Abstract
The high-affinity receptors for human granulocyte-macrophage colony-stimulating factor (GM-CSF), interleukin-3 (IL-3), and IL-5 are heterodimeric complexes consisting of cytokine-specific subunits and a common signal-transducing β subunit (hβc). We have previously demonstrated the oncogenic potential of this group of receptors by identifying constitutively activating point mutations in the extracellular and transmembrane domains of hβc. We report here a comprehensive screen of the entire hβc molecule that has led to the identification of additional constitutive point mutations by virtue of their ability to confer factor independence on murine FDC-P1 cells. These mutations were clustered exclusively in a central region of hβc that encompasses the extracellular membrane-proximal domain, transmembrane domain, and membrane-proximal region of the cytoplasmic domain. Interestingly, most hβc mutants exhibited cell type-specific constitutive activity, with only two transmembrane domain mutants able to confer factor independence on both murine FDC-P1 and BAF-B03 cells. Examination of the biochemical properties of these mutants in FDC-P1 cells indicated that MAP kinase (ERK1/2), STAT, and JAK2 signaling molecules were constitutively activated. In contrast, only some of the mutant β subunits were constitutively tyrosine phosphorylated. Taken together, these results highlight key regions involved in hβc activation, dissociate hβc tyrosine phosphorylation from MAP kinase and STAT activation, and suggest the involvement of distinct mechanisms by which proliferative signals can be generated by hβc.
© 1998 by The American Society of Hematology.
THE HIGH-AFFINITY RECEPTORS for human granulocyte-macrophage colony-stimulating factor (GM-CSF; hGMR), interleukin-3 (IL-3; hIL-3R), and IL-5 (hIL-5R) are composed of cytokine-specific α subunits (hGMRα, hIL-3Rα, and hIL-5Rα) associated with a common signal-transducing β subunit (hβc).1-5 Each receptor subunit belongs to the cytokine receptor family, members of which are characterized by an extracellular cytokine receptor module (CRM) containing several conserved sequence elements, including the distinctive WSXWS (Trp-Ser-Xaa-Trp-Ser) motif, and a cytoplasmic domain that lacks any intrinsic enzymatic activity associated with signal transduction (reviewed in Mui and Miyajima6). Despite the absence of intrinsic tyrosine kinase activity in the α and β subunits of the hGMR, hIL-3R, and hIL-5R, cytokine binding to these receptors results in the induction of several cellular responses, such as tyrosine phosphorylation of intracellular substrates, including the β subunit itself, and activation of the Ras-Raf-MAP kinase and JAK2-STAT5 signaling pathways.5 7-11
Although the definitive mechanisms underlying both cytokine receptor activation and the subsequent activation of signaling pathways in response to ligand binding have not been fully elucidated, an essential event in cytokine receptor activation is the ligand-induced multimerization of receptor subunits. Direct evidence for this has been provided by the crystal structure of the ligand-bound human growth hormone receptor complex in which a receptor homodimer is bound by one ligand molecule.12 The isolation of constitutively active cytokine receptor mutants has also provided a useful tool for examining the normal activation process of some receptors, because these mutant receptors most likely mimic the structure of the normal cytokine-activated receptors. For example, constitutive point mutations that replace specific residues with cysteines in the extracellular region of the receptors for erythropoietin and thrombopoietin (c-Mpl) result in constitutive disulphide-linked receptor homodimerization, suggesting that ligand-induced homodimerization is required for signaling by the normal receptors.13-15 Indeed, the recently published crystal structure of an erythropoietin receptor homodimer bound to a peptide agonist also provides strong evidence for the involvement of homodimerization in erythropoietin receptor activation.16
In the case of the normal hGMR, hIL-3R, and hIL-5R, there is increasing evidence that the formation of active hGMR and hIL-3R complexes involves ligand-induced α and β subunit heterodimerization,17,18 although the precise stoichiometry of receptor subunits in the active complexes remains unresolved. Chimeric receptors containing the hβc cytoplasmic domain fused to the extracellular domains of hGMRα or hIL-5Rα have been reported to confer cytokine-dependent growth on hematopoietic cells, suggesting that dimerization of the hβc cytoplasmic domain is sufficient for cellular proliferation.17,19,20 Consistent with a role for β subunit dimerization in receptor activation, it has been shown that β subunit homodimers are found in active hGMR complexes.21 More recently, it was observed that the functional hGMR complex may contain at least two α subunits.22 In the context of receptor stoichiometry, these results suggest that the α and β subunits of these receptors may form higher order complexes.22 23
We have previously combined polymerase chain reaction (PCR)-based random mutagenesis with retroviral expression cloning to screen for constitutive point mutations within a membrane-spanning region of hβc. This led to the identification of two mutations, one of which is located in the transmembrane domain of hβc (V449E) and is able to confer factor independence on FDC-P1 and BAF-B03 cells.24This mutation is similar to a constitutive mutation in theneu/erbB-2 oncogene25,26 and, by analogy, has been proposed to act by inducing constitutive hβc homodimerization.24 The other constitutive point mutation lies in the extracellular region of hβc (I374N) and confers factor independence on FDC-P1 cells, but not BAF-B03 cells, suggesting that there are alternate mechanisms, possibly involving cell type-specific signaling molecules, by which hβc can be activated.24 27
In addition to the above-mentioned I374N and V449E mutants, we have recently used a site-directed mutagenesis approach to identify amino acid substitutions at two other residues in the extracellular region of hβc, Leu356, and Trp358 that constitutively activate hβc in FDC-P1 cells.28 Interestingly, the Leu356 and Trp358 residues lie within the same membrane-spanning region of hβc that was screened for constitutive point mutations, raising the possibility that other constitutive point mutations were missed in this screen. To address this issue, we report here a comprehensive screen of the entire hβc molecule for constitutive point mutations by combining random mutagenesis with a simplified retroviral expression strategy and a more sensitive screen. The efficiency of this strategy was demonstrated by the identification of several novel constitutive point mutations in hβc, most of which exhibit cell type-specific differences in constitutive activity. Interestingly, these mutations are clustered exclusively in the extracellular membrane-proximal domain, transmembrane domain, and membrane-proximal region of the cytoplasmic domain of hβc, reflecting, we suggest, key sites involved in normal GMR/IL-3R/IL-5R activation. We have also initiated an investigation into the effect these constitutively active mutants have on certain intracellular signaling events in factor-independent FDC-P1 cells. Surprisingly, examination of the tyrosine phosphorylation state of the mutant β subunits indicated that only some mutants were constitutively tyrosine phosphorylated. In contrast, ERK1/2 MAP kinase and STAT signaling molecules involved in the Ras-Raf-MAP kinase and JAK-STAT pathways, respectively, were constitutively activated by all mutants.
MATERIALS AND METHODS
Cell Lines
BOSC 23 retroviral packaging cells29 and Ψ2 cells producing wild-type hGMRα retrovirus24 were maintained as described previously.28 The mouse IL-3/GM-CSF–dependent myeloid cell line, FDC-P1,30 and the BAF-B03 subline31 of the mouse IL-3–dependent pro-B–cell line, Ba/F3, were maintained as described previously.24
Site-Directed Mutagenesis and Construction of Expression Plasmids
The hβc cDNA used here was that described by Barry et al32; amino acids are numbered from the initiating codon. Site-directed mutagenesis was performed on double-stranded DNA with mutagenic oligonucleotides using the Altered Sites in vitro mutagenesis system (Promega, Madison, WI) in accordance with the manufacturer’s instructions. All mutations were confirmed by DNA sequencing, after which mutant hβc cDNAs were subcloned between theBamHI and HindIII restriction sites of the pRUFNeo retroviral expression vector.33
PCR Mutagenesis and Construction of Point-Mutated hβc cDNA Libraries
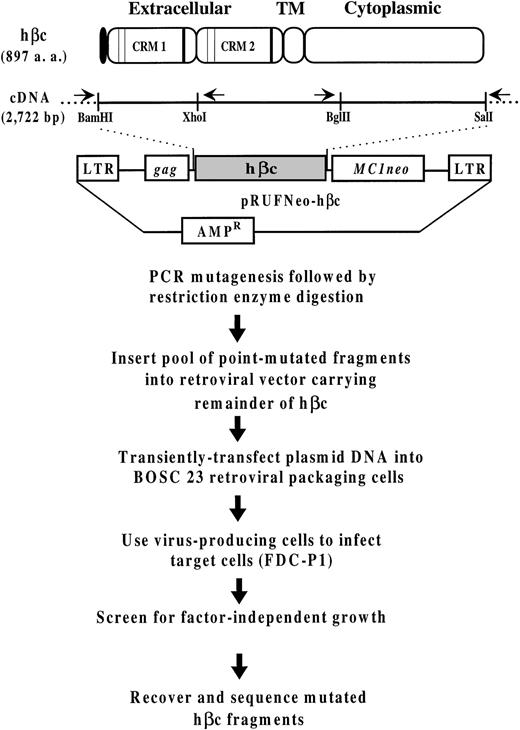
PCR mutagenesis was performed on the pRUFNeo-hβc plasmid in which aXho I site was silently introduced into wild-type hβc to facilitate cloning of PCR products (Fig 1). Random point mutations were introduced into the N-terminal 770-bpBamHI/Xho I segment, bases 1-770, and the C-terminal 1,017-bp Bgl II/Sal I segment, bases 1705-2722, of the hβc cDNA2 (sequence accession no. M38275) at a mutation frequency of approximately 0.3% (1/350) under the mutagenic reaction conditions described by Jenkins et al.24 The primers used for amplification of the N-terminal region were the RCF1 primer,33 corresponding to the gag sequence in the pRUFNeo vector, and an internal hβc primer (5′-AGCTGGCCACCTCCTTCCTCACCT-3′, bases 839-816) defining a 937-bp fragment. The primers used for amplification of the C-terminal region were an internal hβc primer (5′-CCCCAAGCATGTCTGTGATCCACC-3′, bases 1651-1674) and the RCR1 primer,33 corresponding to the MC1Neo sequence in the pRUFNeo vector, defining a 1,105-bp fragment. After digestion with the appropriate restriction enzymes, mutant fragments were agarose gel-purified and ligated directionally into pRUFNeo-hβc from which the BamHI/Xho I or Bgl II/Sal I segment of hβc had been excised. After transformation of Escherichia coli (DH10B), the resultant point-mutated hβc cDNA libraries of approximately 3.5 × 104 (BamHI/Xho I) and approximately 9.5 × 104 (BglII/Sal I) independent plasmid clones were further amplified as described previously.33
Outline of the strategy used to generate and express hβc mutants. Schematic illustration of hβc showing the signal sequence (shading), the two cytokine receptor modules (CRMs72) containing the conserved cysteine residues (thin vertical lines) and the characteristic WSXWS motifs (thick vertical lines; see Bazan73 for a description of these elements), and the transmembrane and cytoplasmic domains. Also included is a schematic diagram of the pRUFNeo retroviral expression vector containing the hβc cDNA. The positions in the hβc cDNA of theBamHI, Xho I, Bgl II, and Sal I restriction sites that define the regions subjected to random mutagenesis are shown underneath. The arrows above the cDNA represent the primers used for PCR amplification/mutagenesis of the N-terminal and C-terminal hβc fragments; they lie just outside the restriction sites defining the mutagenized regions (see Materials and Methods).
Outline of the strategy used to generate and express hβc mutants. Schematic illustration of hβc showing the signal sequence (shading), the two cytokine receptor modules (CRMs72) containing the conserved cysteine residues (thin vertical lines) and the characteristic WSXWS motifs (thick vertical lines; see Bazan73 for a description of these elements), and the transmembrane and cytoplasmic domains. Also included is a schematic diagram of the pRUFNeo retroviral expression vector containing the hβc cDNA. The positions in the hβc cDNA of theBamHI, Xho I, Bgl II, and Sal I restriction sites that define the regions subjected to random mutagenesis are shown underneath. The arrows above the cDNA represent the primers used for PCR amplification/mutagenesis of the N-terminal and C-terminal hβc fragments; they lie just outside the restriction sites defining the mutagenized regions (see Materials and Methods).
The construction of the Xho I/Bgl II point-mutated hβc cDNA library has been described previously.24
Infection of Hematopoietic Cells With Mutant hβc Retroviruses
High-titer retroviruses carrying mutant hβc cDNAs were generated by transiently transfecting BOSC 23 retroviral packaging cells with retroviral DNA essentially as described by Jenkins et al.28
For the generation of retroviruses representing the three point-mutated hβc libraries, one 60-mm dish (BamHI/Xho I library), five 60-mm dishes (Xho I/Bgl II library), and two 60-mm dishes (Bgl II/Sal I library) were each seeded with 2 × 106 BOSC 23 cells and transfected with 20 μg of the appropriate retroviral DNA. Infections of FDC-P1 cells were performed by cocultivation as described previously,28 with 2.5 × 105 FDC-P1 cells added to each dish. FDC-P1 cells from each dish were harvested, washed, and selected for factor-independent growth in 24-well multidishes (84 wells forBamHI/Xho I library, 204 wells for XhoI/Bgl II library, and 102 wells for Bgl II/SalI library, each seeded with 2 × 104 cells) in liquid culture medium without mouse (m) GM-CSF. Factor-independent cells were expanded in liquid culture to 25-cm2 flasks for further analysis. FDC-P1 cell plating in soft agar was performed essentially as described by Johnson34; mGM-CSF (80 U/mL) or G418 (1 mg/mL) was added as required.
Retroviral infection of FDC-P1 and BAF-B03 cells with hβc point mutants generated by site-directed mutagenesis was performed using BOSC 23 cells, and cells were harvested and selected as described previously.28 FDC-P1 and BAF-B03 cells infected with wild-type hGMRα retrovirus were selected as described previously.28
Recovery of Mutant hβc cDNAs From Factor-Independent Cells
Genomic DNA was isolated from cells using a proteinase K/SDS procedure essentially as described by Hughes et al.35 PCR was performed on 100 ng of genomic DNA with Pfu DNA polymerase (Stratagene, La Jolla, CA) under conditions recommended by the manufacturer. The primers and cycling parameters used were those for generating the point-mutated hβc cDNA libraries. PCR products were agarose gel-purified and directly sequenced with the PCR primers using a Taq DyeDeoxy Terminator Cycle Sequencing Kit (Perkin Elmer, Norwalk, CT), followed by analysis on an ABI Prism 377 DNA Sequencer (ABI, Foster City, CA).
Flow Cytometric Analysis of Receptor Subunit Expression
Expression of hβc mutants on the cell surface of infected FDC-P1 or BAF-B03 cells was detected by standard indirect immunofluorescence with the anti-hβc monoclonal antibody 1C118 and fluorescein isothiocyanate (FITC)-conjugated antimouse IgG (Silenus, Hawthorn, Victoria, Australia) followed by flow cytometry on an Epics-Profile II analyser (Coulter, Hialeah, FL). hGMRα expression was analyzed as described above with the 8G6 monoclonal antibody.36
Cell Proliferation Assays
Infected FDC-P1 or BAF-B03 cells were washed twice and triplicate samples of equal cell number (103 or 5 × 103) were cultured in 96-well microtiter plates with or without appropriate growth factor for 72 hours. Cell proliferation was measured by the CellTiter 96 Non-Radioactive Cell Proliferation Assay (Promega).
Antibodies
The monoclonal hβc antibodies, 8E4 and 1C1, were a kind gift from Angel Lopez (Hanson Centre for Cancer Research, Adelaide, Australia). An anti-MAP kinase (ERK1/2) antibody was purchased from Zymed Laboratories (San Francisco, CA), and the antibody specific for the active phosphorylated form of MAP kinases ERK1/2 was purchased from Promega. Horseradish peroxidase-conjugated antiphosphotyrosine antibody, RC20, was purchased from Transduction Laboratories (Lexington, KY). The antiphosphotyrosine antibody, 4G10, and an anti-JAK2 polyclonal antibody were purchased from Upstate Biotechnology (Lake Placid, NY).
Immunoprecipitation and Western Blot Analysis
For stimulation with human GM-CSF (hGM-CSF), cells (2 to 4 × 107) were initially starved for 12 hours in growth media without cytokine. Cells were left unstimulated or were stimulated with hGM-CSF (50 ng/mL) at 37°C for 10 minutes or, in some cases, cells were grown continuously in the presence of 2 ng/mL hGM-CSF. Cells were washed with cold phosphate-buffered saline (PBS) containing 20 mmol/L sodium orthovanadate and lysed on ice in lysis buffer (50 mmol/L HEPES [pH 7.5], 150 mmol/L NaCl, 1% NP-40, 2 mmol/L sodium orthovanadate, 1 mmol/L phenylmethylsulfonyl fluoride, 1 mmol/L EDTA, 1 mmol/L EGTA, 2 mg/mL iodoacetamide, 0.2 mg/mL trypsin inhibitor [Boehringer Mannheim, Indianapolis, IN], and Complete protease inhibitor [Boehringer Mannheim]) for 15 minutes. Insoluble material was removed by centrifugation at 12,000 rpm for 2 minutes and samples were taken for protein estimation (Dc protein assay kit; Bio-Rad Laboratories, Richmond, CA). Cell lysates were incubated with the indicated antibody for 1 hour at 4°C. Immune complexes were precipitated with 75 μL (50% slurry) of protein A-sepharose (Pharmacia) for 1 hour at 4°C, washed three times with lysis buffer, and boiled for 2 minutes in 1× reducing sodium dodecyl sulfate (SDS) sample buffer. In the case of total protein analysis, samples were lysed and insoluble material was removed and boiled for 2 minutes in 1× reducing SDS sample buffer.
JAK2 phosphorylation was detected in cells (1 × 108) treated (±hGM-CSF) as described above, except that 500 nmol/L sodium orthovanadate was added to the medium for 10 minutes before lysis. Cells were lysed on ice in lysis buffer with the addition of 0.1% deoxycholic acid and 0.1% SDS and immunoprecipitated as described.
Proteins were electrophoresed on SDS-polyacrylamide gels and electrophoretically transferred to 0.2-μm nitrocellulose (Schleicher and Schuell, Keene, NH). After blocking with TBS-T (50 mmol/L Tris [pH 7.4], 135 mmol/L NaCl, 0.1% Tween 20) containing 3% bovine serum albumin (BSA; Fraction V; Boehringer Mannheim), membranes were incubated with the primary antibody and washed three times in TBS-T. After incubation with goat antirabbit or antimouse secondary antibodies coupled with horseradish peroxidase (Pierce, Rockford, IL), filters were washed with TBS-T three times and subjected to enhanced chemiluminescence detection (Pierce). Before reprobing with the indicated antibodies, membranes were stripped in 50 mmol/L Tris (pH 7.4), 2% SDS, 100 mmol/L β-mercaptoethanol at 55°C for 20 minutes; washed three times in TBS-T; and blocked in TBS-T containing 3% BSA.
Electrophoretic Mobility Shift Assay (EMSA)
Nuclei from cells washed and lysed as described above were pelleted by centrifugation at 12,000 rpm for 2 minutes. Nuclei were resuspended in lysis buffer (without NP-40) supplemented with 150 mmol/L NaCl and 10% glycerol. After incubation at 4°C for 15 minutes, insoluble material was removed by centrifugation at 12,000 rpm for 5 minutes and samples were stored at −70°C. EMSAs were performed using a β-casein promoter probe that contains a binding site for STAT1, STAT3, and STAT5 essentially as described by Barry et al.37
RESULTS
Isolation of Factor-Independent FDC-P1 Cells Infected With the Membrane-Spanning Xho I/Bgl I Point-Mutated hβc Library
We have previously combined PCR-based random mutagenesis with retroviral expression cloning to identify two distinct constitutive point mutations within a region of hβc, delimited by Xho I and Bgl II sites in hβc cDNA (see Fig 1), by virtue of their ability to confer factor independence on the murine factor-dependent hematopoietic cell line, FDC-P1.24 The retroviral library used in that study was of moderate size, and the selection for factor independence was biased towards strongly activating mutations present in large pools of infected FDC-P1 cells, suggesting that other less frequent or weakly activating point mutations present in this region of hβc may have been missed. Indeed, this possibility has been supported by the recent identification of two other constitutive mutations in hβc that lie within this region.28
To investigate whether other sites for oncogenic activation by point mutation are present in the Xho I/Bgl II region of the hβc cDNA, we devised a strategy, outlined in Fig 1, that involved transient transfection of the Xho I/Bgl II point-mutated hβc expression library24 into BOSC 23 retroviral packaging cells to produce high-titer retroviruses carrying the hβc point mutants. After transfection, FDC-P1 cells were infected by cocultivation with the mutant hβc and, as a control wild-type hβc, virus-producing BOSC 23 cells. FDC-P1 cells were then selected for factor-independent growth in 24-well multidishes, with each well containing 2 × 104 cells in medium without mGM-CSF. After several weeks in culture in the absence of factor, 96/204 wells seeded with FDC-P1 cells infected with the mutant hβc retroviral library contained viable, proliferating cells, whereas no such cells were present in control wells seeded with uninfected FDC-P1 cells or FDC-P1 cells infected with wild-type hβc retrovirus.
Factor-independent FDC-P1 cell populations expressing the previously identified I374N and V449E constitutive hβc mutants were identified by recovery of mutagenized hβc fragments by PCR from genomic DNA, followed by restriction enzyme digestions diagnostic of the I374N (BstYI) and V449E (Bgl II) mutants.24 Of the 96 factor-independent FDC-P1 cell cultures, 23 contained the extracellular I374N mutant and 32 contained the transmembrane domain V449E mutant (data not shown). Conditioned medium from the 41 factor-independent cell cultures containing unidentified constitutive mutations failed to support the growth of uninfected FDC-P1 cells, thus eliminating the possibility of factor independence being due to autocrine growth factor production (data not shown).
Identification of Novel Constitutive hβc Point Mutations in the Factor-Independent FDC-P1 Cell Populations
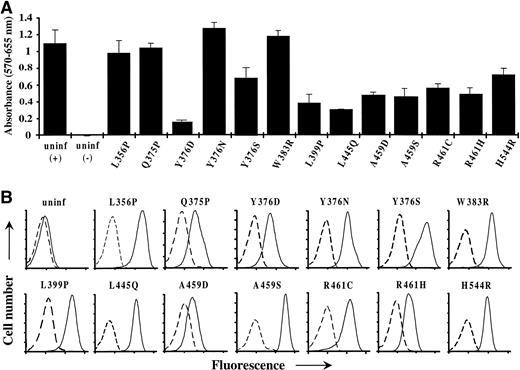
Novel, potentially constitutive hβc point mutations in the 41 factor-independent FDC-P1 cell populations were identified by PCR recovery of the mutated hβc region and sequencing (see Materials and Methods). Mutations of potential interest were selected for further analysis on the basis of (1) being the only mutation in a factor-independent cell population, (2) presence in more than one factor-independent cell population, or (3) proximity to sequences implicated in receptor activation or signaling. To test whether selected mutations could induce constitutive activity, they were recreated independently by site-directed mutagenesis. After insertion into the pRUFNeo retroviral vector and transient transfection into the BOSC 23 retroviral packaging cells, these mutants, as well as wild-type hβc, were introduced into FDC-P1 cells and then selected for either G418-resistance or for growth in factor-free medium. Flow cytometric analysis of G418-resistant cell populations indicated that a substantial proportion (8.5% to 42%) of infected cells expressed each hβc mutant on the cell surface (data not shown). Of the hβc mutants tested, 13 (Table 1 and Fig 8) were able to confer factor-independent growth on FDC-P1 cells (Fig 2), with several mutants containing different amino acid substitutions for the same wild-type residue. Of these mutants, Tyr376 was replaced with Ser, Asp, and Asn; Ala459 with Asp and Ser; and Arg461 with Cys and His. Importantly, the factor independence exhibited by each of the 41 FDC-P1 cell populations could be attributed to at least one of these constitutive mutations.
Summary of Biological and Biochemical Activities Conferred by Constitutively Active hβc Mutants
| Mutant-150 . | Location . | Proliferative Activity in BAF-B03 Cells . | Tyrosine Phosphorylation of hβc-151 . | MAP Kinase Activation-151 . | STAT5 Activation-151 . | JAK2 Activation-151 . | ||
|---|---|---|---|---|---|---|---|---|
| Alone . | +hGMRα and hGM-CSF . | Alone . | +hGMRα and hGM-CSF . | |||||
| L356P | Domain 4, β-strand B | No | No | No | No | Yes | Yes | Yes |
| W358N | Domain 4, β-strand B | No | No | No | No | Yes | Yes | ND |
| I374N | Domain 4, β-strand C | No | Yes | No | Yes | Yes | Yes | Yes |
| Q375P | Domain 4, β-strand C | No | Yes | No | Yes | Yes | Yes | ND |
| Y376N | Domain 4, β-strand C | No | Yes | No | Yes | Yes | Yes | ND |
| W383R | Domain 4, β-strand D | No | Yes | No | Yes | Yes | Yes | ND |
| L399P | Domain 4, β-strand E | No | Yes | No | No | Yes | Yes | Yes |
| L445Q | Transmembrane domain | No | Yes | No | Yes | Yes | Yes | ND |
| V449E | Transmembrane domain | Yes | ND | Yes | Yes | Yes | Yes | Yes |
| A459D | Transmembrane domain | Yes | ND | Yes | Yes | Yes | Yes | ND |
| R461C | Cytoplasmic domain | No | Yes | Yes | Yes | Yes | Yes | ND |
| H544R | Cytoplasmic domain, box 2 | No | Yes | No | No | Yes | Yes | Yes |
| Mutant-150 . | Location . | Proliferative Activity in BAF-B03 Cells . | Tyrosine Phosphorylation of hβc-151 . | MAP Kinase Activation-151 . | STAT5 Activation-151 . | JAK2 Activation-151 . | ||
|---|---|---|---|---|---|---|---|---|
| Alone . | +hGMRα and hGM-CSF . | Alone . | +hGMRα and hGM-CSF . | |||||
| L356P | Domain 4, β-strand B | No | No | No | No | Yes | Yes | Yes |
| W358N | Domain 4, β-strand B | No | No | No | No | Yes | Yes | ND |
| I374N | Domain 4, β-strand C | No | Yes | No | Yes | Yes | Yes | Yes |
| Q375P | Domain 4, β-strand C | No | Yes | No | Yes | Yes | Yes | ND |
| Y376N | Domain 4, β-strand C | No | Yes | No | Yes | Yes | Yes | ND |
| W383R | Domain 4, β-strand D | No | Yes | No | Yes | Yes | Yes | ND |
| L399P | Domain 4, β-strand E | No | Yes | No | No | Yes | Yes | Yes |
| L445Q | Transmembrane domain | No | Yes | No | Yes | Yes | Yes | ND |
| V449E | Transmembrane domain | Yes | ND | Yes | Yes | Yes | Yes | Yes |
| A459D | Transmembrane domain | Yes | ND | Yes | Yes | Yes | Yes | ND |
| R461C | Cytoplasmic domain | No | Yes | Yes | Yes | Yes | Yes | ND |
| H544R | Cytoplasmic domain, box 2 | No | Yes | No | No | Yes | Yes | Yes |
Abbreviation: ND, not determined.
All mutants confer factor-independent proliferation on FDC-P1 cells. Mutants isolated in previous studies are underlined.
Proliferation of factor-independent FDC-P1 cells infected with novel constitutive hβc mutants. (A) Proliferation assay of FDC-P1 cells, infected with the indicated hβc mutants, which had been selected before assay for growth in the absence of factor. Also shown are uninfected cells (uninf) that were washed and assayed in medium with (+) and without (−) mGM-CSF. Cells (103) were plated in triplicate and cell proliferation was measured at 72 hours as described in Materials and Methods. The mean and standard error of each triplicate is shown. (B) Flow cytometric analysis of constitutive hβc mutant expression on the factor-independent FDC-P1 cells depicted in (A). Cells were stained by standard indirect immunofluorescence; dashed lines represent cells stained with an irrelevant control antibody and solid lines indicate staining with an anti-hβc antibody. Cell number and fluorescence are in arbitrary units; the latter is plotted on a logarithmic scale. Also shown are analyses of uninfected FDC-P1 cells. Note that data for only the novel mutants isolated in this study are shown.
Proliferation of factor-independent FDC-P1 cells infected with novel constitutive hβc mutants. (A) Proliferation assay of FDC-P1 cells, infected with the indicated hβc mutants, which had been selected before assay for growth in the absence of factor. Also shown are uninfected cells (uninf) that were washed and assayed in medium with (+) and without (−) mGM-CSF. Cells (103) were plated in triplicate and cell proliferation was measured at 72 hours as described in Materials and Methods. The mean and standard error of each triplicate is shown. (B) Flow cytometric analysis of constitutive hβc mutant expression on the factor-independent FDC-P1 cells depicted in (A). Cells were stained by standard indirect immunofluorescence; dashed lines represent cells stained with an irrelevant control antibody and solid lines indicate staining with an anti-hβc antibody. Cell number and fluorescence are in arbitrary units; the latter is plotted on a logarithmic scale. Also shown are analyses of uninfected FDC-P1 cells. Note that data for only the novel mutants isolated in this study are shown.
Over several independent experiments, the proliferation rates of some infected factor-independent cell populations were consistently different to each other and to that of uninfected FDC-P1 cells grown in the presence of mGM-CSF (Fig 2A). Interestingly, these differences in factor-independent growth rates could not be attributed to corresponding differences in cell-surface expression (Fig 2B). Furthermore, the proliferation rates of factor-independent cell populations grown in the presence of mGM-CSF were similar to those of uninfected FDC-P1 cells, indicating that all factor-independent cell populations retained a similar responsiveness to mGM-CSF–generated mitogenic signals (data not shown). Taken together, these data suggest that some constitutive hβc mutants may be more strongly activating than others, as also seen in a previous study.28
Absence of Constitutive Mutations in the N-TerminalBamHI/Xho I and C-Terminal Bgl II/Sal I Point-Mutated hβc Libraries
Given the efficiency at which constitutive mutations in the XhoI/Bgl II point-mutated hβc library were identified, we decided to use the same retroviral expression cloning strategy to screen for constitutively activating point mutations in the remainder of hβc. Two independent screens were used, one covering a 770-bpBamHI/Xho I segment of the hβc cDNA encoding the first 239 amino acids that contain the N-terminal extracellular cytokine receptor module (CRM) and the other a 1,017-bp BglII/Sal I segment of the hβc cDNA encoding the C-terminal 337 amino acids of the cytoplasmic domain (Fig 1). Point mutations were introduced into both hβc regions at a rate of approximately 0.3% (1 in 350 bp), and libraries of approximately 3.5 × 104(BamHI/Xho I) and approximately 9.5 × 104 (Bgl II/Sal I) plasmid clones representing hβc cDNAs bearing point mutations in the targeted regions were generated. Considering that there was no overwhelming mutational bias in these procedures (data not shown), the libraries should adequately represent all of the possible point mutations in both hβc segments.
In separate experiments, retroviruses generated by transient transfection of BOSC 23 cells with the BamHI/Xho I andBgl II/Sal I point-mutated hβc libraries were used to infect FDC-P1 cells by cocultivation with infection frequencies of 38% and 25%, respectively. Again, parallel cocultivations were performed in both experiments with untransfected BOSC 23 cells and BOSC 23 cells producing wild-type hβc retrovirus. After several weeks in liquid culture in the absence of factor, only wells seeded with FDC-P1 cells infected with the mutant hβc retroviruses (2/84 forBamHI/Xho I library and 3/102 forBgl II/Sal I library) contained viable, proliferating cells. Conditioned medium from these cultures again demonstrated that factor independence was not due to autocrine growth factor production (data not shown). However, retesting of β subunits bearing mutations identified in hβc PCR fragments recovered from these factor-independent cell populations in FDC-P1 cells failed to generate factor-independent cells (data not shown). Sequencing of these PCR fragments showed that the factor-independent cell populations isolated from each screen contained identical, full-length hβc integrants, suggesting that these populations may have originated from single infected FDC-P1 cells that acquired factor independence without constitutive mutations in hβc.
Differential Tyrosine Phosphorylation of hβc Mutants
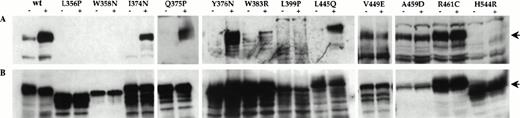
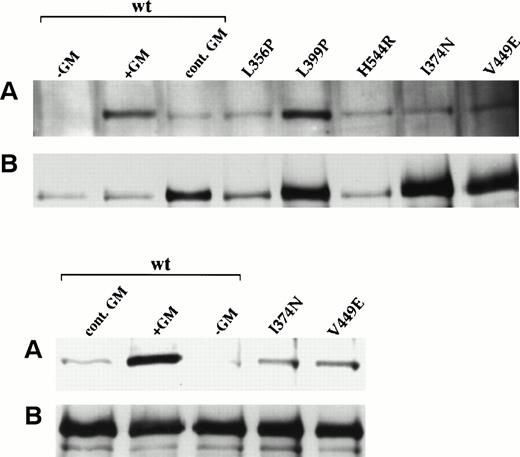
Previous studies have demonstrated that one of the early biochemical events in response to hGM-CSF stimulation is tyrosine phosphorylation of hβc.5 7 To assess the tyrosine phosphorylation state of mutant β subunits expressed in the factor-independent FDC-P1 cell populations, cell lysates were subjected to immunoprecipitation with an anti-hβc antibody and immunoblotting with an antiphosphotyrosine antibody. As shown in Fig 3A, only V449E, A459D, and R461C mutants were constitutively tyrosine phosphorylated in the absence of factor, despite the fact that immunoblotting with an anti-hβc antibody (Fig 3B) indicated that all immunoprecipitates contained readily detectable levels of β subunits. We also determined whether β subunit tyrosine phosphorylation could be detected in all cell lines by coexpressing the hGMRα subunit with mutant β subunits (and wild-type, as a control) in each FDC-P1 cell population. Only I374N, Q375P, Y376N, W383R (weakly), L445Q, and, as expected, wild-type β subunits demonstrated an increase in phosphorylation on tyrosine residues upon stimulation with hGM-CSF (Fig 3A), despite the fact that hGMRα expression could be detected on the surface of all cell lines (data not shown).
Tyrosine phosphorylation of constitutive hβc mutants in factor-independent FDC-P1 cells. FDC-P1 cells coexpressing hGMR and the indicated β subunits were incubated without (−) or with (+) 50 ng/mL hGM-CSF for 10 minutes. Whole cell lysates were immunoprecipitated with an anti-hβc antibody and immunoblotted using (A) an antiphosphotyrosine antibody and (B) an anti-hβc antibody. The position of hβc in each panel is indicated by an arrow.
Tyrosine phosphorylation of constitutive hβc mutants in factor-independent FDC-P1 cells. FDC-P1 cells coexpressing hGMR and the indicated β subunits were incubated without (−) or with (+) 50 ng/mL hGM-CSF for 10 minutes. Whole cell lysates were immunoprecipitated with an anti-hβc antibody and immunoblotted using (A) an antiphosphotyrosine antibody and (B) an anti-hβc antibody. The position of hβc in each panel is indicated by an arrow.
Constitutive Activation of the Ras-Raf-MAP Kinase and JAK2-STAT5 Signaling Pathways by hβc Mutants
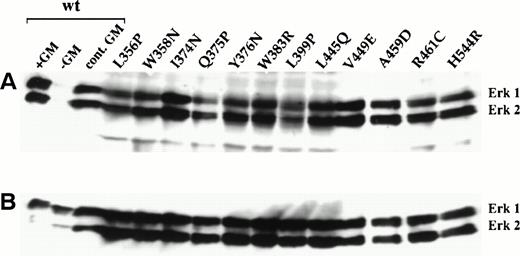
One of the major signaling pathways activated in response to cytokines, including GM-CSF, is the Ras-Raf-MAP kinase pathway.10,38Tyrosine phosphorylation and activation of MAP kinase is dependent on the sequential activation of upstream Ras, Raf-1, and MEK-1 effector molecules (reviewed in Marshall39). We therefore examined the levels of tyrosine phosphorylation on ERK1 and ERK2 MAP kinases in factor-independent FDC-P1 cells expressing the mutant β subunits. Western blot analysis of cell lysates with an antibody specific for activated, ie, phosphorylated, ERK1/2 MAP kinases showed that both proteins were similarly tyrosine phosphorylated in all cells in the absence of factor (Fig 4A). Furthermore, the extent of phosphorylation was similar to that seen in FDC-P1 cells expressing hGMRα and wild-type hβc (hGMR) in the presence of hGM-CSF (lanes 1 and 3). In contrast, no MAP kinase phosphorylation was detected in cells expressing hGMR in the absence of hGM-CSF (lane 2). As shown in Fig 4B, the levels of total ERK1/2 proteins present in lysates from all cell populations were comparable.
Constitutive tyrosine phosphorylation of ERK1/2 MAP kinases in factor-independent FDC-P1 cells expressing constitutive hβc mutants. Whole cell lysates from factor-independent FDC-P1 cells were analyzed by immunoblotting using (A) an anti-phospho-ERK1/2 antibody and (B) an anti-ERK1/2 antibody. Also shown are analyses of FDC-P1 cells expressing wild-type hGMR that were either starved of growth factor overnight and incubated with (lane 1) or without (lane 2) 50 ng/mL hGM-CSF for 10 minutes or continuously cultured in 1 ng/mL hGM-CSF (lane 3).
Constitutive tyrosine phosphorylation of ERK1/2 MAP kinases in factor-independent FDC-P1 cells expressing constitutive hβc mutants. Whole cell lysates from factor-independent FDC-P1 cells were analyzed by immunoblotting using (A) an anti-phospho-ERK1/2 antibody and (B) an anti-ERK1/2 antibody. Also shown are analyses of FDC-P1 cells expressing wild-type hGMR that were either starved of growth factor overnight and incubated with (lane 1) or without (lane 2) 50 ng/mL hGM-CSF for 10 minutes or continuously cultured in 1 ng/mL hGM-CSF (lane 3).
Another major pathway that has been implicated in cytokine receptor signaling is the recently identified JAK-STAT pathway (reviewed in Ihle40). Indeed, several studies have shown that GM-CSF induces tyrosine phosphorylation and activation of the JAK2 protein tyrosine kinase.11,41-43 Activation of this kinase results in the subsequent phosphorylation and activation of STAT5 and, in some cases, other members of the STAT family of transcription factors.41,44 45 Activation of STAT proteins is reflected in their ability to dimerize and translocate to the nucleus, where they stimulate gene transcription by binding to specific DNA sequences. We therefore examined nuclear extracts from the factor-independent FDC-P1 cells for the presence of STAT DNA-binding activity by performing EMSAs. All of these extracts contained a protein complex that specifically bound to a β-casein oligonucleotide probe containing a DNA-binding motif for STAT1, STAT3 and STAT5 (Fig 5). As expected, this DNA-binding activity was also induced in cells expressing wild-type hGMR that had been stimulated with hGM-CSF (lanes 1 and 3), but was absent in unstimulated cells (lane 2). Furthermore, the level of induction in hGM-CSF–stimulated cells expressing the hGMR was similar to that seen in factor-independent cells expressing the constitutively active mutants.
Constitutive STAT activation in factor-independent FDC-P1 cells expressing constitutive hβc mutants. Nuclear extracts prepared from factor-independent FDC-P1 cells expressing the indicated hβc mutants were subjected to EMSA using a β-casein promoter oligonucleotide probe. Also shown are FDC-P1 cells expressing wild-type hGMR that were either continuously cultured in 1 ng/mL hGM-CSF (lane 1) or starved of growth factor overnight and incubated without (lane 2) or with (lane 3) 50 ng/mL hGM-CSF for 10 minutes. The DNA-binding complexes are marked by an arrow.
Constitutive STAT activation in factor-independent FDC-P1 cells expressing constitutive hβc mutants. Nuclear extracts prepared from factor-independent FDC-P1 cells expressing the indicated hβc mutants were subjected to EMSA using a β-casein promoter oligonucleotide probe. Also shown are FDC-P1 cells expressing wild-type hGMR that were either continuously cultured in 1 ng/mL hGM-CSF (lane 1) or starved of growth factor overnight and incubated without (lane 2) or with (lane 3) 50 ng/mL hGM-CSF for 10 minutes. The DNA-binding complexes are marked by an arrow.
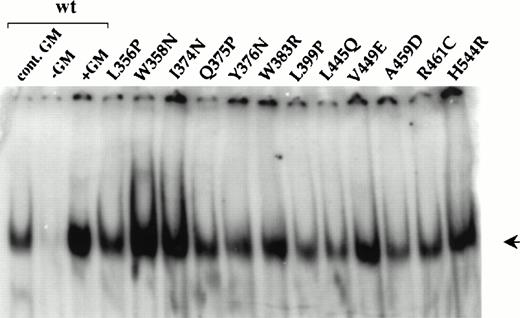
Constitutive activation of STAT DNA-binding activity by cytokine receptors is believed to be a direct consequence of phosphorylation by activated JAKs.46 Thus, the constitutive STAT activation seen in FDC-P1 cells expressing each of the mutants (Fig 5) implies that JAK2 is also constitutively activated. To confirm this, we analyzed JAK2 tyrosine phosphorylation in cells expressing each of five different mutants (I374N, V449E, L356P, L399P, and H544R) and, as a control, wild-type hβc plus hGMRα. These mutants were chosen as they represent different classes based on their location within hβc, tyrosine phosphorylation, and responsiveness to hGM-CSF plus hGMRα (see also Table 1). Detection of JAK2 tyrosine phosphorylation proved technically difficult in these cells and required the use of large numbers of cells, inhibition of cellular phosphatase activity by vanadate pretreatment, and a very sensitive two-layer immunodetection protocol. As shown in Fig 6, JAK2 phosphorylation was undetectable in factor-deprived cells expressing wild-type hβc plus α, but was readily detected in response to acute or continuous stimulation by hGM-CSF. Figure 6 also shows that, in cells expressing each of the five mutants tested, JAK2 was constitutively tyrosine phosphorylated in the absence of GM-CSF.
Activation of JAK2 by wild-type and constitutively active hβc mutants. Lysates from FDC-P1 cells expressing wild-type hβc plus hGMR or the indicated constitutive mutants were subject to immunoprecipitation with anti-JAK2 antibodies followed by immununoblotting with (A) antiphosphotyrosine or (B) anti-JAK2 antibodies. Cells expressing wild-type hβc plus hGMR were either factor-deprived (−GM), restimulated for 10 minutes (+GM), or grown continuously in hGM-CSF (cont. GM). Because the loading of JAK2 for the I374N and V449E mutants was higher than that for the other samples in the upper panels, a second experiment showing these two mutants (plus wild-type controls) is shown in the lower panels.
Activation of JAK2 by wild-type and constitutively active hβc mutants. Lysates from FDC-P1 cells expressing wild-type hβc plus hGMR or the indicated constitutive mutants were subject to immunoprecipitation with anti-JAK2 antibodies followed by immununoblotting with (A) antiphosphotyrosine or (B) anti-JAK2 antibodies. Cells expressing wild-type hβc plus hGMR were either factor-deprived (−GM), restimulated for 10 minutes (+GM), or grown continuously in hGM-CSF (cont. GM). Because the loading of JAK2 for the I374N and V449E mutants was higher than that for the other samples in the upper panels, a second experiment showing these two mutants (plus wild-type controls) is shown in the lower panels.
Biological Activity of hβc Mutants in BAF-B03 Cells
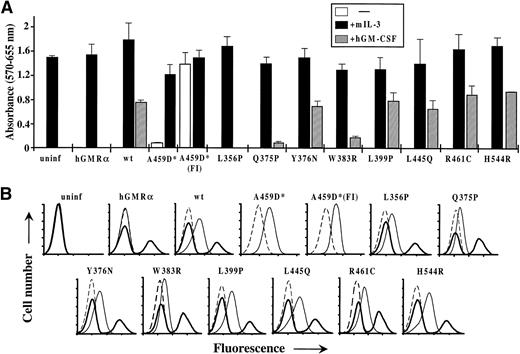
We have previously shown that extracellular hβc mutants, while constitutively active in FDC-P1 cells, are unable to confer factor independence on murine IL-3–dependent BAF-B03 cells,24,28whereas the V449E transmembrane domain mutant confers factor independence on both cell types.24 To examine whether constitutive activation of the extracellular, transmembrane, and cytoplasmic domain mutants identified in this report was also cell type-specific, retroviruses encoding the wild-type and mutant forms of hβc were used to infect BAF-B03 cells. For mutants containing multiple amino acid substitutions at a given residue, the more strongly activating mutant, as determined by the growth rates of factor-independent FDC-P1 cells (see Fig 2A), was used. Figure 7A shows that only the transmembrane domain A459D mutant could confer factor independence on BAF-B03 cells, as indicated by proliferation assays and prolonged monitoring of liquid cultures without growth factor (data not shown), even though each mutant was expressed on the surface of infected cells (Fig 7B). Conditioned medium from these cells again contained no detectable autocrine growth factor (data not shown).
Biological activity of hβc mutants in BAF-B03 cells. (A) Factor-dependent and -independent proliferation of BAF-B03 cells infected with retroviruses encoding the indicated β subunits and subsequently superinfected with a retrovirus encoding the hGMR subunit. Proliferation assays were performed using triplicates of 5 × 103 cells in the presence of mIL-3 or hGM-CSF or in the absence of either factor, as indicated. The asterisk by A459D indicates that, in this case, the cells were not superinfected with the hGMR retrovirus. A459D(FI) represents a population of A459D-infected cells that were selected for factor-independent growth before analysis. For comparison, assays of uninfected BAF-B03 cells and cells infected with the hGMR virus alone are shown. (B) Flow cytometric analysis of hβc and hGMR expression on the BAF-B03 cells used in (A). In each case, the cells infected with the indicated β subunits were stained with an irrelevant control antibody (dashed line), an anti-hβc antibody (thin solid line), and an anti-hGMR antibody (thick solid line) by standard indirect immunofluorescence. Axes are as for Fig2B.
Biological activity of hβc mutants in BAF-B03 cells. (A) Factor-dependent and -independent proliferation of BAF-B03 cells infected with retroviruses encoding the indicated β subunits and subsequently superinfected with a retrovirus encoding the hGMR subunit. Proliferation assays were performed using triplicates of 5 × 103 cells in the presence of mIL-3 or hGM-CSF or in the absence of either factor, as indicated. The asterisk by A459D indicates that, in this case, the cells were not superinfected with the hGMR retrovirus. A459D(FI) represents a population of A459D-infected cells that were selected for factor-independent growth before analysis. For comparison, assays of uninfected BAF-B03 cells and cells infected with the hGMR virus alone are shown. (B) Flow cytometric analysis of hβc and hGMR expression on the BAF-B03 cells used in (A). In each case, the cells infected with the indicated β subunits were stained with an irrelevant control antibody (dashed line), an anti-hβc antibody (thin solid line), and an anti-hGMR antibody (thick solid line) by standard indirect immunofluorescence. Axes are as for Fig2B.
The I374N mutant, although not constitutively active in BAF-B03 cells, is still able to form a high-affinity receptor and deliver a proliferative signal, in the presence of hGM-CSF, when coexpressed with the hGMRα subunit in BAF-B03 cells.28 In contrast, we previously described two other extracellular mutants, L356N and W358N, which, although also constitutively active in FDC-P1 cells, failed to form a high-affinity receptor complex with hGMRα in BAF-B03 cells.28 To examine the ability of the novel hβc mutants to generate a proliferative signal as part of the hGMR complex, BAF-B03 cells expressing these mutants were superinfected with a retrovirus carrying the wild-type hGMRα subunit. After selection for puromycin-resistant hGMRα infectants, flow cytometric analysis indicated that infected cells efficiently coexpressed both subunits (Fig 7B). All mutants, except for the L356P mutant, were able to deliver a proliferative signal in response to 1 ng/mL hGM-CSF, as shown by proliferation assays (Fig 7A), or prolonged monitoring of liquid cultures in 0.1 or 1 ng/mL hGM-CSF (data not shown). We believe the failure of L356P to allow hGM-CSF–dependent growth is probably due to an inability to interact productively with hGM-CSF, because we have previously shown that another mutant with a substitution at the same site (L356N) neither responds to hGM-CSF nor forms a high-affinity receptor complex.28
DISCUSSION
Location of Constitutive Mutations in hβc
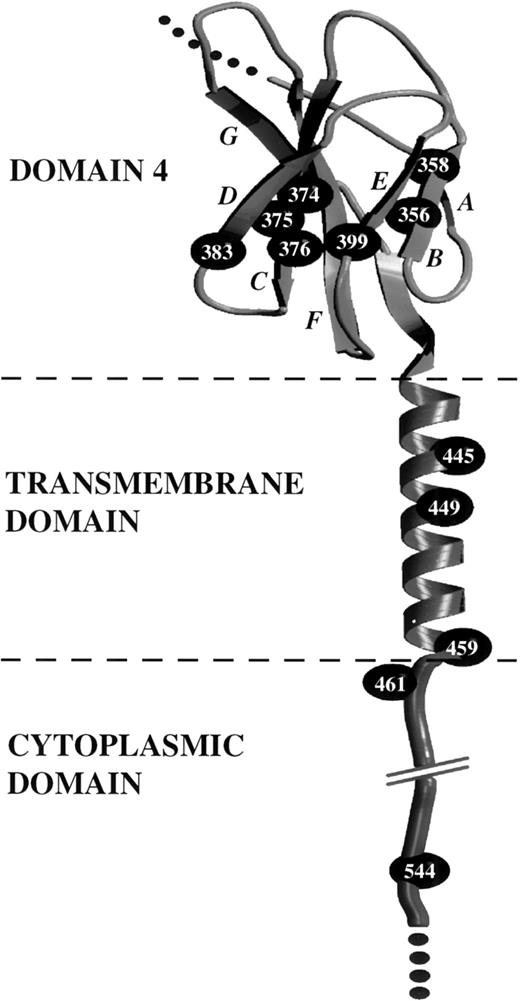
Sequence analysis indicated that the constitutive mutations are located exclusively in a central region of hβc that spans the extracellular, transmembrane, and cytoplasmic domains (Table 1 and Fig 8). The extracellular mutations reported here and in our previous studies24,28 are all clustered in the membrane-proximal domain (domain 4) of hβc, with several resulting in amino acid substitutions at residues in the B (L356P) and C (Q375P and Y376N,D,S) β-strands of domain 4, and two other mutations affecting residues in the D (W383R) and E (L399P) β-strands (Table 1 and Fig 8). Notably, our previous sequence alignment of domain 4 of hβc with the corresponding domain of other cytokine receptors indicates that the residues in hβc targetted for oncogenic activation are highly conserved (see Fig 1B in Jenkins et al28), suggesting that the homologous residues in other cytokine receptors may be targets for constitutively activating mutations.
Location of constitutive mutations in the extracellular domain 4, transmembrane domain, and cytoplasmic domain of hβc. Determination of a molecular model of domain 4 of hβc has been described previously.28 The transmembrane sequence of hβc was joined manually and an Indigo2 computer (Silicon Graphics, Mountain View, CA) was used to run the molecular modelling programs Insight II and Discover (Molecular Simulations Inc, San Diego, CA). Manual and automated methods were used to select an appropriate -helical conformation for the transmembrane region, and the model was evaluated for stereochemical parameters. The model of the hβc domain 4 and transmembrane domain is presented in cartoon form, using Molscript74 and Raster3D.75 β-strands are indicated by arrowed ribbons and italicized letters. The cytoplasmic domain is depicted in an arbitrary conformation and is shown only to illustrate the location of cytoplasmic mutations. -Carbon atoms of residues targetted for constitutive mutations are represented by CPK spheres.
Location of constitutive mutations in the extracellular domain 4, transmembrane domain, and cytoplasmic domain of hβc. Determination of a molecular model of domain 4 of hβc has been described previously.28 The transmembrane sequence of hβc was joined manually and an Indigo2 computer (Silicon Graphics, Mountain View, CA) was used to run the molecular modelling programs Insight II and Discover (Molecular Simulations Inc, San Diego, CA). Manual and automated methods were used to select an appropriate -helical conformation for the transmembrane region, and the model was evaluated for stereochemical parameters. The model of the hβc domain 4 and transmembrane domain is presented in cartoon form, using Molscript74 and Raster3D.75 β-strands are indicated by arrowed ribbons and italicized letters. The cytoplasmic domain is depicted in an arbitrary conformation and is shown only to illustrate the location of cytoplasmic mutations. -Carbon atoms of residues targetted for constitutive mutations are represented by CPK spheres.
Within the remainder of hβc, two transmembrane domain residues, in addition to the previously identified Val449 residue (V449E24), were identified as targets for constitutive mutations. Leu445 was replaced by Gln (L445Q), and Ala459 was replaced by Asp or Ser (A459D,S). Finally, mutations at two positions within the cytoplasmic domain of hβc, Arg461, replaced by Cys and His (R461C,H), and His544, replaced by Arg (H544R), also resulted in constitutive activity. The locations of the cytoplasmic mutations are likely to indicate key roles for the respective regions in receptor signaling, whereas the transmembrane mutants may be somewhat more adventitious and reflect a role for receptor dimerization (see below).
Mechanisms of Activation
Extracellular mutations.
The observation that all extracellular mutants identified in this study, as well as previous studies,24,28 are unable to confer factor independence on BAF-B03 cells (Fig 7) is consistent with a common mechanism of activation by these extracellular mutations. Although this mechanism is not clearly understood, we have previously suggested that the constitutive activity of extracellular hβc mutants may be dependent on the presence of cell type-specific signaling molecules.24 27 Our recent studies have shown that the murine GMRα subunit is one such molecule and, when introduced into BAF-B03 or CTLL-2 cells along with the I374N mutant, allows factor-independent proliferation (B.J.J., F. Le, and T.J.G., manuscript submitted).
Notably, with each of the extracellular mutants, receptor activation has resulted from amino acid substitutions that are predicted to severely disrupt the normal β-sheet structure of domain 4. Indeed, our earlier studies suggested that disruption of interactions between β-strands B and C by constitutive mutations at positions 356, 358 (on strand B), and 374 (on strand C) leads to receptor activation.28 The fact that such mutations at various locations within domain 4 all lead to receptor activation may indicate that activation results from disruption or relaxation of a constrained conformation of domain 4. This, in turn, may relieve an inhibitory intramolecular or intermolecular interaction and allow assembly of an active receptor complex. In agreement with such a model, several studies have demonstrated that the extracellular membrane-proximal domains of other cytokine receptors (equivalent to domain 4 of hβc) play a regulatory role in mediating interactions between receptor subunits.12,47,48 Thus, it is possible that the conformational change induced in domain 4 of hβc by the constitutive extracellular mutations mimicks the effect of ligand binding to the normal receptor. Indeed, crucial regions involved in ligand binding have been identified in domain 4 of hβc (reviewed in Bagley et al23).
Transmembrane mutations.
We have previously isolated a point mutation in the transmembrane domain of hβc (V449E) that confers factor independence on FDC-P1 and BAF-B03 cells.24 The V449E mutation is similar to the constitutive mutation found in the transmembrane domain of the Neu/c-erbB2 receptor tyrosine kinase that stimulates receptor homodimerization.25,26 By analogy, it is likely that V449E generates ligand-independent proliferative signals by inducing constitutive β subunit homodimerization. The exact mechanism by which the mutation in Neu promotes receptor dimerization is still unclear. It has been proposed that introduction of strongly polar or hydrophilic residues within the hydrophobic transmembrane environment induces hydrogen bonding between receptor subunits, thus enhancing receptor dimerization.49 Consistent with this finding, the residues substituted at Leu445 (Gln), Val449 (Glu), and Ala459 (Asp and Ser) contain polar side-chains that could participate in subunit-subunit hydrogen bonding. We might, therefore, expect the constitutive mutations at the Leu445, Val449, and Ala459 residues to act by a similar mechanism. However, the inability of the L445Q mutant to confer factor independence on BAF-B03 cells (Fig 7) suggests that it is activated by a different mechanism, possibly by inducing association with cell type-specific signaling molecules in a manner similar to that proposed for the extracellular mutants.24,27 28 Alternatively, the inactivity of L445Q could be attributed to a quantitative phenomenon whereby dimerization, and therefore the subsequent signal, is weaker than that of the other mutants and below a threshold required for proliferation in BAF-B03 cells, but not FDC-P1 cells.
Cytoplasmic mutations.
To our knowledge, this is the first report of mutations within the cytoplasmic domain of a cytokine receptor that results in constitutive activation. Accordingly, it is difficult at the moment to envisage how the cytoplasmic mutations in hβc (R461C,H and H544R) activate the receptor. The Arg461 residue was originally assigned to the transmembrane domain of hβc2; however, our own analysis of the hβc sequence with a transmembrane domain prediction program (Tmpred50) indicated that this residue lies outside of the transmembrane domain of hβc and is predicted to be the first cytoplasmic residue. Interestingly, the Cys and His residues substituted at this position are less hydrophilic than the wild-type Arg and, therefore, may only weakly anchor the transmembrane domain in its normal position. In such a scenario, one might speculate that the increased flexibilty or length of the transmembrane domain may relieve an inhibitory constraint that exposes a structure within hβc involved in inter-subunit interactions.
The H544R mutation is located within the box 2 motif that is loosely conserved among cytokine receptors.23,51 The exact role of this motif in mitogenesis is somewhat controversial52-56and possibly reflects the differential requirement of this motif for proliferative signaling among cytokine receptors. Nonetheless, impaired mitogenesis of several cytokine receptors that either lack or contain mutations in this motif correlates with a loss in JAK association and activation,53,56,57 suggesting that this motif plays a role in JAK activation. Given the general requirement of cytokine receptors for JAK association and activation (reviewed in Ihle40), the H544R mutation may alter the conformation of the membrane-proximal region and enhance JAK2 association and/or activation. Alternatively, this mutation may trigger receptor activation by facilitating dimerization of the cytoplasmic domains of β subunits in a manner similar to a mutation (V559G) in the membrane-proximal cytoplasmic domain of the c-kit receptor tyrosine kinase, which stimulates constitutive receptor homodimerization.58
Biochemical Consequence of Constitutive Mutations in FDC-P1 Cells
β subunit phosphorylation.
One of the early events indicative of hGMR activation in response to GM-CSF binding is tyrosine phosphorylation of hβc.5,7Phosphorylation of receptor tyrosine residues facilitates binding and activation of signaling molecules that link the receptor to signaling pathways involved in various cellular responses, including survival and proliferation. For example, mutation of a key site for hβc phosphorylation, Tyr750, severely compromised both tyrosine phosphorylation of hβc and the survival of cells expressing this mutant under low serum conditions.59 Our results suggest that tyrosine phosphorylation of major sites in hβc is not essential for either constitutive or GM-CSF–induced survival and proliferation, at least in serum-containing cultures. None of the extracellular mutant β subunits, or the L445Q transmembrane mutant or the H544R cytoplasmic mutant, were constitutively phosphorylated in the absence of hGM-CSF (Fig 3A). Moreover, only the I374N, Q375P, Y376N, W383R, and L445Q mutants (and, as expected, wt hβc) displayed hGM-CSF–induced tyrosine phosphorylation when coexpressed with hGMRα. This is somewhat surprising, considering that all of these mutants, with the exception of L356P and W358N, can confer hGM-CSF growth responsiveness on BAF-B03 cells when coexpressed with hGMRα (Fig 7 and Jenkins et al28). Both V449E and A459D transmembrane domain mutants, and the R461C cytoplasmic mutant, were constitutively phosphorylated. Interestingly, constitutive phosphorylation of the two transmembrane mutants correlated with their ability to confer factor independence on BAF-B03 cells, thus providing further evidence for a similar mechanism of activation by these two mutants.
MAP kinase, STAT, and JAK activation.
In contrast to the differential tyrosine phosphorylation of the mutant β subunits, all factor-independent FDC-P1 cell lines exhibited a similar level of constitutive ERK1/2 MAP kinase activation that resembled that induced by the wild-type hGMR in response to hGM-CSF (Fig 4A). Moreover, we have shown that, with all of the mutants tested, JAK2 is constitutively tyrosine phosphorylated at a level similar to that seen in cells grown continuously in GM-CSF. These results imply that β subunit phosphorylation at the major site(s) detected is not essential for the activation of the Ras-Raf-MAP kinase pathway, although by analogy with other cytokine receptors, JAK2 activation may be.60 Other mechanisms may therefore exist that link hβc to this pathway in the absence of receptor tyrosine phosphorylation. These could be mediated by JAK2, which can link the receptor to this pathway indirectly through its asssociation with the SHP-2 phosphatase61 or directly through its association with Raf-1,62 thus bypassing the requirement for receptor phosphotyrosines. Alternatively, it is conceivable that mutants which do not exhibit hβc tyrosine phosphorylation instead phosphorylate tyrosine residues on other molecules and that the resultant phosphotyrosines act as binding sites for components of the Ras-Raf-MAP kinase pathway such as Shc or Grb2. It is tempting to speculate that the endogenous murine GMR/IL-3R β subunits might fulfil such a role. However, the ability of the I374N mutant to confer factor independence on CTLL-2 cells in the presence of the murine GMRα subunit, as mentioned above (B.J.J., F. Le, and T.J.G., manuscript submitted), renders this less likely, because CTLL-2 cells do not express any components of the murine GMR/IL3R complex. Furthermore, our results also suggest that activation of MAP kinase may be important for these mutants to stimulate proliferation. Indeed, preliminary data suggest that ERK1/2 MAP kinases are constitutively activated in BAF-B03 cells only by hβc mutants able to confer factor independence on these cells (T.J.B., B.J.J., T.J.G., unpublished data). This is consistent with previous studies that have observed constitutive activation of the Ras-Raf-MAP kinase pathway in spontaneously derived factor-independent variants of human TF-1 cells and factor-independent BAF-B03 cells expressing a constitutively active c-Mpl mutant.63 64
We also demonstrated that all factor-independent FDC-P1 cell populations exhibited constitutive STAT DNA-binding activity (Fig 5), which is both indicative of and consistent with constitutive JAK2 activation. Whereas activation of STAT5 has predominantly been associated with the GMR/IL3R (eg, Mui et al8), only a proportion of each DNA-protein complex was supershifted with an anti-STAT5 antibody, and the extent of these supershifts varied among the cell populations (data not shown). Therefore, it is possible that other STAT proteins may be present in these constitutive DNA-binding complexes. Indeed, activation of STAT1, STAT3, and STAT5 in response to GM-CSF stimulation has been reported in various hematopoietic cells.8,41 65
Recently, conflicting reports have emerged as to the role of STAT5 in cytokine-induced mitogenic signaling.66-71 However, constitutive activation of STAT5 has been observed in factor-independent hematopoietic cell lines.63 Together with our data, this is consistent with a role for STAT5 activation in cell proliferation; on the other hand, constitutive STAT5 activation may simply reflect constitutive JAK2 activation.
Induction of other signals, eg, c-myc, is also likely to be required for a proliferative response. In this respect, we are currently investigating the activation state of other signaling molecules implicated in cytokine-induced proliferation by the hβc mutants.
Conclusions
In this report we have coupled saturation mutagenesis with a simple retroviral expression cloning strategy to comprehensively screen the entire hβc molecule for constitutive mutations. The identification of mutations clustered exclusively within the membrane-proximal extracellular domain, transmembrane domain, and membrane-proximal region of the cytoplasmic domain of hβc provides new insights into the role these regions in hβc and, by analogy, other members of the cytokine receptor family play in receptor activation. We observed that most of these mutants exhibited cell type-specific differences in their constitutive activity, which suggests the involvement of different mechanisms in the activation of hβc. Consistent with this, only some mutants were constitutively tyrosine phosphorylated. Intriguingly, this latter observation also dissociates receptor tyrosine phosphorylation from proliferative signaling, JAK2 activation, ERK MAP kinase, and STAT activation and most likely reflects the emerging theme of redundancy in mechanisms underlying activation of downstream signaling molecules.
ACKNOWLEDGMENT
The authors are grateful to Sun Qiyu and Angel Lopez for supplying their antireceptor antibodies, Chris Bagley for advice and assistance with molecular modelling, and our various colleagues who generously supplied us with growth factors. We thank Angel Lopez, Mathew Vadas, Richard D’Andrea, and Leonie Ashman for critical reading of this manuscript; Alan Bishop and Judy Haywood for assistance with flow cytometry; and Arthur Mangos for automated sequencing analyses.
Supported by a research grant from the National Health and Medical Research Council (NHMRC) of Australia. T.J.G. is a Senior Research Fellow of the NHMRC. B.J.J. is the recipient of a Dawes postgraduate scholarship from the Royal Adelaide Hospital.
Address reprint requests to Thomas J. Gonda, PhD, Hanson Centre for Cancer Research, Institute of Medical and Veterinary Science, Frome Road, Adelaide, South Australia 5000, Australia; e-mail:Tom.Gonda@imvs.sa.gov.au.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal