Abstract
Human herpesvirus 8 (HHV-8) genomic sequences were recently detected by polymerase chain reaction (PCR) and in situ hybridization in bone marrow stromal cells grown from multiple myeloma (MM) patients, but not in cells from control subjects (Rettig et al, Science 276:1851, 1997). We sought to confirm these observations in our own group of MM patients (n = 30). DNA was extracted from adherent stromal cells grown under varying conditions and assayed for HHV-8 sequence using PCR to amplify the orf 26 (KS330) sequence (Chang et al,Science 266:1865, 1997), as initially reported. Samples from human control subjects (n = 25) were concurrently extracted and analyzed. After 30 cycles of amplification, we did not detect any positive samples. In a more sensitive nested PCR, samples from 18 of 30 (60%) MM patients were positive, at about the limit of detection, but orf 26 sequence was also amplified from 11 of 25 (44%) human control samples. However, PCR amplification from other regions of the viral genome (orf 72 and orf 75) was uniformly negative for all MM and control samples, despite equivalent sensitivity. Additionally, all sera from MM patients were negative for HHV-8 IgG by immunofluorescence. Our data do not support a role of HHV-8 in the etiology of MM but may suggest the presence of a related (KS330-containing) virus in MM patients and in some control subjects.
This is a US government work. There are no restrictions on its use.
HUMAN HERPESVIRUS 8 (HHV-8), also know as Kaposi’s sarcoma-associated herpesvirus, is a member of the lymphotropic subgroup of herpesviruses. HHV-8 has been implicated in a variety of disorders, including Kaposi’s sarcoma (KS),1 primary effusion lymphomas,2 and multicentric Castleman’s disease.3 The virus is unusual in that the viral genome encodes a large number of homologs of cellular genes, including genes functioning in cell regulation (cyclin D, IRF-1), control of apoptosis (bcl-2, DED domain proteins), cell-cell interaction (OX2), immunoregulation (CD46/CR1, CR2), and cytokine signaling (CC chemokines, CXC chemokine receptor, interleukin-6 [IL-6]).4 In particular, the viral encoded IL-6 (vIL-6) has been shown to be functional in mouse B-cell proliferation assays, preventing apoptosis in IL-6–dependent cells.5
IL-6 has been hypothesized as a major factor in the pathogenesis or progression of multiple myeloma (MM) by a number of different animal models, in vitro studies, and epidemiologic studies (reviewed in Klein et al6). The most compelling human primary cell experiments showed that myeloma plasma cells induced paracrine production of IL-6 by marrow stromal cells.7,8 Both secretion of factors such as IL-1 and/or tumor necrosis factor-α (TNF-α) by the tumor cells and direct adhesive interactions appeared to be important. Of note, myeloma cells can stimulate excess IL-6 production from stromal cells grown from both myeloma patients and normal donors,7and the IL-6, in turn, stimulates the growth or prevents cell death of the tumor cells. The in vivo role of these interactions is unclear, and anti–IL-6 therapies have proved disappointing.9
Recently, it was reported that dendritic-like bone marrow stromal cells grown from marrow mononuclear cells contained HHV-8 genomic sequences detected by polymerase chain reaction (PCR) in 15 of 15 MM patients, a lower percentage of patients with monoclonal gammopathy of undetermined significance (MGUS), and no healthy controls.10 HHV-8 sequence was also detected in cultured cells by in situ hybridization with an HHV-8–specific biotinylated probe. Although in the initial report prolonged in vitro culture of the bone marrow cells was required, a subsequent publication by the same group has described the detection of HHV-8 sequences in multiple myeloma bone marrow core biopsies by both PCR and in situ hybridization.11 The possible involvement of HHV-8 viral IL-6 in myeloma progression has been postulated, and viral IL-6 transcripts were detected in the marrow stromal cells in the original report.10 We have sought to confirm both these observations in myeloma patients and controls studied at two collaborating institutions, using both PCR analysis for multiple regions of the HHV-8 genome and serologic studies on the same patient cohorts.
MATERIALS AND METHODS
Bone marrow samples.
All patients and volunteers gave their written informed consent before participation in the study. The human studies were approved by the Institutional Review Boards of both collaborating institutions and the animal studies by the Animal Care and Use Committee.
Fresh bone marrow was collected from patients with MM before autologous bone marrow transplantation (n = 15; Table1) and mononuclear cells were fractionated on a ficoll-hypaque density gradient (Lymphocyte Separation Medium; Organon Teknika, Durham, NC). Dexter-type long-term cultures were established as previously described using complete long-term culture media (Stem Cell Technologies, Vancouver, British Columbia, Canada),12 13 with a weekly half-volume media change (demidepopulation), and compared with parallel cultures from normal human volunteers (n = 21), normal rhesus macaques (n = 7), and normal dogs (n = 5). At 21 days, the adherent layers were harvested and an aliquot was cryopreserved for later DNA extraction. Cell culture and DNA extraction were performed at a separate institution from PCR analysis to reduce the risk of contamination with PCR products.
PCR Analysis of Myeloma Patient Stroma Grown in Dexter Cultures and Concurrent Serological Testing for HHV-8 Antibody
| Patient ID . | Disease Status . | Cells Cultured-150 . | PCR Results-151 (no. positive/ no. tested) . | Serum HHV-8 IgG by IFA . |
|---|---|---|---|---|
| T1 | PR | Fresh | 1/3 | Neg |
| T2 | PR | Fresh | 1/3 | Neg |
| T3 | PR | Fresh | 2/3 | Neg |
| T5 | PR | Fresh | 2/2 | NT |
| T6 | PR | Fresh | 1/3 | Neg |
| T7 | PR | Fresh | 2/3 | Neg |
| T8 | PR | Fresh | 2/3 | Neg |
| T9 | PR | Fresh | 3/3 | Neg |
| T10 | PR | Fresh | 2/3 | NT |
| T11 | PR | Fresh | 1/3 | Neg |
| T12 | PR | Fresh | 0/3 | Neg |
| T13 | PR | Fresh | 1/3 | NT |
| T14 | PR | Fresh | 1/3 | NT |
| T15 | PR | Fresh | 2/3 | NT |
| T16 | PR | Fresh | 0/2 | NT |
| Patient ID . | Disease Status . | Cells Cultured-150 . | PCR Results-151 (no. positive/ no. tested) . | Serum HHV-8 IgG by IFA . |
|---|---|---|---|---|
| T1 | PR | Fresh | 1/3 | Neg |
| T2 | PR | Fresh | 1/3 | Neg |
| T3 | PR | Fresh | 2/3 | Neg |
| T5 | PR | Fresh | 2/2 | NT |
| T6 | PR | Fresh | 1/3 | Neg |
| T7 | PR | Fresh | 2/3 | Neg |
| T8 | PR | Fresh | 2/3 | Neg |
| T9 | PR | Fresh | 3/3 | Neg |
| T10 | PR | Fresh | 2/3 | NT |
| T11 | PR | Fresh | 1/3 | Neg |
| T12 | PR | Fresh | 0/3 | Neg |
| T13 | PR | Fresh | 1/3 | NT |
| T14 | PR | Fresh | 1/3 | NT |
| T15 | PR | Fresh | 2/3 | NT |
| T16 | PR | Fresh | 0/2 | NT |
Abbreviations: PR, partial remission; NT, not tested.
Fresh or frozen cells were cultured for 3 weeks under standard Dexter culture conditions and adherent cells were analyzed.
PCR was performed using primers from the orf 26 (KS330) region.
Additionally, stromal cell cultures grown under a variety of conditions from patients with MM at various disease stages (n = 15; Table 2) and control subjects (n = 4) were analyzed for the presence of HHV-8 by PCR. Cryopreserved stromal cells that were processed, grown, and harvested as previously described were initially analyzed and consisted of the adherent stromal cell fraction grown from bone marrow mononuclear cells in Dulbecco’s modified Eagles medium supplemented with 10% fetal calf serum (D10).12Parallel standard stromal cell cultures were established by plating fresh bone marrow mononuclear cells or previously cryopreserved bone marrow mononuclear cells at a density of 1 to 2 × 106cells/mL in either Dulbecco’s modified Eagles medium supplemented with 10% fetal calf serum, 2 mmol/L L-glutamine, 10 μg/mL gentamicin (D10), or Iscove’s modified Dulbecco’s medium, supplemented with 10% fetal calf serum, 10% horse serum, 2 mmol/L L-glutamine, 10 μg/mL gentamicin (long-term culture media [LTCM]; Stem Cell Technologies) and incubated at 37°C in 5% CO2, as previously described.12 The medium was changed at 48 hours and then every 3 to 4 days until the adherent stroma was confluent. The adherent cells were then harvested, washed, and expanded. Finally, stromal cultures from additional subjects were set up under all three conditions (D10, LTCM, and Dexter-type). Cultures were harvested at 3 weeks, and DNA was extracted from the fresh cells for HHV-8.
PCR Analysis of Myeloma Patient Stroma Grown in Various Culture Conditions and Concurrent Serological Testing for HHV-8 Antibody
| Patient ID . | Disease Status . | Cells Cultured* . | PCR Results† . | Serum HHV-8 IgG by IFA . | ||
|---|---|---|---|---|---|---|
| D10 . | LTCM . | Dexter . | ||||
| N1 | PR | Fresh | Neg | NT | NT | Neg |
| N2 | PR | Fresh | Neg | NT | NT | NT |
| N3 | PR | Fresh | Neg | NT | NT | Neg |
| N4 | PR | Fresh | Neg | NT | NT | Neg |
| N5 | PR | Frozen | Neg | Neg | NT | Neg |
| PD-T | Fresh | Neg | Neg | NT | ||
| N6 | PR | Frozen | Pos | NT | NT | NT |
| CR-T | Fresh (d10) | Pos | Neg | NT | Neg | |
| Fresh (d21) | Pos | Neg | NT | |||
| N7 | PR | Fresh | Neg | Neg | NT | Neg |
| N8 | PR | Frozen | Neg | Neg | NT | NT |
| N9 | PR | Fresh | Pos | Neg | NT | NT |
| N10 | PD | Fresh | NT | Neg | NT | NT |
| N11 | PD | Fresh | Pos | Neg | Pos | Neg |
| N12 | PR | Fresh | Neg | Neg | Neg | Neg |
| N13 | PR-T | Fresh | Neg | Pos | Neg | Neg |
| N14 | PR | Fresh | Neg | Pos | Neg | Neg |
| N15 | PR | Fresh | Neg | Neg | Neg | Neg |
| Patient ID . | Disease Status . | Cells Cultured* . | PCR Results† . | Serum HHV-8 IgG by IFA . | ||
|---|---|---|---|---|---|---|
| D10 . | LTCM . | Dexter . | ||||
| N1 | PR | Fresh | Neg | NT | NT | Neg |
| N2 | PR | Fresh | Neg | NT | NT | NT |
| N3 | PR | Fresh | Neg | NT | NT | Neg |
| N4 | PR | Fresh | Neg | NT | NT | Neg |
| N5 | PR | Frozen | Neg | Neg | NT | Neg |
| PD-T | Fresh | Neg | Neg | NT | ||
| N6 | PR | Frozen | Pos | NT | NT | NT |
| CR-T | Fresh (d10) | Pos | Neg | NT | Neg | |
| Fresh (d21) | Pos | Neg | NT | |||
| N7 | PR | Fresh | Neg | Neg | NT | Neg |
| N8 | PR | Frozen | Neg | Neg | NT | NT |
| N9 | PR | Fresh | Pos | Neg | NT | NT |
| N10 | PD | Fresh | NT | Neg | NT | NT |
| N11 | PD | Fresh | Pos | Neg | Pos | Neg |
| N12 | PR | Fresh | Neg | Neg | Neg | Neg |
| N13 | PR-T | Fresh | Neg | Pos | Neg | Neg |
| N14 | PR | Fresh | Neg | Pos | Neg | Neg |
| N15 | PR | Fresh | Neg | Neg | Neg | Neg |
Abbreviations: PR, partial remission; PD, progressive disease; CR-T, complete remission after transplantation; PR-T, partial remission after transplantation; PD-T, progressive disease after transplantation; NT, not tested.
Fresh or frozen bone marrow mononuclear cells were cultured under different parallel conditions (D10, LTCM, and Dexter, see Materials and Methods for details) and adherent cells analyzed at 3 weeks unless otherwise stated. d10, analyzed on day 10; d21, analyzed on day 21.
PCR was performed using primers from the orf 26 (KS330) region.
As a positive control, DNA was extracted from cell lines BCBL-1 (provided by Dr Robert Yarchoan, National Cancer Institute, Bethesda, MD) and BCP-1 (ATCC, Rockville, MD), and Jurkat cell DNA was used as a negative control.
PCR for HHV-8.
DNA was extracted from the stromal samples by proteinase K digestion and phenol:chloroform precipitation or using the QIAamp Tissue kit (Qiagen, Inc, Valencia, CA), according to the manufacturer’s protocol. DNA was extracted from approximately 106 cells and eluted into 200 μL of molecular biology grade water. The concentration and purity of DNA were estimated by measuring the optical densities at 260 and 280 nm (∼1 μg of DNA was obtained from 105 cells).
Amplification was performed using sets of primers from different parts of the HHV-8 genome. Amplification from the orf 26 region (minor capsid gene) was performed using the KS330223 primers KS1 and KS2, as originally described, which amplify a 233-bp product.1PCR reactions were performed with ExTaq DNA polymerase (Takara Biomedicals, Otsu, Japan) according to the manufacturer’s instructions (50 μL final volume, containing 100 to 500 ng of DNA, 80 μmol/L dNTPs, 0.5 mmol/L each primer, and 2 U ExTaq). Although PCR was performed for 30 cycles in the initial experiments, this was increased to 40 cycles of amplification for added sensitivity (denaturation for 1 minute at 94°C, then 40 cycles of 40 seconds at 92°C, 40 seconds at 60°C, and 90 seconds at 75 °C, followed by extension for 5 minutes at 75°C). The products (5 μL) were analyzed by agarose electrophoresis, transferred to a membrane, and hybridized using a specific oligonucleotide-radiolabeled probe.1 A second round of PCR amplification was performed on the (40 cycle) products using primers NS1 and NS2, which amplify a 160-bp product.14 Reactions were as described above (5 μL of amplified product was amplified), with 30 cycles of amplification, and products analyzed by agarose electrophoresis and Southern hybridization.
In an attempt to develop more sensitive assays and to confirm the results obtained using primers from the orf 26 (KS330) region, a second nested PCR amplification was developed with primers from the orf 75 region (membrane antigen; KS631Bam2). The outside primers (TAT-TCG-CGG-CCT-TGG-CAA-CC; AAG-ATG-CGC-ACC-GCG-TTG-TC) amplify a 408-bp product, and the internal primers (ACG-TAC-AGC-AGG-CCG-AGA-TG; GGA-GCT-GTC-GCG-ATA-GAG-GT) amplify a 245-bp product. PCR reaction mixes and conditions were as described above, with 30 cycles of amplification for both the outside and internal primer pairs. The nested product was analyzed by Southern hybridization using a specific oligonucleotide-radiolabeled probe (CAG-TCT-GCT-CCA-TCT-CTA-CCA-CTA-CTT-CCA). A third one-stage PCR was performed with primers from the orf 72 region (cyclin D homolog region) that amplify a 106-bp product.15 Forty cycles of amplification were analyzed using the probe previously described.15
To test for the integrity of the DNA and to exclude the presence of PCR inhibitors, PCR amplification with β-actin primers was performed on all samples, as previously described.12 In addition, some repeatedly negative samples were spiked with 3 genome copies of HHV-8 and retested.
To reduce the risk of cross-contamination, all PCR reactions were performed in a completely separate laboratory from the cell culture and DNA extraction. Additional precautions included preparation of a master mix from small working aliquots of commercial stock solutions and molecular grade water, the use of aerosol barrier pipet tips (Molecular Bioproducts, San Diego, CA), and autoclaved master mix and PCR tubes. PCR reactions were set up by first aliquoting the master mix followed by addition of the test DNA samples, the positive control dilutions, Jurkat cell DNA, and finally a reagent control (water used to prepare the master mix).
Sequencing of PCR products.
After amplification, PCR products were ligated into a cloning vector (TA cloning; Invitrogen, Carlsbad, CA) and sequenced either by dideoxynucleotide chain termination using Sequinase II (Amersham, Arlington Heights, IL) or automated cycle sequencing with Taq polymerase (Perkin Elmer, Foster City, CA). The sequences were aligned using DNAStar (DNAStar Inc, Madison, WI).
Serology for HHV-8 antibody.
Sera were tested for HHV-8 IgG antibody by immunofluorescence using the HHV-8 IgG IFA kit (Advanced Biotechnologies, Columbia, MD), according to the manufacturer’s protocol. Briefly, sera were tested at a 1:20 dilution and incubated with the HHV-8 infected (Epstein-Barr virus [EBV] negative) cell line KS.16 Samples were considered positive if there was either diffuse or restricted apple-green fluorescence in the infected cells. Sera from children (provided by Dr Naomi Luban, Children’s National Medical Center, Washington, DC), patients with breast cancer, and patients with Kaposi’s sarcoma were tested in parallel.
RESULTS
Sensitivity of PCR.
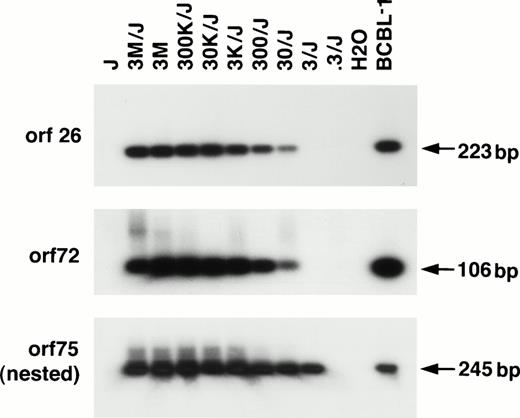
The BCP-1 cell line contains 150 genome copies/cell of HHV-8,17 and we used these cells to calculate the sensitivity of the different PCR reactions. Jurkat cell DNA was spiked with serial dilutions of DNA extracted from BCP-1 cells, and the DNA was analyzed in the different assays. We calculated that the nested PCR reactions (orf 26 region and orf 75 region) could detect less than 3 copies of HHV-8 in 200 ng of cellular DNA, as could the orf 26 (KS330) outside primers alone. The non-nested PCR using the orf 72 primers could detect less than 30 copies of HHV-8 DNA (Fig 1).
Sensitivity curves for three HHV-8 PCR assays using primers from different regions of the genome followed by Southern hybridization with specific internal probes. Jurkat cell DNA (J; 2 μg/reaction) was inoculated with a known copy number of BCP-1 HHV-8 DNA starting at 3 million (M) to 0.3 genome copies along with amplification of BCBL-1, another HHV-8–containing cell line. Jurkat cell DNA as well as water (H2O) were simultaneously amplified. Non-nested amplification in the orf 26 and orf 72 region were able to detect 30 genome copies per reaction. With longer exposure, 3 genome copies could be reproducibly detected without nesting using the orf 26 primers. Nested amplification in the orf 75 region could reproducibly detect 3 genome copies per reaction.
Sensitivity curves for three HHV-8 PCR assays using primers from different regions of the genome followed by Southern hybridization with specific internal probes. Jurkat cell DNA (J; 2 μg/reaction) was inoculated with a known copy number of BCP-1 HHV-8 DNA starting at 3 million (M) to 0.3 genome copies along with amplification of BCBL-1, another HHV-8–containing cell line. Jurkat cell DNA as well as water (H2O) were simultaneously amplified. Non-nested amplification in the orf 26 and orf 72 region were able to detect 30 genome copies per reaction. With longer exposure, 3 genome copies could be reproducibly detected without nesting using the orf 26 primers. Nested amplification in the orf 75 region could reproducibly detect 3 genome copies per reaction.
To test that our primers were not specific for HHV-8 sequence in cell lines only, DNA was extracted from fresh Kaposi sarcoma biopsies (3 individuals, 6 samples), 10-fold serial dilutions of the DNA were prepared, and PCRs were performed. HHV-8 sequence was detected with all three PCR reactions, with detection end points within 1 dilution of each other (data not shown).
Detection of orf 26 (KS330) from stromal cultures.
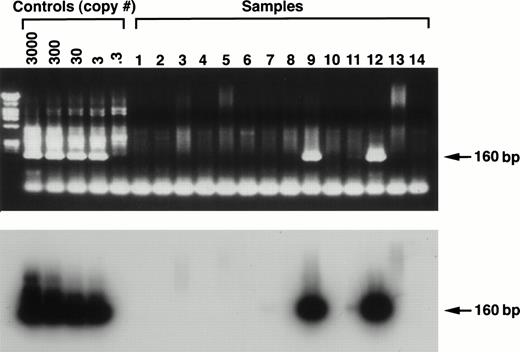
Stromal samples grown under standard long-term culture conditions for 3 weeks and then frozen were examined from 15 MM patients and 20 normal controls and 1 patient with breast cancer. Using the outside primers from the orf 26 (KS330) region, initial experiments with only 30 cycles of amplification were uniformly negative, but after 40 cycles of amplification HHV-8 was initially detected in 9 of 15 myeloma patient samples and 5 of 21 controls. Testing of separate aliquots of DNA at two institutions showed that 2 of 15 myeloma samples were consistently positive, and 13 of 15 samples were positive in at least one assay (Table 1). Nested PCR in the orf 26 region uncovered no additional positive patients (Fig 2). Similarly, after testing separate aliquots of DNA from control patients at both institutions, 2 were positive in both institutions, but a total of 10 of 21 were positive in at least one assay. (Insufficient DNA from control subjects precluded testing the samples a third time.) orf 26 sequence was also detected in a total of 6 of of the rhesus samples, but in none of 5 canine cultures.
Ethidium bromide staining (upper panel) and Southern hybridization (lower panel) of a representative PCR using the nested orf 26 (KS330) primer set. The sensitivity curve (consisting of a known copy number of HHV-8 from BCP-1 inoculated into 2 μg of Jurkat cell DNA) shows the limit of detection to be 3 genome copies per reaction by the presence of bands at the predicted 160-bp position (arrow) by both ethidium staining and Southern hybridization. Numbers 1 through 14 represent patient samples and show scattered positives.
Ethidium bromide staining (upper panel) and Southern hybridization (lower panel) of a representative PCR using the nested orf 26 (KS330) primer set. The sensitivity curve (consisting of a known copy number of HHV-8 from BCP-1 inoculated into 2 μg of Jurkat cell DNA) shows the limit of detection to be 3 genome copies per reaction by the presence of bands at the predicted 160-bp position (arrow) by both ethidium staining and Southern hybridization. Numbers 1 through 14 represent patient samples and show scattered positives.
In an attempt to enhance viral detection, stromal samples from an additional 15 myeloma patients at differing disease stages were grown in parallel under a variety of conditions, including those described in the original report by Rettig et al,10 and the DNA was analyzed (Table 2). orf 26 (KS330) HHV-8 sequence was detected in 5 of 15 patients under at least one culture condition. Samples from no additional patients were positive when the PCR products were amplified in the nested PCR. Stromal samples were also examined from 4 control patients (2 normal, 1 MGUS, and 1 breast cancer patient), and 1 normal donor was positive after nested PCR only.
The 160-bp product was sequenced from 2 patients, 2 control subjects, and the two positive control cell lines BCP-1 and BCBL-1. Although there was greater than 96% similarity, none of the products from patients or controls was identical to the published or cell line sequences and varied between 1 and 4 bp from the published HHV-8 sequence.
Detection of HHV-8 (orf 75, orf 72) from stromal cultures.
Every sample was also tested by the nested PCR from the orf 75 (KS651Bam) region. However, despite equal sensitivity and repetitive testing, no sample was ever positive (MM = 30, controls = 25). In addition, samples from 26 patients and 2 controls were amplified using primers from the orf 72 region. Again, no sample was positive.
Serology for HHV-8.
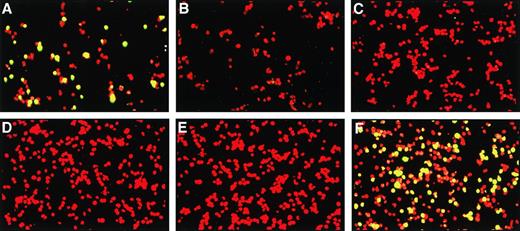
Serum samples were examined from 33 patients with MM (including 21 patients also analyzed by PCR [Table 1] and 12 additional patients), 6 patients with KS, 6 nontransfused children, and 10 patients with breast cancer. All samples were coded and scored for fluorescence by two independent observers before the code was broken. Serum from all KS patients were positive with apple green fluorescence in both the nucleus and cytoplasm (Fig 3). In contrast, none of the 6 children, 10 breast cancer patients, or 33 myeloma patients showed positive fluorescence.
Representative serum indirect immunofluorescence assay. Sera were tested in a 1:20 dilution by incubation on an HHV-8–containing cell line (KS-1); approximately 30% of the KS-1 cells express HHV-8 proteins, whereas the remainder act as internal controls. (A) represents a positive control, (B) a negative control, (C and D) two MM patients, (E) a breast cancer patient, and (F) an AIDS patient with KS. Positivity, green fluorescence restricted to approximately 30% of the cells is seen in the positive control and the AIDS patient only.
Representative serum indirect immunofluorescence assay. Sera were tested in a 1:20 dilution by incubation on an HHV-8–containing cell line (KS-1); approximately 30% of the KS-1 cells express HHV-8 proteins, whereas the remainder act as internal controls. (A) represents a positive control, (B) a negative control, (C and D) two MM patients, (E) a breast cancer patient, and (F) an AIDS patient with KS. Positivity, green fluorescence restricted to approximately 30% of the cells is seen in the positive control and the AIDS patient only.
DISCUSSION
HHV-8 was first identified in acquired immunodeficiency syndrome (AIDS)-associated KS lesions by Chang et al1 using representational difference analysis. Since then, a great deal of work has implicated the virus as critical in the development of KS; however, investigators are not in complete agreement that the virus is causative, and controversy exists as to whether HHV-8 is ubiquitous or disease restricted.4 Seroprevalence studies support the link between HHV-8 and KS and suggest that this human herpesvirus is not ubiquitous in healthy adults.18 Antibodies to HHV-8 were infrequently found (1% to 2%) in the US or UK blood donor population, hemophiliacs, and intravenous drug users and were more common in human immunodeficiency virus (HIV)-infected homosexual men without KS and in persons living in areas of Africa endemic for classic KS. These results support the view that HHV-8 infection is largely confined to those persons at risk for the development of KS.
Based on the presence of a biologically active human IL-6 homolog gene encoded by HHV-8 and the previously recognized production of IL-6 from the adherent cell fraction of bone marrow cultures from myeloma patients, Rettig et al10 evaluated bone marrow stromal cells grown from marrow mononuclear cells and detected HHV-8 genomic sequences by PCR using primers amplifying in the orf 26 region in 15 of 15 MM patients, but not in normal control stromal cells. Reverse transcription-PCR (RT-PCR) for viral IL-6 message was positive in 3 of 3 myeloma patient-derived stromal cultures assayed but in none of 2 controls. In situ hybridization for HHV-8–specific sequences was also positive only in the cultured cells from myeloma patients. Although in situ hybridization showed signal in 100% of the cultured cells, PCR amplification for viral DNA (orf 26 region) required 45 cycles, and no attempt was made to quantitate genome number. Transmission electron microscopy of the cultured cells showed ultrastructural characteristics of macrophages, yet viral structures were not shown. Evaluation of core bone marrow biopsy samples by in situ hybridization suggested that HHV-8 was present in the stromal cell compartment.11 After the original report, preliminary results in the form of “technical comments” appeared: a group in France reported detection of orf 26 (KS330) HHV-8 sequences in 18 of 20 MM bone marrow biopsies after 60 cycles of PCR, but in 0 of 20 control biopsies19; however, two other groups could not detect HHV-8 sequences in any of 40 or 5 bone marrow samples of MM patients, respectively.20,21 Additionally, clinical grade functionally characterized dendritic cells derived from the blood of patients with MM have been shown recently by two groups not to contain HHV-8 sequences by sensitive PCR assays.22 23
However, our data using very sensitive nested PCR methodology do support that sequence from the orf 26 region of HHV-8 is detectable in many myeloma patient-derived stromal cell cultures, but at least 50% of both human and nonhuman primate control cultures also contained orf 26 sequence. The inability to detect viral sequence in any sample after amplification with 30 cycles of PCR despite amplification of the control DNA dilutions suggests that the DNA exists at a very low level in the cultured cells. Additionally, the different results on repeat assays suggest that the orf 26 DNA exists at the limit of detection by PCR that in our assay correlates with 3 genome copies per 200,000 cells. To rule out an inhibitor of the PCR reaction present in our samples, 2 persistently negative samples from MM patients were amplified after the addition of 3 HHV-8 genome copies, yielding positive results.
Unexpectedly, when other areas of the viral genome (orf 72, orf 75) were amplified from the same DNA samples with an equally sensitive PCR assay (limit of detection <3 genome copies per reaction), no positives were detected in either the patient or control samples, even with repetitive testing, despite the ability to detect HHV-8 sequences in KS tissue. Furthermore, screening for serum antibodies to HHV-8 in our myeloma patient cohort using an indirect immunofluorescence technique did not suggest a previous exposure to the virus. Whereas MM patients (n = 33), breast cancer patients (n = 10), and normal children (n = 6) were all negative by serology, HHV-8 antibodies were demonstrated in all of 6 AIDS patients with KS. These results are in agreement with those of several other groups have investigated the seroprevalence of HHV-8 in MM patients and controls by assaying for serum antibodies to HHV-8.20,21,24-26 All groups were unable to confirm an association between HHV-8 and MM. The absence of antibodies to HHV-8 could not be explained by a generalized defective immunity to herpesviruses as antibody testing for other herpesviruses by two groups yielded their predicted freqencies.24,25Additionally, epidemiologically, there is no increased incidence of myeloma in regions of the world with a high seroprevalence for HHV-8.26,27 Whereas strong epidemiologic, serologic, virologic, and molecular (including sequence detection from various regions of the genome) evidence supports the role of HHV-8 in KS, multicentric Castleman’s disease, and primary effusion lymphoma, the finding of orf 26 sequence in both our patient cohort and controls is not dissimilar to the published literature describing detection of sequence in a variety of diseased and nondiseased tissues. orf 26 HHV-8 sequences have been reported to be present in a number of skin conditions, including Bowen’s disease, squamous cell carcinoma, actinic keratosis, leukoplakia, as well as pemphigus vulgaris and pemphigus foliaceous.28,29 Although not restricted, HHV-8 sequences have also been detected by PCR in cerebral tissue from patients with multiple sclerosis as well as cerebral tissue from stillborn control samples.30 Additionally, HHV-8 has been implicated in 3 patients with unexplained encephalitis who were found to have viral sequence present by PCR in brain biopsy specimens.31
The background prevalence of HHV-8 also remains unresolved. A group in Italy assayed urogenital tract specimens by PCR (orf 26) and reported a very high prevalence, with 30 of 33 or 91% of ejaculates from healthy men undergoing varicocele correction positive for viral sequences.14 In Sicily, where KS is endemic, only 13% of semen from HIV-negative heterosexuals and 10% of AIDS patients without KS could be found by PCR to contain HHV-8 sequences.32Similarly, a group in the United States examined semen specimens by PCR for HHV-8 in 99 HIV-infected men (95% of whom reported previous homosexual contact), with only 1 of 99 positive. HHV-8 seropositivity in this HIV-infected population was similar to other published reports at 27%, and the only PCR-positive semen specimen was obtained from an HHV-8 seropositive patient with active KS.33 Another group was also unable to confirm the presence of HHV-8 sequences in urogenital specimens obtained from both the United States and Italy using a sensitive nested PCR capable of detecting less than 3 genome copies per reaction.34
Even in healthy children and adults, a group in Japan reported a very high prevalence of HHV-8 sequences, again orf 26, when PCR was performed on peripheral blood mononuclear cells (35/56 [64%] and 12/15 [80%], respectively), although serologic studies were not performed.35 Another group of investigators using a different primer set evaluated peripheral blood mononuclear cells from children and adults with AIDS but without KS.36 None of 51 children weas positive, and 9 of 33 (27%) adults had detectable HHV-8 sequences in their peripheral blood. Serology for HHV-8 was also performed in this study and correlated with the PCR results.
These reports have left the issue of disease restriction versus ubiquitous distribution for HHV-8 largely unresolved. In general, when highly sensitive assays for the orf 26 region have been used in both diseased and nondiseased tissues, a high percentage of positive samples have been reported; however, less sensitive PCR assays amplifying the orf 26 region or sensitive PCR assays of other regions of the viral genome have largely negative. Our results are in keeping with these findings; the majority of both myeloma and control stromal cultures contained detectable orf 26 sequences, albeit at low levels; HHV-8 sequence from other regions of the genome could not be amplified; and antibodies to HHV-8 could not be detected in the serum of MM patients.
One possible explanation for these discrepancies is PCR contamination with the orf 26 product. However, contamination is unlikely in our study given the repeated negativity of the same samples on multiple occasions in both the nonnested and nested assay in the orf 26 region, similar results in assays run at two separate institutions, and the fact that the number of positives remained constant throughout the study period. Sequencing of several of the products yielded orf 26 HHV-8 sequence with greater than 96% similarity, yet none of the products was identical to sequences derived from either of the two positive control cell lines (nevertheless, the degree of sequence variability is not sufficient to conclusively rule out contaminating exogenous DNA given the error rate inherent to both the PCR and sequencing procedures). Additionally, no samples were ever positive using primers in the orf 72 or orf 75 region despite repetitive testing with simultaneous amplification of the control BCP-1 DNA dilution in every reaction, giving ample opportunity to contaminate the samples for these regions in the same way.
Another possible explanation for these discrepancies is the presence of a related human herpesvirus that retains sequence homology in the orf 26 region but not in the other areas assayed. Precedence for the existence of a related virus exists in the nonhuman primate, because limited DNA sequence for two herpesviruses closely related to HHV-8 have been reported in Macaca mulatta (rhesus)37,38 and a third in Macaca nemestrina.38 A related rhesus virus almost certainly explains the positive results seen in our rhesus derived stromal cultures: in the immunofluorescence assay, 70% of our rhesus sera were positive for HHV-8 antibodies (data not shown). Although we were unable to confirm the recently proposed association between MM and HHV-8 by either PCR for the viral genome or by seroprevalence in our patient cohort, the presence of two HHV-8–like viruses within the same nonhuman primate species, along with our data and those of others, raises the possibility of a common related (KS330-containing) virus in the human population more easily detected in the immunosuppressed host.
Address reprint requests to Kevin E. Brown, MD, Bldg 10/Room 7C218, National Institutes of Health, 9000 Rockville Pike, Bethesda, MD 20892-1652; e-mail: brownk@gwgate.nhlbi.nih.gov.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal