Abstract
Cytogenetics is considered one of the most valuable prognostic determinants in acute myeloid leukemia (AML). However, many studies on which this assertion is based were limited by relatively small sample sizes or varying treatment approach, leading to conflicting data regarding the prognostic implications of specific cytogenetic abnormalities. The Medical Research Council (MRC) AML 10 trial, which included children and adults up to 55 years of age, not only affords the opportunity to determine the independent prognostic significance of pretreatment cytogenetics in the context of large patient groups receiving comparable therapy, but also to address their impact on the outcome of subsequent transplantation procedures performed in first complete remission (CR). On the basis of response to induction treatment, relapse risk, and overall survival, three prognostic groups could be defined by cytogenetic abnormalities detected at presentation in comparison with the outcome of patients with normal karyotype. AML associated with t(8;21), t(15;17) or inv(16) predicted a relatively favorable outcome. Whereas in patients lacking these favorable changes, the presence of a complex karyotype, −5, del(5q), −7, or abnormalities of 3q defined a group with relatively poor prognosis. The remaining group of patients including those with 11q23 abnormalities, +8, +21, +22, del(9q), del(7q) or other miscellaneous structural or numerical defects not encompassed by the favorable or adverse risk groups were found to have an intermediate prognosis. The presence of additional cytogenetic abnormalities did not modify the outcome of patients with favorable cytogenetics. Subgroup analysis demonstrated that the three cytogenetically defined prognostic groups retained their predictive value in the context of secondary as well as de novo AML, within the pediatric age group and furthermore were found to be a key determinant of outcome from autologous or allogeneic bone marrow transplantation (BMT) in first CR. This study highlights the importance of diagnostic cytogenetics as an independent prognostic factor in AML, providing the framework for a stratified treatment approach of this disease, which has been adopted in the current MRC AML 12 trial.
PRESENTATION CYTOGENETICS is widely recognized as one of the most important prognostic determinants in acute myeloid leukemia (AML). However, many studies on which this assertion is based were limited by consideration of relatively small numbers of patients or were confounded by amalgamation of groups receiving widely differing treatment protocols.1-9 This has in a number of cases led to conflicting data regarding the prognostic implications of specific cytogenetic abnormalities. The Medical Research Council (MRC) AML 10 trial for children and younger adults with AML, which was designed to evaluate the role of bone marrow transplantation (BMT) in first complete remission (CR), affords the opportunity to determine the independent prognostic significance of cytogenetics at diagnosis in the context of a large group of patients, who apart from the transplant randomization, received equivalent induction and consolidation therapy. Furthermore, this study also enables one to determine the relative impact of pretreatment cytogenetics on the outcome of subsequent transplant procedures. Overall 1,938 children and adults with de novo or secondary AML were recruited to the trial; the present study considers the prognostic implications of pretreatment cytogenetics in 1,612 patients in whom karyotype analysis was successful.
MATERIALS AND METHODS
Patients.
The MRC AML 10 trial began in May 1988 and closed in April 1995, having accrued 1,966 patients, including 364 children (<15 years) and 1,602 adults, mostly up to 55 years of age. A total of 1,797 were registered as having de novo AML (337 children, 1,460 adults), 141 cases of secondary AML were entered (22 children, 119 adults), while the remaining 28 trial patients were excluded from further analysis, as they were subsequently found not to have AML. Cases of AML were classified as secondary on the basis of a history of previous exposure to chemotherapy or radiotherapy or of an antecedent hematologic condition including myelodysplasia and myeloproliferative disorders.
Therapy.
The trial, which sought to determine the relative efficacy of two different induction protocols and also to establish whether there is a role for allogeneic or autologous BMT in the treatment of patients in first CR, has been fully described previously.10 Briefly, patients were randomized to receive induction therapy with two courses of DAT (daunorubicin, Ara-C, 6-thioguanine: course 1, DAT 3 + 10; course 2, DAT 3 + 8) or ADE (Ara-C, daunorubicin, etoposide: course 1, ADE 10 + 3 + 5; course 2, ADE 8 + 3 + 5). From January 1993, those with a clinical diagnosis of acute promyelocytic leukemia (APL) were eligible for the MRC ATRA trial whereby 75 patients were randomized to receive either short or extended courses of all-trans retinoic acid (ATRA), in addition to the AML 10 chemotherapy protocol, as previously described.11 A further six APL patients also received ATRA, but were not entered into the MRC ATRA trial. The third and fourth courses of consolidation chemotherapy of AML 10 comprised MACE (m-amsacrine, Ara-C, etoposide) and MIDAC (mitozantrone, Ara-C), respectively, with bone marrow harvest being scheduled between the third and fourth courses, provided morphologic CR at this stage was confirmed. Patients achieving CR were subsequently scheduled to proceed to allogeneic BMT if a matched sibling donor was available; patients lacking a suitable donor could be randomized to receive an autograft or no further therapy. No significant difference was found in either CR rate, relapse risk, or overall survival between patients randomized to DAT or ADE induction.10 Overall 1,365 of 1,612 patients (85%) achieved CR, of which 428 (31%) received BMT in first CR (211 sibling allo BMT, 199 autologous BMT, six matched unrelated donor, five autologous peripheral blood stem cells [PBSC], three allogeneic PBSC, two mismatched and two syngeneic). In the remaining 937 patients (69%), consolidation was with chemotherapy alone. There were no significant differences in the treatment received according to the cytogenetic abnormality detected at diagnosis.
Cytogenetics.
The majority of cytogenetic analyses were performed at 41 local laboratories, subject to monitoring by a central quality control scheme (UK NEQAS, National External Quality Assessment Schemes). Where no local cytogenetics service was available, examinations were undertaken at the central MRC AML trials cytogenetics laboratory at University College Hospital, London (n = 180). Bone marrow for cytogenetic analysis was cultured according to standard methods; 20 or more cells were fully analyzed to exclude clonal abnormalities, which were defined in accordance with International System for Human Cytogenetic Nomenclature (ISCN) guidelines.12 For patients with a detectable clonal abnormality, at least 10 metaphases were examined to exclude secondary changes in accordance with NEQAS guidelines for clinical cytogenetics. Complex karyotype was defined by the presence of a clone with at least five unrelated cytogenetic abnormalities, as we found that the outcome of these patients was worse than that of patients with fewer clonal abnormalities. A successful analysis was available for 1,612 patients, representing 83% of cases of AML in the trial. A diagnostic result was not available in 326 cases either because cytogenetic studies were not performed (n = 94) or failed (n = 136), while the reason was unknown in the remainder. Failure rates were greater among samples analyzed centrally in comparison with examinations performed at local laboratories (27%v 5%), most likely reflecting sample deterioration during transit. We report here an analysis of the more frequently observed abnormalities, ie, those found in 20 or more patients. Initially, we sought to determine the prognostic impact of each specific abnormality taken in isolation; hence in these analyses, patients may be counted more than once due to the presence of multiple cytogenetic changes (Figs 2, 3A and B, and Tables 1 and 2). These analyses led to the development of a hierarchical classification based on recognized commonly recurring primary abnormalities (Tables 4-6). In these analyses, patients are defined by the presence of primary abnormalities and hence are counted only once.
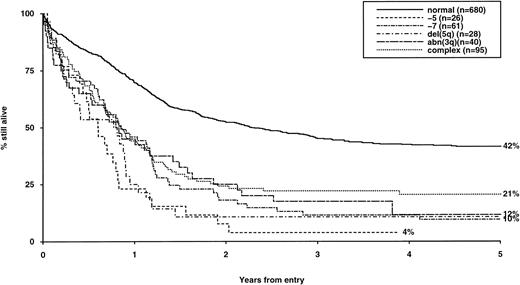
Overall survival of patients with adverse cytogenetic abnormalities, irrespective of the presence of additional abnormalities. The group with normal karyotype is included for comparison.
Overall survival of patients with adverse cytogenetic abnormalities, irrespective of the presence of additional abnormalities. The group with normal karyotype is included for comparison.
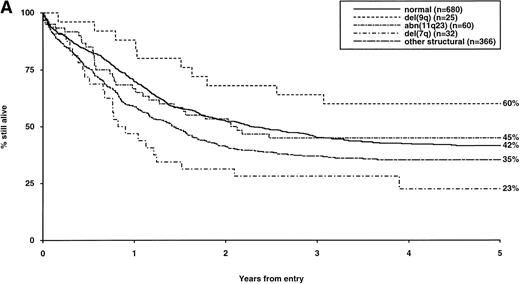
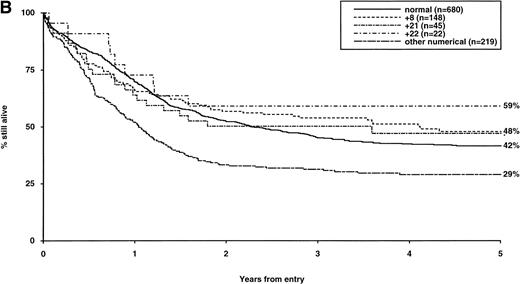
Overall survival of patients with intermediate structural (A) or numerical (B) cytogenetic abnormalities, irrespective of the presence of additional abnormalities. The group with normal karyotype is included for comparison.
Overall survival of patients with intermediate structural (A) or numerical (B) cytogenetic abnormalities, irrespective of the presence of additional abnormalities. The group with normal karyotype is included for comparison.
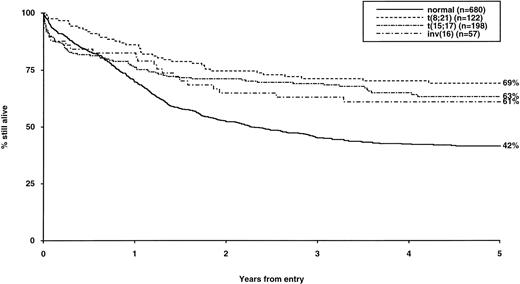
Overall survival of patients with favorable cytogenetic abnormalities, irrespective of the presence of additional abnormalities. The group with normal karyotype is included for comparison.
Overall survival of patients with favorable cytogenetic abnormalities, irrespective of the presence of additional abnormalities. The group with normal karyotype is included for comparison.
Cytogenetic Risk Groups
| Risk Group . | Abnormality . | Comment . |
|---|---|---|
| Favorable | t(8;21) t(15;17) inv(16) | Whether alone or in conjunction with other abnormalities. |
| Intermediate | Normal +8 +21 +22 del(7q) del(9q) Abnormal 11q23 All other structural/numerical abnormalities | ie, Cytogenetic abnormalities not classified as favorable or adverse. Lack of additional favorable or adverse cytogenetic changes. |
| Adverse | −5 −7 del(5q) Abnormal 3qComplex | Whether alone or in conjunction with intermediate-risk or other adverse-risk abnormalities. |
| Risk Group . | Abnormality . | Comment . |
|---|---|---|
| Favorable | t(8;21) t(15;17) inv(16) | Whether alone or in conjunction with other abnormalities. |
| Intermediate | Normal +8 +21 +22 del(7q) del(9q) Abnormal 11q23 All other structural/numerical abnormalities | ie, Cytogenetic abnormalities not classified as favorable or adverse. Lack of additional favorable or adverse cytogenetic changes. |
| Adverse | −5 −7 del(5q) Abnormal 3qComplex | Whether alone or in conjunction with intermediate-risk or other adverse-risk abnormalities. |
Hierarchical prognostic classification, derived taking into consideration the influence of additional cytogenetic abnormalities on outcome, and used for directing treatment approach in the current MRC AML 12 trial.
Definitions of endpoints.
A normocellular bone marrow aspirate containing less than 5% blast cells and showing evidence of normal maturation of other marrow elements was the criterion for the achievement of CR. The persistence of myelodysplastic features did not exclude the diagnosis of CR. Full recovery of normal peripheral blood counts was not required to define CR, which might contribute to the relatively favorable CR rates observed in this study. Remission failures were classified by the referring clinician as due either to induction death (ID), ie, related to treatment and/or hypoplasia, or as resistant disease (RD), ie, related to the failure of therapy to eliminate the disease (including partial remissions with 5% to 20% blasts). Where the clinician's evaluation was not available, deaths within 30 days of entry were classified as ID and deaths at more than 30 days as RD. The following definitions are also used: overall survival (OS) is the time from entry to death: for remitters, the relapse risk (RR) is the cumulative probability of relapse, ignoring (ie, censoring at) death in CR.
Statistical methods.
The Mantel-Haenszel test for trend and Wilcoxon two sample test were used to test for associations with age, type of AML, and white blood cell count (WBC) at presentation. Remission rates and reasons for failure to achieve CR were compared using the Fisher exact test. Kaplan-Meier life-tables were constructed for survival data and were compared by means of the log-rank test, with surviving patients being censored on August 1, 1997, when follow-up was up-to-date for over 98% of patients (the small number of patients lost to follow-up are censored at the date they were last known to be alive). Median follow-up was 61 months (1 to 111 months). All P values are two-tailed; because of the large number of significance tests performed and the associated increased probability of obtaining conventionally significant (P < .05) results by chance, only Pvalues < .01 are quoted.
RESULTS
Incidence of specific cytogenetic abnormalities in AML and their prognostic significance.
The frequency of the most common cytogenetic abnormalities detected at diagnosis among 1,612 patients with AML and their associated clinical features are presented in Table 1. On the basis of response to induction therapy, RR, and OS, three prognostic groups were retrospectively distinguished by the presence of specific abnormalities, initially irrespective of additional cytogenetic changes, (Table 2). Patients with t(8;21), t(15;17), and inv(16) comprised a group with relatively favorable prognosis, characterized by low rates of primary drug resistance and superior OS associated with a reduced RR (Table 2, Fig 1). The outcome of the subgroup of APL cases with the t(15;17) who were not treated with ATRA (n = 117) was also found to be significantly better (CR 88%, RR at 5 years 41%;P < .01, OS at 5 years 61%; P < .001) than patients with normal cytogenetics, thereby justifying inclusion of t(15;17) in the favorable cytogenetic risk category. Virtually all patients with t(8;21) achieved CR (98%); CR rates for those with t(15;17) and inv(16) did not differ from patients with normal karyotype due to ID, reflecting the associated hemorrhagic diathesis and propensity to high presentation WBC, respectively (Table 1). In contrast, the presence of complex cytogenetic changes, −5/del(5q), 3q abnormalities, or −7 was found to predict a significantly poorer outcome than patients with normal cytogenetics. Patients within this adverse cytogenetics group were significantly less likely to achieve CR associated with higher rates of primary RD and/or ID; furthermore they had poorer OS reflecting increased risk of death on induction and/or relapse (Table 2, Fig 2). The remaining patients were placed within the intermediate risk group (Table 2, Fig 3A and B). For the purposes of this analysis, cases with del(9q) were included in this group, despite their relatively favorable outcome due to the frequent association with t(8;21) (Table 3). Similarly, cases with del(7q) or other structural and numerical changes were also included in the intermediate group, although they exhibited a poorer outcome than patients with normal cytogenetics due to an association with abnormalities within the adverse risk category (Table 3). However, in the absence of associated favorable or adverse cytogenetic features, the outcome of patients with del(9q), del(7q), or those with other structural or numerical abnormalities did not differ significantly from patients with normal cytogenetics (Table 3), thereby justifying their present inclusion in the intermediate risk category. Stratification by age, WBC at presentation, and type of leukemia (de novo/secondary) confirmed diagnostic cytogenetics as an independent prognostic factor in AML (Table 2). Indeed, subgroup analysis confirmed t(8;21) as a favorable prognostic factor in pediatric as well as adult AML. Of 41 children with t(8;21), 98% achieved CR associated with an OS of 83% at 3 years (cf OS of 59% for children with normal cytogenetics, P < .01). Differences in outcome between cytogenetic risk groups could not be accounted for by significant variation in deaths in remission (Table 2) or postremission therapy.
Frequency and Percentage of Cytogenetic Abnormalities
| Abnormality . | All Patients No. (%) . | Age Group (yr) . | Median Age (yr) . | Median Initial WBC-150 . | Type of AML . | |||
|---|---|---|---|---|---|---|---|---|
| 0-14 No. (%) . | 15-34 No. (%) . | 35+ No. (%) . | De Novo No. (%) . | Secondary No. (%) . | ||||
| Overall | 1,612 | 340 | 461 | 811 | 35.0 | 12.6 | 1,493 | 119 |
| No abnormality | 680 (42) | 91 (27) | 177 (38) | 412 (51) | 40.0 | 16.4 | 639 (43) | 41 (34) |
| t(15;17) | 198 (12) | 31 (9) | 87 (19) | 80 (10)-151 | 29.0-151 | 3.3-151 | 192 (13) | 6 (5) |
| +8 | 148 (9) | 46 (14) | 47 (10) | 55 (7)-151 | 27.5-151 | 7.0-151 | 137 (9) | 11 (9) |
| t(8;21) | 122 (8) | 41 (12) | 28 (6) | 53 (7)-151 | 29.5-151 | 10.7-152 | 118 (8) | 4 (4) |
| Complex | 95 (6) | 19 (6) | 29 (6) | 47 (6) | 34.0 | 7.2-151 | 84 (6) | 11 (9) |
| −7 | 61 (4) | 12 (4) | 16 (3) | 33 (4) | 37.0 | 7.0 | 48 (3) | 13 (11)-151 |
| 11q23 | 60 (4) | 26 (8) | 21 (5) | 13 (2)-151 | 17.0-151 | 17.6 | 59 (4) | 1 (1) |
| inv(16) | 57 (4) | 16 (5) | 26 (6) | 15 (2)-151 | 26.0-151 | 44.2-151 | 53 (4) | 4 (3) |
| +21 | 45 (3) | 20 (6) | 13 (3) | 12 (1)-151 | 20.0-151 | 9.7 | 39 (3) | 6 (5) |
| abn(3q) | 40 (3) | 6 (2) | 15 (3) | 19 (2) | 33.5 | 14.5 | 34 (2) | 6 (5) |
| del(7q) | 32 (2) | 7 (2) | 8 (2) | 17 (2) | 37.5 | 7.4 | 28 (2) | 4 (3) |
| del(5q) | 28 (2) | 4 (1) | 5 (1) | 19 (2) | 46.0 | 10.5 | 24 (2) | 4 (3) |
| −5 | 26 (2) | 2 (1) | 8 (2) | 16 (2) | 40.0 | 5.7 | 24 (2) | 2 (2) |
| del(9q) | 25 (2) | 12 (4) | 5 (1) | 8 (1)-151 | 19.0-152 | 13.0 | 23 (2) | 2 (2) |
| +22 | 22 (1) | 4 (1) | 9 (2) | 9 (1) | 32.0 | 11.8 | 20 (1) | 2 (2) |
| Other numerical | 219 (14) | 61 (18) | 64 (14) | 94 (12)-151 | 29.0-151 | 10.9 | 199 (13) | 20 (17) |
| Other structural | 366 (23) | 108 (32) | 86 (19) | 172 (21)-151 | 32.0-151 | 11.9 | 323 (22) | 43 (36)-151 |
| Abnormality . | All Patients No. (%) . | Age Group (yr) . | Median Age (yr) . | Median Initial WBC-150 . | Type of AML . | |||
|---|---|---|---|---|---|---|---|---|
| 0-14 No. (%) . | 15-34 No. (%) . | 35+ No. (%) . | De Novo No. (%) . | Secondary No. (%) . | ||||
| Overall | 1,612 | 340 | 461 | 811 | 35.0 | 12.6 | 1,493 | 119 |
| No abnormality | 680 (42) | 91 (27) | 177 (38) | 412 (51) | 40.0 | 16.4 | 639 (43) | 41 (34) |
| t(15;17) | 198 (12) | 31 (9) | 87 (19) | 80 (10)-151 | 29.0-151 | 3.3-151 | 192 (13) | 6 (5) |
| +8 | 148 (9) | 46 (14) | 47 (10) | 55 (7)-151 | 27.5-151 | 7.0-151 | 137 (9) | 11 (9) |
| t(8;21) | 122 (8) | 41 (12) | 28 (6) | 53 (7)-151 | 29.5-151 | 10.7-152 | 118 (8) | 4 (4) |
| Complex | 95 (6) | 19 (6) | 29 (6) | 47 (6) | 34.0 | 7.2-151 | 84 (6) | 11 (9) |
| −7 | 61 (4) | 12 (4) | 16 (3) | 33 (4) | 37.0 | 7.0 | 48 (3) | 13 (11)-151 |
| 11q23 | 60 (4) | 26 (8) | 21 (5) | 13 (2)-151 | 17.0-151 | 17.6 | 59 (4) | 1 (1) |
| inv(16) | 57 (4) | 16 (5) | 26 (6) | 15 (2)-151 | 26.0-151 | 44.2-151 | 53 (4) | 4 (3) |
| +21 | 45 (3) | 20 (6) | 13 (3) | 12 (1)-151 | 20.0-151 | 9.7 | 39 (3) | 6 (5) |
| abn(3q) | 40 (3) | 6 (2) | 15 (3) | 19 (2) | 33.5 | 14.5 | 34 (2) | 6 (5) |
| del(7q) | 32 (2) | 7 (2) | 8 (2) | 17 (2) | 37.5 | 7.4 | 28 (2) | 4 (3) |
| del(5q) | 28 (2) | 4 (1) | 5 (1) | 19 (2) | 46.0 | 10.5 | 24 (2) | 4 (3) |
| −5 | 26 (2) | 2 (1) | 8 (2) | 16 (2) | 40.0 | 5.7 | 24 (2) | 2 (2) |
| del(9q) | 25 (2) | 12 (4) | 5 (1) | 8 (1)-151 | 19.0-152 | 13.0 | 23 (2) | 2 (2) |
| +22 | 22 (1) | 4 (1) | 9 (2) | 9 (1) | 32.0 | 11.8 | 20 (1) | 2 (2) |
| Other numerical | 219 (14) | 61 (18) | 64 (14) | 94 (12)-151 | 29.0-151 | 10.9 | 199 (13) | 20 (17) |
| Other structural | 366 (23) | 108 (32) | 86 (19) | 172 (21)-151 | 32.0-151 | 11.9 | 323 (22) | 43 (36)-151 |
All patients with a specific abnormality are considered, irrespective of the presence of additional/secondary cytogenetic changes. The +21 cytogenetic group in this table and all subsequent analyses exclude 14 patients with Down's syndrome; in each of these cases, additional cytogenetic changes accompanying the +21 constitutional abnormality were used for assignment to prognostic risk group. Among 119 patients classified as secondary AML with available cytogenetics, 26 had previously been exposed to chemotherapy and/or radiotherapy, while the remainder had an antecedent hematologic disorder including 69 with documented myelodysplasia. The majority of cases with 11q23 abnormalities had balanced translocations, breakpoints on rearranged partner chromosomes were as follows: 1p32 (n = 2), 3q21 (n = 1), 4p16 (n = 1), 6q27 (n = 4), 8? (n = 1), 9p22 (n = 18), 10p12 (n = 12), 10q22 (n = 3), 17q21 (n = 5), 19p13 (n = 7), 20? (n = 1), Xq22 (n = 1), Xq24 (n = 1). In one case, a three-way rearrangement was observed (t(9;11;15)(p13;q23;q22)), while in the two remaining cases, the nature of the rearrangement was not characterized. Percents are column percents.
Ten patients did not have initial WBC recorded.
P < .001: P values are for Mantel-Haenszel test for trend in age (grouped), Wilcoxon 2-sample test in age (continuous), and for Fisher exact test in type of AML and initial WBC comparing each abnormality with normal karyotype (ie, no abnormality). Percentages may not add to 100 because of rounding.
P < .01.
CR Rates, Survival, and Relapse Risk by Individual Abnormalities
| Abnormality . | Total No. . | CR and Reason for Failure . | Deaths in Remission % (SE) . | Relapse Risk at 5 yr % (SE) . | Overall Survival at 5 yr % (SE) . | ||
|---|---|---|---|---|---|---|---|
| CR Rate % . | Induction Deaths % . | Resistant Disease % . | |||||
| Overall | 1,612 | 85 | 8 | 8 | 14 (1.1) | 49 (1.5) | 44 (1.3) |
| Favorable | |||||||
| t(15;17) | 198 | 87 | 11 | 2 | 13 (3.1) | 37 (4.1)* | 63 (3.6)* |
| t(8;21) | 122 | 98* | 2 | 0† | 15 (3.4) | 29 (4.5)* | 69 (4.2)* |
| inv(16) | 57 | 88 | 12 | 0 | 9 (4.3) | 42 (7.4) | 61 (6.5)† |
| Intermediate | |||||||
| No abnormality | 680 | 88 | 6 | 6 | 15 (1.7) | 53 (2.3) | 42 (1.9) |
| +8 | 148 | 84 | 7 | 8 | 12 (2.9) | 44 (4.8) | 48 (4.3) |
| 11q23 | 60 | 87 | 7 | 7 | 9 (4.6) | 47 (7.2) | 45 (6.4) |
| +21 | 45 | 80 | 7 | 13 | 11 (6.2) | 50 (9.0) | 47 (7.7) |
| del (7q) | 32 | 75 | 6 | 19 | 19 (10.9) | 59 (10.5) | 23 (8.1) |
| del (9q) | 25 | 100 | 0 | 0 | 9 (5.8) | 39 (10.1) | 60 (9.8) |
| +22 | 22 | 91 | 5 | 5 | 13 (8.6) | 51 (12.4) | 59 (10.5) |
| Other numerical | 219 | 76* | 11 | 14* | 19 (3.4) | 60 (4.2) | 29 (3.1)* |
| Other structural | 366 | 76* | 9 | 14* | 14 (2.5) | 51 (3.2) | 35 (2.5)† |
| Adverse | |||||||
| Complex | 95 | 67* | 13 | 20* | 12 (4.9) | 68 (6.2)* | 21 (4.2)* |
| −7 | 61 | 54* | 16† | 30* | 8 (8.0) | 80 (7.1)* | 10 (3.8)* |
| abn(3q) | 40 | 63* | 23† | 15 | 20 (11.9) | 85 (7.8)* | 12 (6.2)* |
| del(5q) | 28 | 57* | 14 | 29* | 14 (9.1) | 85 (9.5)* | 11 (5.8)* |
| −5 | 26 | 42* | 12 | 46* | 12 (11.7) | 90 (9.8)† | 4 (3.8)* |
| Abnormality . | Total No. . | CR and Reason for Failure . | Deaths in Remission % (SE) . | Relapse Risk at 5 yr % (SE) . | Overall Survival at 5 yr % (SE) . | ||
|---|---|---|---|---|---|---|---|
| CR Rate % . | Induction Deaths % . | Resistant Disease % . | |||||
| Overall | 1,612 | 85 | 8 | 8 | 14 (1.1) | 49 (1.5) | 44 (1.3) |
| Favorable | |||||||
| t(15;17) | 198 | 87 | 11 | 2 | 13 (3.1) | 37 (4.1)* | 63 (3.6)* |
| t(8;21) | 122 | 98* | 2 | 0† | 15 (3.4) | 29 (4.5)* | 69 (4.2)* |
| inv(16) | 57 | 88 | 12 | 0 | 9 (4.3) | 42 (7.4) | 61 (6.5)† |
| Intermediate | |||||||
| No abnormality | 680 | 88 | 6 | 6 | 15 (1.7) | 53 (2.3) | 42 (1.9) |
| +8 | 148 | 84 | 7 | 8 | 12 (2.9) | 44 (4.8) | 48 (4.3) |
| 11q23 | 60 | 87 | 7 | 7 | 9 (4.6) | 47 (7.2) | 45 (6.4) |
| +21 | 45 | 80 | 7 | 13 | 11 (6.2) | 50 (9.0) | 47 (7.7) |
| del (7q) | 32 | 75 | 6 | 19 | 19 (10.9) | 59 (10.5) | 23 (8.1) |
| del (9q) | 25 | 100 | 0 | 0 | 9 (5.8) | 39 (10.1) | 60 (9.8) |
| +22 | 22 | 91 | 5 | 5 | 13 (8.6) | 51 (12.4) | 59 (10.5) |
| Other numerical | 219 | 76* | 11 | 14* | 19 (3.4) | 60 (4.2) | 29 (3.1)* |
| Other structural | 366 | 76* | 9 | 14* | 14 (2.5) | 51 (3.2) | 35 (2.5)† |
| Adverse | |||||||
| Complex | 95 | 67* | 13 | 20* | 12 (4.9) | 68 (6.2)* | 21 (4.2)* |
| −7 | 61 | 54* | 16† | 30* | 8 (8.0) | 80 (7.1)* | 10 (3.8)* |
| abn(3q) | 40 | 63* | 23† | 15 | 20 (11.9) | 85 (7.8)* | 12 (6.2)* |
| del(5q) | 28 | 57* | 14 | 29* | 14 (9.1) | 85 (9.5)* | 11 (5.8)* |
| −5 | 26 | 42* | 12 | 46* | 12 (11.7) | 90 (9.8)† | 4 (3.8)* |
The prognostic significance of specific cytogenetic abnormalities is considered, irrespective of the presence of additional/secondary cytogenetic changes. On the basis of response to therapy, relapse risk, and overall survival, three prognostic groups were defined.
P < .001: P values are for Fisher exact test (CR and reasons for failure) or log rank test (deaths in remission, relapse risk, and overall survival) comparing each abnormality with normal karyotype (ie, no abnormality). All P values remain significant when stratified by age, type of leukemia (de novo or secondary) and WBC at presentation, except for inv(16) where the P value for survival becomes P = .1 when stratified by age. Percentages may not add to 100 because of rounding.
P < .01.
Influence of Additional Cytogenetic Abnormalities on Outcome in AML
| Abnormality . | No. of Patients . | CR Rate % . | Relapse Risk at 3 yr % (SE) . | Overall Survival at 3 yr % (SE) . |
|---|---|---|---|---|
| Normal | 680 | 88 | 49 (2.2) | 45 (1.9) |
| t(15;17) | 198 | |||
| Alone | 137 | 88 | 30 (4.5) | 69 (3.9) |
| +8 | 31 | 84 | 40 (9.9) | 71 (8.3) |
| Other | 30 | 90 | 32 (10.1) | 66 (8.7) |
| t(8;21) | 122 | |||
| Alone | 49 | 96 | 31 (7.3) | 67 (6.7) |
| −X/−Y | 57 | 98 | 25 (6.1) | 77 (5.6) |
| Other | 16 | 100 | 30 (12.6) | 63 (12.1) |
| inv(16) | 57 | |||
| Alone | 35 | 89 | 43 (9.0) | 63 (8.2) |
| Other | 22 | 86 | 24 (10.5) | 64 (10.3) |
| +8 | 148 | |||
| Alone | 48 | 83 | 42 (8.4) | 42 (7.1) |
| Favorable | 43 | 88 | 32 (7.7) | 77 (6.5)* |
| Adverse | 20 | 70 | 50 (13.45) | 50 (11.2) |
| Other | 43 | 91 | 44 (8.8) | 51 (7.6) |
| 11q23 | 60 | |||
| t(9;11)(p22;q23) | 18 | 89 | 35 (12.6) | 50 (11.8) |
| t(10;11)(p12;q23) | 12 | 83 | 77 (14.3) | 17 (10.8) |
| Other | 30 | 87 | 43 (9.8) | 53 (9.1) |
| del(7q) | 32 | |||
| Adverse | 17 | 65 | 69 (14.7) | 24 (10.3) |
| Alone/other | 15 | 87 | 50 (14.4) | 32 (12.2) |
| del(9q) | 25 | |||
| Alone | 6 | 100 | 37 (21.3) | 50 (20.4) |
| t(8;21) | 9 | 100 | 12 (11.7) | 89 (10.5) |
| Other | 10 | 100 | 60 (15.5) | 50 (15.8) |
| +22 | 22 | |||
| Alone | 1 | 100 | 100 (0.0) | 100 (0.0) |
| Adverse | 4 | 100 | 75 (21.7) | 25 (21.7) |
| Other | 17 | 88 | 29 (12.5) | 65 (11.6) |
| −7 | 61 | |||
| Alone | 15 | 60 | 70 (16.4) | 13 (8.8) |
| Other adverse | 30 | 50 | 87 (8.8) | 7 (4.6) |
| Other | 16 | 56 | 78 (13.9) | 19 (9.8) |
| Other numerical | 219 | |||
| Alone | 35 | 80 | 43 (11.0) | 40 (8.3) |
| Favorable | 25 | 92 | 32 (10.8) | 64 (9.6) |
| Adverse | 102 | 65 | 74 (5.7)* | 18 (3.8) |
| Other | 66 | 85 | 51 (7.2) | 39 (6.0) |
| Other structural | 366 | |||
| Alone | 117 | 76 | 45 (5.5) | 43 (4.6) |
| Favorable | 47 | 89 | 34 (7.7) | 66 (6.9)* |
| Adverse | 115 | 65 | 71 (5.5)† | 19 (3.7)† |
| Other | 95 | 84 | 50 (6.0) | 40 (5.1) |
| abn(3q) | 40 | |||
| Alone | 14 | 79 | 80 (12.6) | 29 (12.1) |
| −7 | 11 | 45 | 100 (0.0) | 0 (0.0) |
| Other | 15 | 60 | 84 (14.2) | 20 (10.3) |
| del(5q) | 28 | |||
| Alone | 5 | 100 | 100 (0.0) | 0 (0.0) |
| Complex | 16 | 50 | 85 (13.8) | 6 (6.1) |
| Other | 7 | 43 | 67 (27.2) | 29 (17.1) |
| −5 | 26 | |||
| Alone | 0 | — | — | — |
| Other adverse | 22 | 41 | 87 (12.1) | 5 (4.4) |
| Other | 4 | 50 | 100 (0.0) | 0 (0.0) |
| Abnormality . | No. of Patients . | CR Rate % . | Relapse Risk at 3 yr % (SE) . | Overall Survival at 3 yr % (SE) . |
|---|---|---|---|---|
| Normal | 680 | 88 | 49 (2.2) | 45 (1.9) |
| t(15;17) | 198 | |||
| Alone | 137 | 88 | 30 (4.5) | 69 (3.9) |
| +8 | 31 | 84 | 40 (9.9) | 71 (8.3) |
| Other | 30 | 90 | 32 (10.1) | 66 (8.7) |
| t(8;21) | 122 | |||
| Alone | 49 | 96 | 31 (7.3) | 67 (6.7) |
| −X/−Y | 57 | 98 | 25 (6.1) | 77 (5.6) |
| Other | 16 | 100 | 30 (12.6) | 63 (12.1) |
| inv(16) | 57 | |||
| Alone | 35 | 89 | 43 (9.0) | 63 (8.2) |
| Other | 22 | 86 | 24 (10.5) | 64 (10.3) |
| +8 | 148 | |||
| Alone | 48 | 83 | 42 (8.4) | 42 (7.1) |
| Favorable | 43 | 88 | 32 (7.7) | 77 (6.5)* |
| Adverse | 20 | 70 | 50 (13.45) | 50 (11.2) |
| Other | 43 | 91 | 44 (8.8) | 51 (7.6) |
| 11q23 | 60 | |||
| t(9;11)(p22;q23) | 18 | 89 | 35 (12.6) | 50 (11.8) |
| t(10;11)(p12;q23) | 12 | 83 | 77 (14.3) | 17 (10.8) |
| Other | 30 | 87 | 43 (9.8) | 53 (9.1) |
| del(7q) | 32 | |||
| Adverse | 17 | 65 | 69 (14.7) | 24 (10.3) |
| Alone/other | 15 | 87 | 50 (14.4) | 32 (12.2) |
| del(9q) | 25 | |||
| Alone | 6 | 100 | 37 (21.3) | 50 (20.4) |
| t(8;21) | 9 | 100 | 12 (11.7) | 89 (10.5) |
| Other | 10 | 100 | 60 (15.5) | 50 (15.8) |
| +22 | 22 | |||
| Alone | 1 | 100 | 100 (0.0) | 100 (0.0) |
| Adverse | 4 | 100 | 75 (21.7) | 25 (21.7) |
| Other | 17 | 88 | 29 (12.5) | 65 (11.6) |
| −7 | 61 | |||
| Alone | 15 | 60 | 70 (16.4) | 13 (8.8) |
| Other adverse | 30 | 50 | 87 (8.8) | 7 (4.6) |
| Other | 16 | 56 | 78 (13.9) | 19 (9.8) |
| Other numerical | 219 | |||
| Alone | 35 | 80 | 43 (11.0) | 40 (8.3) |
| Favorable | 25 | 92 | 32 (10.8) | 64 (9.6) |
| Adverse | 102 | 65 | 74 (5.7)* | 18 (3.8) |
| Other | 66 | 85 | 51 (7.2) | 39 (6.0) |
| Other structural | 366 | |||
| Alone | 117 | 76 | 45 (5.5) | 43 (4.6) |
| Favorable | 47 | 89 | 34 (7.7) | 66 (6.9)* |
| Adverse | 115 | 65 | 71 (5.5)† | 19 (3.7)† |
| Other | 95 | 84 | 50 (6.0) | 40 (5.1) |
| abn(3q) | 40 | |||
| Alone | 14 | 79 | 80 (12.6) | 29 (12.1) |
| −7 | 11 | 45 | 100 (0.0) | 0 (0.0) |
| Other | 15 | 60 | 84 (14.2) | 20 (10.3) |
| del(5q) | 28 | |||
| Alone | 5 | 100 | 100 (0.0) | 0 (0.0) |
| Complex | 16 | 50 | 85 (13.8) | 6 (6.1) |
| Other | 7 | 43 | 67 (27.2) | 29 (17.1) |
| −5 | 26 | |||
| Alone | 0 | — | — | — |
| Other adverse | 22 | 41 | 87 (12.1) | 5 (4.4) |
| Other | 4 | 50 | 100 (0.0) | 0 (0.0) |
OS and RR are quoted at 3 years, as relatively small numbers restrict the reliability of 5-year values.
P < .01.
P < .001: P values are for log rank test, comparing the outcome of groups in the presence or absence of an additional cytogenetic abnormality. Outcome is also shown for patients with the two most common translocations involving 11q23, as well as for the remaining patients with other rearrangements disrupting 11q23, irrespective of the presence of additional cytogenetic changes.
Influence of additional cytogenetic abnormalities on outcome in AML.
Additional cytogenetic abnormalities, irrespective of the nature or complexity, were found not to have a deleterious effect on the outcome of patients with the t(8;21), t(15;17), or inv(16) (Table 3 and Fig 4). Coexistence of abnormalities associated with the favorable and adverse risk categories was associated with a favorable outcome, whereas the presence of adverse abnormalities in patients with intermediate risk changes had a deleterious effect on outcome (Table 3 and Fig 4). Subgroup analysis of patients with 11q23 abnormalities suggested a poorer outcome among those with t(10;11)(p12;q23), compared with patients with t(9;11)(p22;q23), although this needs to be confirmed in a much larger patient group (Table 3).
Influence of additional cytogenetic abnormalities on overall survival in AML.
Prognostic value of a hierarchical cytogenetic classification in newly diagnosed AML; importance in predicting outcome following postremission BMT.
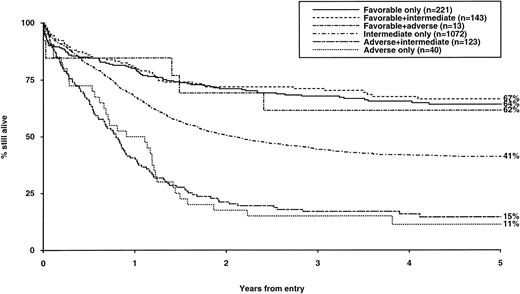
Consideration of the influence of additional cytogenetic abnormalities on outcome permitted a more refined hierarchical prognostic classification as shown in Table 4. This revised classification can distinguish groups with highly significant differences in CR rates, RR, and OS (summarized in Table 5) and has been adopted to direct treatment approach in the current MRC AML 12 trial. The hierarchical classification was subsequently evaluated in a variety of clinical contexts and was found to retain its predictive value in all age groups examined, in both de novo and secondary AML, in patients treated with chemotherapy alone, and among those receiving autologous or allogeneic BMT (Table 6). Furthermore, stratified log rank tests showed that cytogenetic risk group (P < .001) was the most important predictor of relapse risk after BMT; while cytogenetics (P < .001) and age (P = .004) were the most important predictors of posttransplant survival. While we have shown that the three cytogenetic risk groups were independent of the postremission therapy received in the context of the AML 10 protocol, applying whether or not BMT was performed, it should be noted that the analysis presented here cannot be interpreted as indicating that BMT is beneficial. Relapse rates after BMT will be lower in all cytogenetic risk groups when compared with relapse rates after CR for nontransplanted patients, as the median times from CR to allogeneic and autologous BMT were 157 and 171 days, respectively. Thus, patients receiving BMT will already have an improved prognosis by virtue of having remained in CR long enough to reach transplant. This selection factor will apply especially to poor risk patients who have a very high early relapse rate, so those who reach BMT represent a better risk subset within this group.
CR Rates, Reasons for Failure, Relapse Risk, and Survival by Hierarchical Cytogenetic Risk Group
| Group . | No. of Patients . | CR (%) . | ID (%) . | RD (%) . | Relapse Risk at 5 yr % (SE) . | Survival at 5 yr % (SE) . |
|---|---|---|---|---|---|---|
| Favorable | 377 | 91* | 8 | 1* | 35 (2.8)* | 65 (2.5)* |
| Intermediate | 1,072 | 86 | 6 | 8 | 51 (1.8) | 41 (1.5) |
| Adverse | 163 | 63 | 14 | 23 | 76 (4.5) | 14 (2.8) |
| Group . | No. of Patients . | CR (%) . | ID (%) . | RD (%) . | Relapse Risk at 5 yr % (SE) . | Survival at 5 yr % (SE) . |
|---|---|---|---|---|---|---|
| Favorable | 377 | 91* | 8 | 1* | 35 (2.8)* | 65 (2.5)* |
| Intermediate | 1,072 | 86 | 6 | 8 | 51 (1.8) | 41 (1.5) |
| Adverse | 163 | 63 | 14 | 23 | 76 (4.5) | 14 (2.8) |
*P < .001, P values are for Mantel-Haenszel (CR and reasons for failure) or log rank (relapse risk and overall survival) test for trend.
Evaluation of the Prognostic Value of Hierarchical Cytogenetic Risk Group Classification in Newly Diagnosed AML and in BMT in First Remission
| . | Outcome . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of Patients Hierarchical Risk Group . | CR Rate Hierarchical Risk Group . | Relapse Risk at 3 yr % (SE) Hierarchical Risk Group . | Survival at 3 yr % (SE) Hierarchical Risk Group . | |||||||||
| Favorable . | Intermediate . | Adverse . | Favorable . | Intermediate . | Adverse . | Favorable . | Intermediate . | Adverse . | Favorable . | Intermediate . | Adverse . | |
| Age group | ||||||||||||
| 0-14 yr | 88 | 219 | 33 | 94 | 92 | 88 | 32 (5.4)5-150 | 40 (3.6) | 61 (9.2) | 78 (4.4)5-151 | 55 (3.4) | 42 (8.6) |
| 15-34 yr | 141 | 273 | 47 | 905-150 | 91 | 70 | 32 (4.3)5-151 | 45 (3.4) | 72 (8.8) | 66 (4.0)5-151 | 51 (3.0) | 19 (5.8) |
| 35+ yr | 148 | 580 | 83 | 895-151 | 81 | 49 | 31 (4.4)5-151 | 53 (2.5) | 92 (4.5) | 65 (3.9)5-151 | 37 (2.0) | 5 (2.4) |
| Type of leukemia | ||||||||||||
| De novo | 363 | 996 | 134 | 915-151 | 87 | 66 | 32 (2.7)5-151 | 47 (1.8) | 78 (4.7) | 69 (2.4)5-151 | 46 (1.6) | 17 (3.3) |
| Secondary | 14 | 76 | 29 | 935-150 | 72 | 52 | 19 (2.7)5-151 | 62 (1.8) | 67 (12.2) | 79 (11.0)5-150 | 25 (5.0) | 14 (6.4) |
| Treatment in first remission5-152 | ||||||||||||
| Chemotherapy | 242 | 626 | 69 | 38 (3.3)5-151 | 55 (2.1) | 76 (5.6) | 76 (2.7)5-151 | 48 (2.0) | 25 (5.4) | |||
| Allogeneic BMT | 50 | 148 | 13 | 8 (4.3)5-151 | 18 (3.6) | 77 (14.1) | 62 (9.6)5-151 | 65 (4.5) | 13 (9.0) | |||
| Autologous BMT | 50 | 131 | 18 | 20 (6.0)5-150 | 35 (4.5) | 65 (11.8) | 78 (8.3)5-151 | 56 (4.6) | 46 (12.1) | |||
| . | Outcome . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of Patients Hierarchical Risk Group . | CR Rate Hierarchical Risk Group . | Relapse Risk at 3 yr % (SE) Hierarchical Risk Group . | Survival at 3 yr % (SE) Hierarchical Risk Group . | |||||||||
| Favorable . | Intermediate . | Adverse . | Favorable . | Intermediate . | Adverse . | Favorable . | Intermediate . | Adverse . | Favorable . | Intermediate . | Adverse . | |
| Age group | ||||||||||||
| 0-14 yr | 88 | 219 | 33 | 94 | 92 | 88 | 32 (5.4)5-150 | 40 (3.6) | 61 (9.2) | 78 (4.4)5-151 | 55 (3.4) | 42 (8.6) |
| 15-34 yr | 141 | 273 | 47 | 905-150 | 91 | 70 | 32 (4.3)5-151 | 45 (3.4) | 72 (8.8) | 66 (4.0)5-151 | 51 (3.0) | 19 (5.8) |
| 35+ yr | 148 | 580 | 83 | 895-151 | 81 | 49 | 31 (4.4)5-151 | 53 (2.5) | 92 (4.5) | 65 (3.9)5-151 | 37 (2.0) | 5 (2.4) |
| Type of leukemia | ||||||||||||
| De novo | 363 | 996 | 134 | 915-151 | 87 | 66 | 32 (2.7)5-151 | 47 (1.8) | 78 (4.7) | 69 (2.4)5-151 | 46 (1.6) | 17 (3.3) |
| Secondary | 14 | 76 | 29 | 935-150 | 72 | 52 | 19 (2.7)5-151 | 62 (1.8) | 67 (12.2) | 79 (11.0)5-150 | 25 (5.0) | 14 (6.4) |
| Treatment in first remission5-152 | ||||||||||||
| Chemotherapy | 242 | 626 | 69 | 38 (3.3)5-151 | 55 (2.1) | 76 (5.6) | 76 (2.7)5-151 | 48 (2.0) | 25 (5.4) | |||
| Allogeneic BMT | 50 | 148 | 13 | 8 (4.3)5-151 | 18 (3.6) | 77 (14.1) | 62 (9.6)5-151 | 65 (4.5) | 13 (9.0) | |||
| Autologous BMT | 50 | 131 | 18 | 20 (6.0)5-150 | 35 (4.5) | 65 (11.8) | 78 (8.3)5-151 | 56 (4.6) | 46 (12.1) | |||
OS and RR are quoted at 3 years, as relatively small numbers in some subgroups would restrict the reliability of 5-year values.
P < .01.
P < .001: P values are for log rank test (OS and RR) or Mantel Haenszel test for trend (CR rate) in effect of hierarchical risk group within age group, type of leukemia, or treatment in first remission.
Chemotherapy patients are censored at BMT. Survival is from the date of BMT in the transplanted groups and from CR in the chemotherapy group. Percentages may not add to 100 because of rounding.
DISCUSSION
For well over a decade, it has been appreciated that diagnostic cytogenetics provides one of the most valuable prognostic indicators in AML.1-9 However, many studies on which such conclusions were drawn were compromised to a variable extent either by relatively small sample size or by inconsistency of treatment approach. These limitations have in a number of instances resulted in contradictory data regarding the prognostic implications of specific cytogenetic abnormalities, undermining employment of karyotype at diagnosis as a means of directing treatment strategy. Nevertheless, the majority of studies associate inv(16) with a relatively favorable outcome and −7, −5/del(5q) and complex cytogenetic abnormalities with an adverse prognosis, suggesting that in many cases, cytogenetic abnormalities reflect basic differences in leukemia biology that transcend the relative sensitivity to a particular treatment approach. However, there has been little consensus as to the prognostic significance of a number of frequently recurring abnormalities: for example, t(8;21) and t(15;17) have been variably assigned to favorable and intermediate risk groups, while +8 and 11q23 abnormalities have fluctuated between intermediate and adverse-risk categories. The MRC AML 10 trial affords the opportunity to resolve such issues in the context of large patient groups receiving equivalent therapy.
On the basis of response to induction therapy, RR, and OS, three prognostic groups could be defined by cytogenetic abnormalities detected at presentation in comparison with cases with normal karyotype as summarized in Tables 4 and 5. Patients with AML associated with t(8;21), t(15;17) and inv(16) were found to comprise a group with relatively favorable outcome (Table 2). Conversely, detection of complex cytogenetic changes, −5, del(5q), −7, or abnormalities of 3q defined a group with adverse prognosis (Table 2). Bivariate analysis showed that adverse prognosis previously ascribed to del(7q) was due to a close association with complex cytogenetic abnormalities. Indeed, for patients with del(7q) in the absence of cytogenetic features associated with adverse-risk, outcome did not differ significantly from the group with normal karyotype, although this analysis was based on small numbers (Table 3). This latter result is consistent with the outcome of the Fourth International Workshop on Chromosomes in Leukemia, which found del(7q) without concurrent abnormality of chromosome 5 to be associated with a relatively favorable outcome.7 In addition to normal karyotype and del(7q), the intermediate prognostic category included patients with 11q23 abnormalities, +21, +8, +22, and del(9q). It is worth noting that the latter three abnormalities are frequent secondary changes in t(15;17), inv(16), and t(8;21) associated AML, respectively. However, a recent study suggests that the relatively good prognosis of +8, +22, and del(9q) even in the absence of overt t(15;17), inv(16), or t(8;21) cannot be accounted for by cryptic rearrangements of their respective fusion genes.13 The intermediate prognostic category also incorporated a miscellaneous group of other structural and numerical changes not encompassed by the other two risk groups, which were too infrequent to be confidently assigned a prognostic significance in their own right. Similarly, prognostic implications of 11q23 abnormalities detected in individual patients could not be reliably established, due to considerable molecular heterogeneity within this cytogenetic category leading to relatively small sample sizes. 11q23 abnormalities typically disrupt the MLL gene,14which has a plethora of potential fusion partners (see Waring and Cleary15 for review and references therein). In the present study, t(9;11)(p22;q23) and t(10;11)(p12;q23) were the most common abnormalities detected, associated with MLL fusion toAF916 and AF1017 genes, respectively. Patients with t(9;11) were found to have a relatively favorable outcome compared with those with t(10;11) (Table 3). Although this analysis was based on small numbers and the difference did not reach statistical significance, it is in accordance with previous reports concerning the prognostic significance of 11q23 abnormalities in children18 and adults.19
Recently, there has been increasing interest to determine whether the presence of additional cytogenetic abnormalities, particularly in the context of the favorable prognosis group, influences outcome. Previous smaller studies have provided conflicting data as to the significance of additional changes in the presence of the t(15;17),20-22while a study that included seven patients with del(9q) advocated that this additional abnormality predicts a poor prognosis in patients with t(8;21).23 The MRC AML 10 trial affords the opportunity to address these issues in much larger groups of patients. Additional cytogenetic abnormalities, including those associated with the adverse-risk group were found to have no significant effect on CR rates, RR, or OS in patients with t(15;17), t(8;21), or inv(16); indeed the group with t(8;21),del(9q) exhibited the most favorable survival (Table 3). While the number of patients with t(8;21), del(9q) in the present study was too small to confidently attach prognostic significance to this specific abnormality, this result renders the previous suggestion that this karyotype is associated with poor risk somewhat questionable. Furthermore, in our study, the presence of adverse risk abnormalities in patients with intermediate risk changes was found to have a deleterious effect on outcome (Fig 4). On this basis, a hierarchical system of karyotype classification was developed (Table 4) and has been used in the subsequent AML 12 trial to define prognostic groups and determine treatment approach. This classification was evaluated in a variety of clinical contexts in AML 10 and found to retain its predictive value in all age groups examined, in both de novo and secondary AML, in patients treated with chemotherapy alone, and among those receiving autologous or allogeneic BMT (Table 6), indeed cytogenetic risk group was the most important determinant of outcome following BMT in first CR. The present study confirms previous reports demonstrating that pretreatment cytogenetics retains its prognostic significance in the context of BMT in first CR.24-26 The relative value of each treatment modality to each cytogenetic risk group requires careful prospective analysis in an intention to treat manner. We report here the impact of cytogenetics on treatment delivered, in patients who were all given the same or equivalent chemotherapy before transplantation.
While it is clear that conventional cytogenetic assessment can assign patients to distinct prognostic groups in the context of modern chemotherapy treatment protocols, the challenges of future studies are to determine whether prognostic significance can be confidently ascribed to extremely rare cytogenetic abnormalities by consideration of larger data sets and as to whether targeted molecular screening and novel techniques such as spectral karyotyping27 may further enhance determination of risk groups and in particular achieve stratification within the heterogeneous intermediate group. For many years progress has been severely hampered by paucity of information regarding the nature of critical genes disrupted within the adverse prognostic category, notably those associated with del(5q) and monosomy 5 and 7. However, genes implicated in 3q21 and 3q26 defects have recently been delineated (reviewed by Nucifora and Rowley,28 Lopingco and Perkins,29 Zent et al,30 and references therein). The most common reported abnormality, inv(3)(q21q26),31 is associated with overexpression of the zinc-finger transcription factor EVI1, postulated to lead to deregulation of hematopoiesis. EVI1 overexpression has also been reported in AML cases apparently lacking 3q abnormalities32; as to whether more widespread determination of EVI1 expression would be of value to identify individual patients with poor prognosis among the favorable and intermediate risk groups remains to be determined.
Recent studies have confirmed MLL as a relatively frequent target of cryptic rearrangements,33,34 spawning considerable interest in a multiplex polymerase chain reaction (PCR) approach to identify an array of potential fusion partners in the expectation of establishing clinically relevant prognostic differences over and above those already demonstrated for patients with overt, cytogenetically established 11q23 abnormalities. Indeed, preliminary data suggests that among patients with normal karyotype, the presence of cryptic MLL rearrangements predicts a worse prognosis.35 Cryptic rearrangements of genes associated with the favorable risk cytogenetic group have also been identified.11,36-43 Analysis of material derived from patients entered into the MRC AML trials has demonstrated that up to 4% harbor CBFβ/MYH11 rearrangements in the absence of inv(16) by conventional cytogenetics43; while 8% to 9% of French-American-British (FAB) AML M2 have molecular evidence for AML1/ETO fusion without the t(8;21).41,42 It remains to be established as to whether patients in whom there is solely molecular evidence for rearrangements associated with favorable outcome fare as well as those confirmed by conventional cytogenetics. However, preliminary evidence in relation to patients with morphologic APL indicates that cases lacking the t(15;17) who have molecular evidence for a PML/RARα rearrangement share the favorable prognosis of patients with the t(15;17).44 This is particularly pertinent bearing in mind that previous work has suggested the merit of adopting differing treatment approaches according to diagnostic karyotype. In particular, both high dose daunorubicin45 and ATRA in combination with chemotherapy46,47 have been found to confer significant survival advantage in patients with the t(15;17), while consolidation with high dose Ara-C has been reported to be particularly beneficial for patients with inv(16) and t(8;21).48
The MRC AML 10 study has clearly established diagnostic karyotype as one of the most important determinants of outcome in children and younger adults with AML. While cytogenetic analysis provides a framework that can clearly distinguish groups of patients with differing response to treatment and likelihood of relapse suitable for directing treatment strategy, it lacks the ability to define outcome in individual patients or to distinguish between cases particularly within the heterogeneous intermediate risk group, which accounts for 55% of patients entered into AML 10. There has been increasing interest over the last few years in attempting to identify further parameters that might be of independent prognostic value, including immunophenotype, identification of myelodysplastic features, in vitro growth characteristics of leukemic blasts, and involvement of molecular pathways implicated in leukemogenesis such as the presence of ras mutations, or involved in response to therapy, eg, expression of the multidrug resistance glycoprotein MDR 1 (reviewed by Rowe and Liesveld49). However, it is likely that many such factors are inextricably linked to karyotype. It remains the goal of future trials to determine whether analysis for various such factors in addition to targeted screening for cryptic gene rearrangements might complement diagnostic cytogenetics, thereby providing more accurate risk assessment, which may ultimately permit a more refined treatment approach.
ACKNOWLEDGMENT
We thank all of the clinicians participating in the MRC trials, previously listed in Hann et al,10 and the cytogeneticists involved in performing the karyotype analyses. The following cytogenetics laboratories participated in the study: EIRE, Dept of Genetics, Trinity College, Dublin and University College Hospital, Galway; ENGLAND, Hospital cytogenetic laboratories participating included: Birmingham Maternity and Heartlands; Southmead, Bristol; Addenbrooke's, Cambridge; St Richard's, Chichester; Queen's, Croydon; Northwick Park, Harrow; Ipswich; St James', Leeds; Liverpool Women's; Christie, Manchester; Middlesbrough General; Norfolk and Norwich; Nottingham City; Hammersmith, King's College, Royal Free, St Mary's and University College, London; Churchill, Oxford; Salisbury District and Royal Marsden, Sutton in addition to Geoffrey Schofield Laboratories, British Nuclear Fuels plc, Cumbria; Leicester Royal Infirmary; Department of Human Genetics, University of Newcastle and Centre for Human Genetics, Sheffield; NEW ZEALAND, the following centres participated: Auckland, Christchurch, Dunedin, Palmerston, Waikato, Wellington. NORTHERN IRELAND, Department of Medical Genetics, Belfast City Hospital. SCOTLAND, Medical Genetics Laboratories, Aberdeen; Ninewells Hospital Medical School, Dundee; MRC Human Genetics Unit, Western General Hospital, Edinburgh; Duncan Guthrie Institute of Medical Genetics, Yorkhill, Glasgow; Royal Northern Infirmary, Inverness. WALES, Department of Haematology and Institute of Medical Genetics, University Hospital of Wales, Cardiff. We are grateful to Stephen Langabeer for critical reading of the manuscript and to Michael Neat for helpful discussions. Finally, we thank Kate Grimwade for all her support.
D.G. was supported by a MRC clinical training fellowship and subsequently by the Imperial Cancer Research Fund. We are also indebted to the Kay Kendall Leukaemia Fund for supporting the MRC trials cytogenetics database.
Address reprint requests to Dr Anthony Goldstone, FRCP, FRC Path, Department of Haematology, University College Hospital, Gower St, London, WC1E 6AU, UK.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal