the distal end of chromosome 4p results in the well-described Wolf-Hirschhorn syndrome (WHS). WHS patients are reported to have severe growth deficiency, profound mental retardation, convulsions, microcephaly, sacral dimples, characteristic facial signs, a variety of midline defects (cleft lip, hypospadias, and cryptorchidism), skeletal defects, heart defects, hemangiomas, and eye defects.28,29Recently the critical region deleted in this syndrome has been mapped by genetic linkage and shown to fall within a 165-kb area (from 190b4 going centromeric),30 which we now demonstrate overlaps theMMSET gene. Although WHS is believed to be a contiguous gene syndrome, it may be predicted that patients with this syndrome have only one copy of MMSET. Based on the mapping data and the trx homology, it is a likely candidate to contribute to this phenotype.
The IgH locus has been shown to contain two kinds of enhancers: (1) an Eμ intronic enhancer that is located downstream of JH and upstream of switch μ sequences, and (2) stronger 3′ enhancers (perhaps with locus control region activity) that are located downstream of the α 1 and α 2 constant region genes.31 Both kinds of enhancers are on der(14) when a translocation occurs upstream of a JH sequence. By contrast, a reciprocal translocation into any of the switch regions separates the two kinds of enhancers, with at least one 3′ enhancer on der(14) and the Eμ intronic enhancer on the other derivative chromosome (see Fig 3). The t(4;14) translocation into an IgH switch region results in dysregulated expression of MMSET on der(4) and FGFR3 on der(14). As summarized above, the dysregulation of MMSET in MM, together with the homology of MMSET to the Hrx gene at 11q23 that is dysregulated by chromosome translocation in acute leukemia, suggests an oncogenic role of MMSET in MM tumors with a t(4;14) translocation. The simultaneous dysregulation of FGFR3 by this translocation may provide a survival or proliferative signal. Alternatively, it is possible that the overexpression of FGFR3 has no immediate effect on transformation of MM cells, but that the occurrence of a kinase-activating mutation in the dysregulated FGFR3 gene contributes to tumor progression. It remains to be determined how the simultaneous dysregulation of MMSET and FGFR3 contribute to MM oncogenesis. However, regardless of the mechanisms involved, this appears to be the first example of an IgH translocation that simultaneously dysregulates two genes with oncogenic potential: FGFR3 on der(14) and MMSET on der(4).
NOTE ADDED IN PROOF
Following submission of this manuscript, the partial genomic structure and developmental expression of the same gene, named WHSC1, was reported by Stec et al.32
M.C. and E.N. contributed equally to this work.
Supported by the Howard Temin Award from the National Cancer Institute (CA74265).
Address reprint requests to P. Leif Bergsagel, MD, 525 E 68th St, Room C609, New York, NY 10021; e-mail: plbergsa@mail.med.cornell.edu.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal