Abstract
The Notch signaling system regulates proliferation and differentiation in many tissues. Notch is a transmembrane receptor activated by ligands expressed on adjacent cells. Hematopoietic stem cells and early progenitors express Notch, making the stromal cells which form cell-cell contacts with progenitor cells candidate ligand-presenting cells in the hematopoietic microenvironment. Therefore, we examined primary stromal cell cultures for expression of Notch ligands. Using reverse transcription-polymerase chain reaction, in situ hybridization, immunohistochemistry, and Western blotting, we demonstrate expression of Jagged 1 in primary stromal cultures. To investigate if the stromal expression of Jagged 1 has functional effects on hematopoietic progenitors, we cultured CD34+, c-kit+ hematopoietic progenitor cells derived from the aorto gonadal mesonephros region of day 11 mouse embryos on the Jagged 1− stromal cell line S17 and on S17 cells engineered to express Jagged 1. The presence of Jagged 1 increased the number of colonies formed in subsequent methylcellulose culture fourfold. Larger increases in colony numbers were observed under the same culture conditions with CD34+, c-kit+ hematopoietic progenitor cells derived from d11 fetal liver. These results obtained in vitro table Jagged 1 as a candidate regulator of stem cell fate in the context of stromal microenvironments in vivo.
© 1998 by The American Society of Hematology.
THE NOTCH SIGNALING SYSTEM is highly conserved from Drosophila to vertebrates and regulates cell differentiation and proliferation during development and in proliferating adult tissues.1 The Notch receptor is a transmembrane glycoprotein that binds to ligands expressed on adjoining cells. Once ligand is bound, signal transduction takes place, resulting in the transcription of nuclear target genes, including transcription factors. The net effect of Notch activation is that the cell becomes refractory to differentiation signals. This effect is transient and reversible; once Notch activation ceases the cell is able to differentiate.2
Four different Notch genes have been identified in mice and humans, two of which are known to be expressed in the hematopoietic system.3-7 Notch 1 has been shown to be expressed in CD34+ lineage-negative hematopoietic stem cells in humans.8 Expression has also been found in CD34+ early progenitor cells expressing lineage markers,8 in the spleen, on thymic T cells and peripheral blood lymphocytes, and in a B-cell lymphoma line, FL18.3,4,9 Notch 2 is expressed in the spleen of adult mice and a limited analysis of hematopoietic cell lines has shown expression in the multilineage hematopoietic progenitor cell line FDCP-mix A4, and the myeloid progenitor cell line 32D.7 10
Dysregulation of the balance between self-renewal and differentiation in hematopoiesis is an essential feature of leukemogenesis. In adult T-cell lymphoblastic leukemia a t(7;9)(q34;q34.3) chromosomal translocation has been found to result in the expression of a truncated Notch 1 comprising the transmembrane and cytoplasmic domains. Similar Notch 1 mutations have been found in transgenic mice that develop spontaneous T-cell lymphoma,11 and feline leukemia virus isolates from thymic lymphomas in cats contain truncated Notch 2.12 Such truncated Notch mutants (Nintra) have been shown to be dominant activators of Notch signaling inDrosophila and Caenorhabditis elegans.13The introduction of Nintra into hematopoietic progenitors results in T-cell lymphoma when the cells are engrafted into lethally irradiated mice14 and expression of Nintra in T cells in transgenic mice results in altered T-cell differentiation.15 16 The effects of Nintra are not confined to T cells; it is also able to block cytokine-induced differentiation when introduced into the 32D myeloid progenitor cell line in vitro, with constitutively active Notch 1 and Notch 2 blocking differentiation in response to granulocyte colony-stimulating factor (G-CSF) and granulocyte-macrophage (GM)-CSF, respectively (ref 10 and references therein).
The role of Notch signaling in regulating the differentiation of normal hematopoietic stem cells and early progenitors remains unclear. The regulation of Notch activation in vivo is at least in part achieved by restricted expression of Notch ligand.1 The ligands described in mouse and human are Delta 1, highly homologous toDrosophila Delta, and Jagged 1 and 2, the homologues ofDrosophila Serrate.17-19 These transmembrane proteins have different patterns of expression in development and in adult tissues. Although many cells in a tissue may express Notch, only those adjoining cells expressing Notch ligand show activated Notch signaling.20 21
Hematopoietic stem cells and progenitors form cell-cell contacts with bone marrow (BM) stromal cells in vivo.22,23 In vitro such contacts appear essential for the maintenance of long-term BM cultures.24 Expression of ligand in stromal cells may provide one means of activation of the Notch receptor in the progenitor cell populations that are associated with stromal cells. Therefore, we examined the expression of Notch ligands in normal murine BM stromal cultures. We found Jagged 1 was expressed in murine stroma and went on to investigate if stromal expression of Jagged 1 is able to modulate the functional output of murine fetal hematopoietic progenitor cells in vitro.
MATERIALS AND METHODS
Cell culture.
Mouse femoral BM was obtained from 6- to 8-week-old C57/Bl6 mice and cultured for 6 to 8 weeks in α-minimal essential medium (GIBCO-BRL, Paisley UK) supplemented with 10% fetal bovine serum (FBS; GIBCO), 10% horse serum (TCS, Buckingham, UK), and 10−7mol/L hydrocortisone (Sigma, Poole, Dorset, UK).25 Cultures were maintained until no hematopoietic colonies were visible on examination with phase microscopy. Stromal cell lines PA6 and S1726,27 were cultured in α-minimal essential medium supplemented with 20% FBS. Cells were grown in a humidified 5% CO2 incubator at 37°C.
Antibodies and probes.
cDNA probes for Jagged 1 and Delta 1 cloned into Bluescript were a gift from Domingos Henrique and David Ish-Horowicz (Imperial Cancer Research Fund, London, UK). For immunohistochemistry, rat polyclonal antiserum raised against the cytoplasmic tail of human Jagged 1 (kind gift of Spyros Artavanis Tsakonas, Bayer Center for Molecular Medicine, Yale University, New Haven, CT) was used at a dilution of 1:50. A cocktail of rat IgG1, IgG2, and IgM immunoglobulins (Serotec, Kidlington, Oxford, UK) was used as a negative control at a concentration of 5 μg/mL. Biotinylated anti-rat IgG (Vector, Peterborough, UK) was used as a second layer antibody at a concentration of 5 μg/mL. For Western blotting a goat polyclonal antibody raised against amino acids 1200-1219 of the carboxy terminus of rat Jagged 1 was used at a concentration of 0.5 μg/mL (Santa Cruz Biotechnology, Autogen Bioclear UK, Carne, Wiltshire, UK); horseradish peroxidase–conjugated anti-goat secondary antibody was used at a concentration of 0.08 μg/mL (Santa Cruz).
Polymerase chain reaction (PCR) primers.
The forward and reverse primers used for PCR were as follows: Delta 1, CTGAGGTGTAAGATGGAAGCG and caactgtccatagtgcaatgg; Jagged 1, TGCAGCTGTCAATCACTTCG and CAGAATGACGCTTCCTGTCG; Jagged 2, AGAAGACTGCAACAGCTGCC and AACAGACCTGTGGAAGAGCC; β actin TGTATGCCTCTGGTCGTACC and CAACGTCACACTTCATGATGG. Primers for mouse β actin and mouse Delta 1 were designed from Genbank sequences to amplify bases 505 to 942 and 2172 to 2416, respectively. The mouse Jagged 1 primers were obtained by sequencing the partial cDNA fragment of mouse Jagged 1. The product corresponds to bases 1761 to 2122 of the rat sequence. Primers for Jagged 2 were designed to amplify bases 2523 to 3119 of the published rat sequence (U70050); the mouse PCR product displayed over 90% sequence identity with the corresponding region of rat Jagged 2.
Reverse transcription (RT)-PCR.
Total RNA was isolated using an RNeasy kit (Qiagen, Crawley, West Sussex, UK), incubated with DNAse I (Boehringer Mannheim, Lewes, East Sussex, UK), and reverse transcribed using AMV reverse transcriptase (Boehringer Mannheim) with an oligo dT primer. As a positive control, cDNA was also prepared from polyA RNA from a day 13.5 mouse embryo (gift from A Zelent, Institute of Cancer Research, London, UK). cDNA from 100 ng of total RNA or 10 ng polyA embryo RNA was amplified through 37 cycles comprising 94°C, for 20 seconds, 60°C for 30 seconds, and 72°C for 90 seconds in Perkin Elmer PCR buffer II with 2.0 mmol/L added MgCl2, with Amplitaq Gold DNA polymerase (Perkin Elmer, Warrington, UK), on a GeneAmp 9600 or 9700 thermal cycler (Perkin Elmer). Products were analyzed by electrophoresis through 2% agarose gels with ethidium bromide. Representative samples of each PCR product were gel purified using a Qiaquick gel isolation kit (Qiagen) and then direct sequenced using the appropriate PCR primer and an Amplitaq FS DNA sequencing kit (Perkin Elmer). Sequences were identified using Blastn searches at the National Center for Biotechnology Information (NCBI) database and in all cases corresponded with the predicted product.
In situ hybridization.
Radioactive in situ hybridization was performed essentially as described.28 Briefly, mouse stromal cells were cultured on glass slides (Lab-Tek TC; Nunc, Paisley, UK) as described above, until no hematopoietic colonies remained as assessed by phase microscopy. Cells were washed in phosphate-buffered saline (PBS), fixed in acetone, air dried, and stored at −70°C. Before hybridization, cells were fixed in 4% paraformaldehyde in PBS for 2 minutes at room temperature, washed in PBS, treated with 1 μg/mL proteinase K for 10 minutes at 37°C, acetylated, washed twice in 1× sodium citrate buffer (SSC), dehydrated through graded alcohols, and air dried.
Sense and antisense probes for Delta 1, Serrate 1, and β actin, labeled with [α35S] CTP (800 Ci/mmol/L; Amersham International, Amersham, Bucks, UK) were synthesized using a Maxiscript T3/T7 Kit (Ambion; AMS Biotechnology, Witney, Oxon, UK). Probes were dissolved in hybridization buffer at 3 × 104 counts per minute; hybridization buffer consisted of 1× salts (0.3 mol/L NaCl, 0.02 mol/L Tris pH 6.8, 5 mmol/L EDTA, 1 mmol/L sodium phosphate buffer pH 6.8, and 1× Denhardt’s solution [Sigma]), 10% dextran sulfate (Sigma), 50% deionized formamide (Sigma), 20 mmol/L dithiothreitol (DTT; IBI Kodak Ltd, Cambridge, UK), and 500 μg/mL yeast tRNA (Boehringer Mannheim). Twenty microliters of probe solution was applied to each slide and allowed to hybridize overnight at 55°C. Slides were washed with 1× salts, 50% formamide, 10 mmol/L DTT for 15 minutes and then 1 hour at 55°C, treated with 20 μg/mL RNAse A (Sigma) in 0.5 mol/L sodium chloride, 10 mmol/L Tris pH 7.5, 5 mmol/L EDTA for 30 minutes at 37°C, washed twice in 2× SSC at room temperature for 15 minutes per wash, once in 0.2× SSC at 50°C for 15 minutes, and then washed in 3 L of 0.2× SSC for 30 minutes at room temperature. After dehydration through graded alcohols containing 0.3 mol/L ammonium acetate, slides were dipped in emulsion (LM-1; Amersham) and then exposed for 2 to 3 weeks at 4°C. After development slides were stained with nuclear fast red (Vector).
Immunohistochemistry.
Cultured stromal cells were detached from the culture dish using cell dissociation buffer (Sigma), washed in PBS, resuspended in α-medium with 10% FBS, and then plated onto superfrost plus slides (BDH) previously coated with rat tail type I collagen (Sigma) by incubating the slides for 2 hours at 37°C with a 100 μg/mL solution of collagen type I in PBS. Cells were allowed to adhere for 6 hours at 37°C. Nonadherent cells were removed by washing with PBS. After fixation in 1% paraformaldehyde (Sigma) at room temperature for 10 minutes, slides were washed in PBS, incubated with 0.1% Triton X-100 (Sigma) in PBS for 10 minutes at room temperature, washed in PBS, incubated for 30 minutes at room temperature with PBS containing 0.1% hydrogen peroxide, washed in PBS, and incubated for 1 hour at room temperature with PBS containing 5% heat-inactivated sheep serum (PBS-HS). Cells were incubated overnight at 4°C with primary antibody diluted in PBS-HS, washed three times for 10 minutes each in PBS, incubated with secondary antibody diluted in PBS-HS for 1 hour at room temperature, and washed as before. Slides were developed using a Vector Stain ABC Elite kit with diaminobenzidine according to the manufacturer’s instructions. Slides were counterstained with hematoxylin.
Western blotting.
Cultured mouse stromal cells were boiled in sample buffer (10% glycerol, 1% sodium dodecyl sulfate [SDS], 5% β-mercaptoethanol, 50 mmol/L Tris pH 6.8), and electrophoresed through a 3% to 12% gradient SDS gel and transferred to Immobilon-P membrane (Millipore). The blot was then blocked with 5% milk in Tris-buffered saline containing 0.1% Tween 20 (TBS-T) for 2 hours at room temperature, incubated with primary anti-Jagged 1 antibody overnight at 4°C overnight, washed five times in TBS-T, incubated with secondary antibody for 1 hour at room temperature, washed five times in TBS-T, and visualized with an ECL or an ECL Plus kit (Amersham International) using Biomax MR film (Kodak).
Introduction of Jagged 1 into 3T3 and S17 cells.
Full-length human Jagged 1 cDNA in pBluescript (a gift from Spyros Artavanis Tsakonas, Bayer Center for Molecular Medicine, Yale University, New Haven, CT) was released by digestion with Not 1 and EcoR1 and inserted into the retroviral vector p50-M-X-neo (gift of J. Hanneman, Institute of Cancer Research). S17 and NIH 3T3 cells were stably transfected with this construct as follows. Cells were trypsinized, pelleted, and 107 cells resuspended in 0.4-mL culture medium. Cells were then incubated with 40 μg linearized DNA for 10 minutes at room temperature, electroporated at 260 V, 960 μF using a Biorad Gene Pulser (pulse time 27 ms) transferred into 30 mL culture media and split between three 9-cm dishes. After 20 hours the medium was replaced with fresh medium containing 1 mg/mL Geneticin (GIBCO-BRL). After 10 days’ culture, each plate contained 50 to 100 colonies. Plates were maintained independently to provide three pools of stably transfected cells. Expression of Jagged 1 was confirmed by Western blot as described above.
Sorting and culture of murine fetal hematopoietic progenitors.
Single-cell suspensions were prepared from aorto gonadal mesonephros (AGM) and fetal liver, dissected from day 11 postcoitum CBA × C57/BL10 embryos, stained for c-kit and CD34 and CD34+, c-kit+ cells sorted using a FACStar cell sorter (Beckton Dickinson). Fifty cells per well were plated onto irradiated stromal layers comprising either untransfected S17 cells or S17 cells transfected with Jagged 1 (pool 1). After 1 week of culture in the presence of interleukin-7 (IL-7), c-kit ligand, and 2-mercaptoethanol as previously described,29 all cells from each well were counted and passaged into three methlyl cellulose cultures (methocult M3430; Stem Cell Technologies Inc, Northampton, UK). The morphology and number of colonies comprising more than 50 cells was scored at 1, 2, 3, and 4 weeks.
RESULTS
Expression of Notch ligands in primary murine stromal cell cultures.
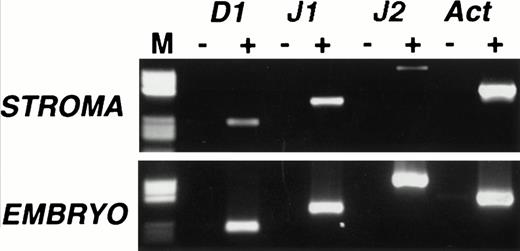
We began our study by examining Notch ligand expression in cultures of mouse primary stromal cells. Mouse femoral BM from 6- to 8-week-old mice was cultured for 6 to 8 weeks until no hematopoietic colonies were visible on examination with phase microscopy. Two independent stromal cultures were analyzed for expression of the Notch ligands Delta 1, Jagged 1, and Jagged 2 by RT-PCR, using conditions similar to those previously described for the analysis of Notch ligand expression.17 We used day 13.5 mouse embryo cDNA—in which all three ligands are known to be expressed—as a positive control, and the identity of all PCR products was confirmed by sequencing. Delta 1 and Jagged 1 mRNAs were detectable by RT-PCR in both stromal cultures, one of which is shown in Fig 1 (top panel). Trace levels of Jagged 2 were also detected in this culture (Fig 1) but absent from the other (not shown). The presence of Jagged 2 may therefore be due to contamination with hematopoietic cells because Jagged 2 is readily detectable in a variety of immunopurified myeloid and erythroid cells (P. Jones, unpublished observations, July 1997).
Expression of Notch ligands in primary mouse stromal cell cultures. RT-PCR analysis of Delta 1 (D1), Jagged 1 (J1), and Jagged 2 (J2) mRNAs in primary murine stromal cell culture. RT-PCR was performed on cultures that contained no visible hematopoietic elements (top panel) and on day 13.5 mouse embryo RNA (bottom panel), with (+) or without (−) reverse transcription. Products were analyzed on a 2% agarose gel. Lane M contains a pBR322 HaeIII marker (Sigma), the visible fragment sizes are 587, 540, 504, 458, 434, 267, 234, 213, 192, and 184 bp. All RT-PCR products were of the predicted size and their identity was further confirmed by sequencing.
Expression of Notch ligands in primary mouse stromal cell cultures. RT-PCR analysis of Delta 1 (D1), Jagged 1 (J1), and Jagged 2 (J2) mRNAs in primary murine stromal cell culture. RT-PCR was performed on cultures that contained no visible hematopoietic elements (top panel) and on day 13.5 mouse embryo RNA (bottom panel), with (+) or without (−) reverse transcription. Products were analyzed on a 2% agarose gel. Lane M contains a pBR322 HaeIII marker (Sigma), the visible fragment sizes are 587, 540, 504, 458, 434, 267, 234, 213, 192, and 184 bp. All RT-PCR products were of the predicted size and their identity was further confirmed by sequencing.
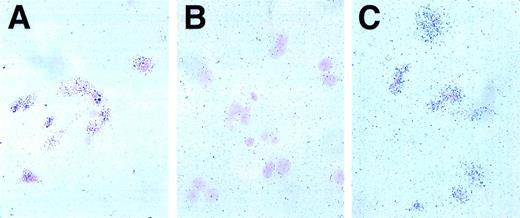
We next used radioactive in situ hybridization to gain an appreciation of the level and cellular distribution of Jagged 1 and Delta 1 expression in the primary stromal cell cultures. With an antisense Jagged 1 probe, there was a clear increase in grain deposition over the majority of cells (Fig 2C) whereas a sense (control) probe gave no such increase (Fig 2B). An antisense β actin probe provides a positive control for the technique (Fig 2A). There was no difference in the pattern of grain deposition between sense and antisense probes for Delta 1 (data not shown). Thus, Jagged 1 is transcribed in the majority of stromal cells in primary culture while Delta 1 transcription appears to be at a low level or confined to very few cells in the stromal population.
In situ hybridization for Jagged 1 in mouse stromal cell cultures. Radioactive in situ hybridization of primary mouse stromal cell cultures prepared as described in the text. The probe is visualized by grain deposition in the emulsion layer overlying the cells, which have been counterstained with nuclear fast red. (A) Positive control anti-sense actin probe, 1 week exposure. Grain deposition over cells in excess of background indicates RNA is intact. (B) Negative control, sense Jagged 1 probe, 3-week exposure. Grain deposition over cells is not increased over background, indicating no nonspecific hybridization of control probe. (C) Experimental sample, Jagged 1 anti-sense probe, 3-week exposure. Grain deposition over cells indicates Jagged 1 RNA is transcribed by the majority of stromal cells.
In situ hybridization for Jagged 1 in mouse stromal cell cultures. Radioactive in situ hybridization of primary mouse stromal cell cultures prepared as described in the text. The probe is visualized by grain deposition in the emulsion layer overlying the cells, which have been counterstained with nuclear fast red. (A) Positive control anti-sense actin probe, 1 week exposure. Grain deposition over cells in excess of background indicates RNA is intact. (B) Negative control, sense Jagged 1 probe, 3-week exposure. Grain deposition over cells is not increased over background, indicating no nonspecific hybridization of control probe. (C) Experimental sample, Jagged 1 anti-sense probe, 3-week exposure. Grain deposition over cells indicates Jagged 1 RNA is transcribed by the majority of stromal cells.
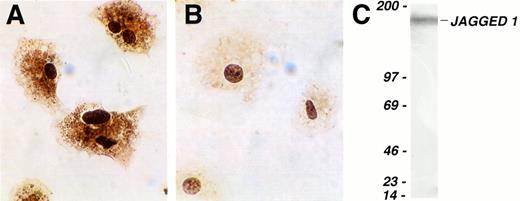
Finally, we investigated whether Jagged 1 mRNA was translated into protein in primary cultures of stromal cells. Immunohistochemistry using a polyclonal antibody raised against the cytoplasmic domain of human Jagged 1 gave staining clearly in excess of control in stromal cells (Fig 3A and B). However, because such staining may be partially caused by binding of epitopes in proteins other than Jagged 1, we confirmed Jagged 1 protein expression in extracts of mouse stromal cultures by Western blotting using a second polyclonal anti-Jagged 1 antibody. A single band was seen running at the predicted molecular weight for Jagged 1 of 170 kD (Fig 3C).
Expression of Jagged 1 protein in cultured mouse stromal cells. (A and B) Cultured primary mouse stromal cells were allowed to adhere to slides coated with type I collagen and then stained immunohistochemically with a rat anti–Jagged 1 polyclonal antiserum (A) or with control anti-rat Ig (B). Antibody binding was visualized by staining with diaminobenzidine, and the cells were counterstained with hematoxylin. (C) Western blot of cultured primary mouse stromal cell extract, which was separated on a 3% to 12% gradient SDS-polyacrylamide gel electrophoresis under reducing conditions, transferred, and probed with a goat anti-Jagged 1 antibody. The lefthand column shows the position of molecular weight markers (Amersham); the figures are molecular weights in kilodaltons. The single band detected at approximately 170 to 180 kD corresponds to Jagged 1.
Expression of Jagged 1 protein in cultured mouse stromal cells. (A and B) Cultured primary mouse stromal cells were allowed to adhere to slides coated with type I collagen and then stained immunohistochemically with a rat anti–Jagged 1 polyclonal antiserum (A) or with control anti-rat Ig (B). Antibody binding was visualized by staining with diaminobenzidine, and the cells were counterstained with hematoxylin. (C) Western blot of cultured primary mouse stromal cell extract, which was separated on a 3% to 12% gradient SDS-polyacrylamide gel electrophoresis under reducing conditions, transferred, and probed with a goat anti-Jagged 1 antibody. The lefthand column shows the position of molecular weight markers (Amersham); the figures are molecular weights in kilodaltons. The single band detected at approximately 170 to 180 kD corresponds to Jagged 1.
In summary, of the three Notch ligands examined, Jagged 1 is the major ligand expressed in primary stromal cells in culture as assessed by RT-PCR, in situ hybridization, immunohistochemistry, and Western blotting.
Functional effects of stromal Jagged 1 expression on murine fetal hematopoietic progenitors.
These results raise the possibility that the Notch receptors expressed on hematopoietic progenitors bind to Jagged on stromal cells, resulting in activation of the Notch signal transduction pathway. In other cell systems, Notch activation results in a transient block to differentiation and, consistent with this, constitutively active mutants of Notch are associated with leukemia and able experimentally to block G-CSF–induced differentiation of granulocytic cell lines (see introduction). We next performed functional experiments to explore the possibility that expression of the Notch ligand Jagged 1 on stromal cells could modulate the behavior of primary hematopoietic progenitor cells cultured in vitro.
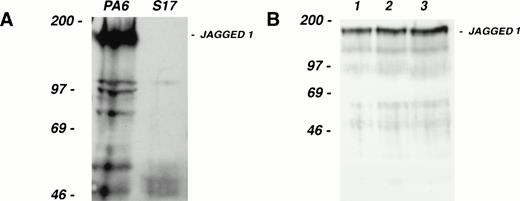
As a source of progenitor cells for our study, we have used CD34+, c-kit+ cells from the AGM region of the developing mouse embryo. This population contains the first detectable stem cells capable of long-term reconstitution of adult irradiated recipients.30 Consistent with this capacity, CD34+, c-kit+ AGM cells are multipotential in vitro (S. Delassus, unpublished observations, April 1997). The in vitro culture system used to support multilineage differentiation of AGM-derived CD34+, c-kit+ cells utilizes the stromal cell line S17 as one of its components.29 S17 do not express Jagged 1 mRNA as judged by Northern blotting (data not shown); Western blotting confirmed the absence of Jagged 1 protein expression in S17 (Fig 4A). We exploited the fact that S17 cells do not express Jagged 1 to assess the role of this Notch ligand in modulating hematopoietic progenitor cell behavior.
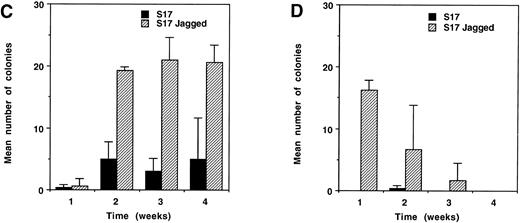
Colony-forming activity of fetal hematopoietic progenitors. (A) Western blot of cell extracts from the stromal cell lines PA6 (lefthand lane) and S17 (righthand lane), separated on a 6% SDS gel under reducing conditions, transferred, and probed with the same anti–Jagged 1 antibody used in Fig 3. Note that while PA6 expresses Jagged 1, S17 cells do not. (B) Western blot of cell extracts from three independent pools (lanes 1 through 3) of S17 cells stably transfected with a human Jagged 1–containing expression vector. (C and D) Colony production by fetal CD34+, c-kit+ hematopoietic progenitor cells from d11 AGM (C) or fetal liver (D) after culture for 1 week on irradiated wild-type or Jagged 1–transfected S17 cells. CD34+, c-kit+ cells from AGM and fetal liver were sorted and cultured on irradiated S17 cells or pool 1 of the Jagged 1–transfected S17 cells (S17 Jagged). The numbers of cells generated after 1 week of stromal culture were as follows: AGM on S17 = 104; AGM S17 Jagged = 104; fetal liver on S17 = 6 × 103; fetal liver on S17 Jagged = 4 × 104. All cells derived from each stromal culture were passaged into methylcellulose suspension cultures as described in the text. Colonies of over 50 cells were counted after 1, 2, 3, and 4 weeks in culture. The mean number of colonies formed per dish from triplicate dishes at each time point is shown; error bars show the standard deviation.
Colony-forming activity of fetal hematopoietic progenitors. (A) Western blot of cell extracts from the stromal cell lines PA6 (lefthand lane) and S17 (righthand lane), separated on a 6% SDS gel under reducing conditions, transferred, and probed with the same anti–Jagged 1 antibody used in Fig 3. Note that while PA6 expresses Jagged 1, S17 cells do not. (B) Western blot of cell extracts from three independent pools (lanes 1 through 3) of S17 cells stably transfected with a human Jagged 1–containing expression vector. (C and D) Colony production by fetal CD34+, c-kit+ hematopoietic progenitor cells from d11 AGM (C) or fetal liver (D) after culture for 1 week on irradiated wild-type or Jagged 1–transfected S17 cells. CD34+, c-kit+ cells from AGM and fetal liver were sorted and cultured on irradiated S17 cells or pool 1 of the Jagged 1–transfected S17 cells (S17 Jagged). The numbers of cells generated after 1 week of stromal culture were as follows: AGM on S17 = 104; AGM S17 Jagged = 104; fetal liver on S17 = 6 × 103; fetal liver on S17 Jagged = 4 × 104. All cells derived from each stromal culture were passaged into methylcellulose suspension cultures as described in the text. Colonies of over 50 cells were counted after 1, 2, 3, and 4 weeks in culture. The mean number of colonies formed per dish from triplicate dishes at each time point is shown; error bars show the standard deviation.
S17 cells were stably transfected with a human Jagged 1–containing expression vector and expression was confirmed by Western blotting (Fig4B). AGM regions were dissected from day 11 mouse embryos and CD34+, c-kit+ cells isolated by fluorescence-activated cell sorting. These AGM cells were cultured on wild-type or Jagged 1–expressing S17 stromal layers for 6 days before replating in methylcellulose cultures. Colony formation was monitored over a period of 4 weeks and the results obtained are presented in Fig4C. After 1 week in methylcellulose culture, few colonies were observed in either S17 or S17-Jagged samples. At the 2-, 3-, and 4-week timepoints, more colonies were observed in the S17-Jagged than the S17 samples. In the case of the 2- and 3-week time points, the increase in colony formation was highly significant (P < .01 by two-tailed t-test). However, at the latest (4-week) time point, the increase seen was less statistically significant (P = .08). The results indicate that colony-forming activity is significantly enhanced in the samples pre-exposed to Jagged 1–expressing stroma. The colonies produced in the methylcellulose cultures were large mixed-lineage and their morphology and kinetics of appearance are consistent with a high proliferative potential–colony-forming cells (HPP-CFC)–like potential31 as might be expected for primitive progenitors of AGM origin.30
Figure 4D shows the results of a similar analysis conducted using CD34+, c-kit+ progenitors derived from fetal liver. Few replated colonies were obtained from the S17 samples at any time point. Pre-incubation on S17-Jagged stroma resulted in considerably increased colony formation. Note that these colonies derived from fetal liver progenitors appeared earlier in the culture than those derived from AGM. The increase in the number of colonies seen after 1 week of methylcellulose culture was highly significant (P < .001 by two-tailed t-test). Enhanced colony formation in the S17-Jagged samples was also seen after longer periods of culture but overall colony numbers were lower, and it should be noted that the increase observed was no longer statistically significant. These kinetics of colony formation are consistent with the less primitive nature of the CD34+, c-kit+ progenitor population isolated from fetal liver.
DISCUSSION
Our results show that ligands for the Notch receptor, in particular Jagged 1, are indeed expressed by primary stromal cells in culture. Furthermore, we have conducted functional experiments which suggest that the stromal presentation of Jagged 1 ligand to hematopoietic progenitors modulates their behavior in vitro.
Confirming heterotypic activation of Notch by Jagged 1 in vivo is complex because of the numerous potential interactions between stromal cells of different types and between hematopoietic progenitors themselves. Given this cellular complexity in the BM it is hard to speculate as to the nature of the cell-cell interactions that might bring the Notch signaling pathway into play and sophisticated in vivo manipulations of these pathways will presumably be required to address this issue.
With regard to the apparent selective expression of Jagged 1 by stroma, it is important to note that the current survey has been limited to Notch ligands for which probes and reagents are readily available. Our results do not exclude the possibility that Notch ligands other than those investigated here may also be expressed by stromal cells. In this context it is worth noting that Jagged 1 mutations have recently been reported to be associated with Alagille syndrome, a rare human inherited disorder characterized by liver, skeletal, facial, eye, and cardiac defects, but apparently normal hematopoiesis.32,33However, the effects of the mutations on Jagged 1 function are unknown, and it is possible that expression of other Notch ligands in stroma compensates for the Jagged 1 deficiency. The recent isolation of the new Notch ligands Delta 2 in Xenopus and Delta 3 in mouse34 35 may be important in this regard.
We show here that culture on Jagged 1–expressing stroma increases colony formation by AGM– and fetal liver–derived hematopoietic progenitors. The exact nature of the hematopoietic progenitors in which Notch signaling is activated by Jagged 1 is not clear from these experiments. Cellular targets for Notch activation may be either the progenitors originally present in the AGM and fetal liver or a later population of progenitors derived from them during the 6 days of stromal culture, or both. The increase in colony-forming activity we observed in these experiments using normal hematopoietic progenitors is broadly speaking consistent with observations obtained using hematopoietic cell lines and published during revision of this manuscript. Thus, Li et al36 have shown that activation of an ectopically expressed Notch receptor on 32D cells by Jagged 1 blocks G-CSF–induced differentiation of this cell line. Also interesting in this context are the recent observations of Moore et al,37who have shown that expression of dlk in stromal cells promotes cobblestone area formation by stem/progenitor cells. Although dlk shares many structural features with Notch ligands such as EGF repeats, it is unlikely that it functions through Notch because it lacks the DSL domain shared by all authentic Notch ligands and thought to be necessary for interaction with the Notch receptor. In conclusion, our results raise the possibility that stromal cells contribute to maintaining hematopoietic cells in an undifferentiated or self-renewing state through activation of Notch signaling via Jagged 1. If so, such an interaction could well be exploited in the context of ex vivo expansion or maintenance of stem cell populations in vitro for clinical purposes.
NOTE ADDED IN PROOF
We would like to draw the reader’s attention to the recently published work of B. Varnum-Finney, et al (Blood 91:4084, 1998).
ACKNOWLEDGMENT
We thank Domingos Henrique, David Ish-Horowicz, and Spyros Artavanis-Tsakonas for reagents.
Supported by the Leukaemia Research Fund and the Institute of Cancer Research:Royal Cancer Hospital. P.J. acknowledges a Senior Clinical Fellowship at the Institute of Cancer Research.
Address correspondence to Tariq Enver, PhD, Section of Gene Function and Regulation, Chester Beatty Laboratories, Institute of Cancer Research, Fulham Rd, London, SW3 6JB, UK; email: <tariq@icr.ac.uk>.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal