Abstract
Leukemic lymphoblasts expressing the E2A-HLF oncoprotein possess wild-type p53 genes, but do not undergo apoptosis in response to DNA damage. Experimentally, E2A-HLF prevents apoptosis due to growth factor deprivation or γ-irradiation in interleukin-3 (IL-3)–dependent murine pro-B cells. To directly test the chimeric protein’s ability to abrogate p53-mediated cell death, we used mouse myeloid leukemia cells (M1p53tsval) that constitutively express a temperature-sensitive (ts) mutant p53 gene and undergo apoptosis when p53 assumes an active wild-type configuration. This effect is blocked by treatment with IL-6, which allows the cells to survive in culture despite wild-type p53 activation. We introduced E2A-HLF into M1p53tsval cells and found that they were resistant to p53-mediated apoptosis and that E2A-HLF effectively substituted for the survival functions of IL-6. The expression of p53-responsive genes such as p21 and Bax was upregulated normally, suggesting that E2A-HLF acts downstream of p53 to block execution of the p53-induced apoptotic program. NFIL3, a growth factor-regulated bZIP protein that binds to the same DNA-consensus site as E2A-HLF, delays apoptosis in IL-3–dependent pro-B cells deprived of growth factor. By contrast, in the present study, enforced expression of NFIL3 failed to protect M1p53tsval cells from p53-dependent apoptosis and actively antagonized the ability of IL-6 to rescue cells from that fate, consistent with its role as either a transcriptional repressor or activator, depending on the cell type in which it is expressed. We conclude that the E2A-HLF chimera abrogates p53-induced apoptosis in leukemic cells, possibly through the transcriptional modulation of cell death pathways that are activated by p53 in response to DNA damage.
© 1998 by The American Society of Hematology.
THE E2A-HLF FUSION gene, first cloned from leukemic pro-B lymphocytes with the t(17;19) translocation, encodes a chimeric protein that includes the 2 transactivation domains of E2A and the basic leucine zipper (bZIP) region of HLF.1,2 Results of programmed expression of a dominant-negative E2A-HLF protein in t(17;19)-positive human leukemic cells (UOC-B1 line) demonstrated that the activity of this fusion protein is required for the survival of these cells.3Similarly, in interleukin-3 (IL-3)–dependent murine pro-B cells (Baf-3 line), E2A-HLF blocked the apoptotic death that otherwise occurs when such cells are deprived of cytokine.3 Although unable to replace the proliferative signals provided by IL-3, E2A-HLF maintained Baf-3 cell viability in the absence of cytokine for up to 2 weeks.
During lymphocyte development, cytokine signals not only support the survival of lymphoid cells, but also regulate their differentiation and proliferation. In the aberrant progression to a leukemic state, lymphoblasts lose normal control of cellular homeostasis at multiple levels.4 One pathway that generally remains intact in childhood acute lymphoblastic leukemias is the p53-mediated response to DNA damage, apparently accounting for the rapid responsiveness of these leukemias to chemotherapy.5,6 However, in leukemias with the t(17;19), the malignant cells respond poorly to treatment, even though p53 is intact.7,8 Additionally, Baf-3 clones expressing E2A-HLF show a decrease in the rate of cell death after γ-irradiation in the absence of IL-3.3 These observations suggest that the E2A-HLF fusion protein interferes with apoptotic pathways mediated by p53.
NFIL3/E4BP4 (hereafter referred to as NFIL3) is a growth factor-regulated transcription factor related to E2A-HLF within the bZIP region that is normally expressed in hematopoietic cells. NFIL3 was initially identified by its ability to recognize a binding site in the adenovirus E4 promoter and independently shown to bind to a similar sequence motif in the human IL-3promoter.9,10 NFIL3 can function as either a repressor or an activator of gene expression, depending on the cellular context.9-11 Endogenous Nfil3 expression was shown to be upregulated at the level of transcription in murine pro-B cells stimulated with IL-3, making it a potential transcriptional effector downstream of IL-3–mediated survival signals in these cells.12 NFIL3 was recently shown to have anti-apoptotic properties similar to those of E2A-HLF in IL-3–dependent pro-B cells deprived of cytokine, although its ability to promote survival was not as pronounced as that of E2A-HLF.12
The M1 myeloid leukemia cell line provides a useful model system for studying the effects of E2A-HLF and NFIL3 on the p53-mediated cell death cascade. Parental M1 cells are null for p53, permitting the generation of a M1p53tsval subline carrying a temperature-sensitive mutant p53 transgene. These cells can be induced to undergo apoptosis when the p53 protein assumes its wild-type configuration at a temperature of 32°C.13 This effect is not preceded by G1-phase arrest and occurs at all phases of the cell cycle.14 The IL-6 cytokine is able to block p53-induced apoptosis in this system, leading us to consider that E2A-HLF might also abrogate the apoptotic effects of p53, as it did in murine pro-B lymphocytes dependent on IL-3.3
We report here that E2A-HLF effectively blocks the p53-mediated cell death cascade in the M1p53tsval murine myeloid leukemia cells. By contrast, NFIL3 had effects opposite to those of E2A-HLF, potentiating p53-induced apoptosis and overriding the protective effect of IL-6 in these cells. Because expression of wild-type p53 results in the activation of the p21 and Bax genes, whether or not E2A-HLF is present, we suggest that E2A-HLF selectively interferes with apoptotic pathways downstream of p53 target sites.
MATERIALS AND METHODS
Cells and culture conditions.
The generation of clones of the M1 murine myeloid leukemia cell line containing p53 temperature-sensitive (M1p53tsval) mutants has been described.13 Parental M1, M1p53tsval, and retrovirally infected cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% horse serum (GIBCO, Grand Island, NY) and grown at 37.5°C in 8% CO2-92% air. In experiments with recombinant IL-6 (Amgen, Thousand Oaks, CA), the cytokine was added at a concentration of 12.5 ng/mL. Viable cell counts were determined by trypan-blue dye exclusion in triplicate assays.
Construction of vectors and retrovirus production.
The human fusion gene E2A-HLF was subcloned into theXho I site of the SRaMSV-TkCD8 retroviral vector.15,16 The human NFIL3 cDNA was subcloned into the BgI II site of the same vector. Expression ofE2A-HLF and NFIL3 was driven by the long terminal-repeat promoter of Moloney murine leukemia virus. The gene encoding the mouse T-cell surface protein CD8 was expressed under control of the thymidine kinase promoter. For virus production, vectors were cotransfected with a plasmid encoding an amphotropic helper virus containing a defective virion-packaging (Ψ2) sequence in 293T cells.16 Culture supernatants containing retroviruses were harvested 48 to 72 hours after transfection and used to infect proliferating M1p53tsval cells. Infected cells were centrifuged and resuspended in 100 μL of phosphate-buffered saline (PBS) containing human γ-globulin (100 mg/mL) and incubated on ice for 30 minutes. After 2 washes with PBS, cells were resuspended in 50 μL of a titered excess of either rat antimouse CD8-monoclonal antibody LY2 (Pharmingen, San Diego, CA) or an isotype-matched control rat Ig IgG2a (Pharmingen) and incubated for 30 minutes on ice. After 2 washes with PBS, cells were resuspended in 50 μL of a titered excess of fluorescein-conjugated goat antirat Ig (BioSource International, Camarillo, CA) and incubated for 30 minutes on ice. After 1 wash with PBS, cells were resuspended in 1 mL medium containing 0.018 mg/mL of propidium iodide (PI). Cells expressing CD8 that excluded PI were sorted under sterile conditions with a FACStar Plus instrument equipped with a Clone-Cyt single-cell sorting attachment (Becton Dickinson, San Jose, CA) that distributed 1 cell into each well of a 96-well plate. Individual clones were then examined for expression of E2A-HLF and NFIL3 by Northern, electromobility shift assay (EMSA), and Western assays.
Terminal deoxynucleotidyl transferase (TdT) assay (TUNEL).
Cells (5 × 106) were fixed with 1% paraformaldehyde/PBS, followed by washing and fixation in cold 70% ethanol, and held overnight at −20°C. Each sample was divided into 2 equal aliquots and washed twice with PBS. Pellets were resuspended in 50 μL of reaction mixture (TdT buffer, CoCl2, and digoxigenin-11-dUTP), either with or without TdT (all reagents from Boehringer Mannheim, Indianapolis, IN), and incubated at 37°C for 30 minutes. Cells were then washed once with cold PBS, resuspended in 100 μL of a 1:10 solution of fluorescein-conjugated antidigoxigenin monoclonal antibody (Boehringer Mannheim) in PBS, and incubated in the dark for 30 minutes at room temperature. One milliliter of PBS/azide/bovine serum albumin (BSA) (2 mmol/L Na azide, 0.35% BSA) was added, and the cells were immediately centrifuged and resuspended in 0.1% Triton X-100/PBS. Cells were then vortexed and resuspended in PBS/azide/BSA/PI containing 62.5 μg/mL PI, DNAse-free RNase was added to each sample (final concentration, 0.05 mg/mL), and samples were incubated at room temperature for 30 minutes. Samples were filtered through 40-μ mesh and analyzed by flow cytometry. The degree of apoptosis in a sample was determined as that portion of cells having fluorescein fluorescence greater than background fluorescence present in the absence of TdT.
EMSA.
Nuclear extracts and gel shift assays were prepared as previously described.17 Anti-HLF-(C) and anti-NFIL3 rabbit sera were prepared from animals injected with recombinant human E2A-HLF or NFIL3 proteins. One microliter of antiserum per lane was used in antibody-perturbed gel shift assays. Signals were quantified with a phosphorimager (Molecular Dynamics, Sunnyvale, CA).
Western blot analysis.
Cells (1 × 107) were lysed in RIPA buffer (150 mmol/L NaCl, 1% NP40, 0.5% deoxycholate (DOC), 1% sodium dodecyl sulfate [SDS], 50 mmol/L Tris, pH 8) in the presence of protease inhibitors, and total cellular proteins were separated by SDS-gel electrophoresis. After wet electrotransfer onto nitrocellulose membranes (Millipore, Bedford, MA), immunoblotting was performed with anti-HLF-(C) antiserum. Blots were incubated with horseradish-conjugated antirabbit Ig secondary antibody and subjected to autoradiography with enhanced chemiluminescence (Amersham Life Sciences, Arlington Heights, IL).
Northern blot analysis.
Twenty micrograms of total RNA was subjected to denaturing agarose electrophoresis containing 1 mol/L formaldehyde and transferred to a nylon membrane (Stratagene, La Jolla, CA). Blots were then hybridized with a 32P-labeled full-length cDNA encoding humanNFIL3, murine Nfil3, Bcl-2, Bax, Bcl-xl, p21WAF1/CIP1, Ei24, or Gapdh. Signals were quantified with a phosphorimager.
Ribonuclease protection assay (RPA).
Twenty micrograms of total RNA isolated from E2A-HLF clone 25 and M1p53tsval control cells was hybridized with32P-labeled mApo-2 and mApo-3 probes (Riboquant Kit; Pharmingen). After overnight hybridization, the protected products were detected by electrophoresis in a 5% polyacrylamide gel. Levels of protected fragments were quantitated by analysis on a phosphorimager (Molecular Dynamics) and corrected for loading errors by comparison to levels of the housekeeping genes, Gapdh and L32.
Statistical analysis.
The results of cell viability assays, presented in Fig 2, were analyzed for statistical significance by use of a Wilcoxon exact test.18 Similar determinations were performed for the flow cytometric TUNEL assay with a Pearson’s χ2 test. Differences were considered significant if the P values were less than .017 (0.05/3), in that a Bonferroni adjustment was made to account for multiple comparisons with the same control group.
RESULTS
E2A-HLF blocks p53-mediated apoptosis.
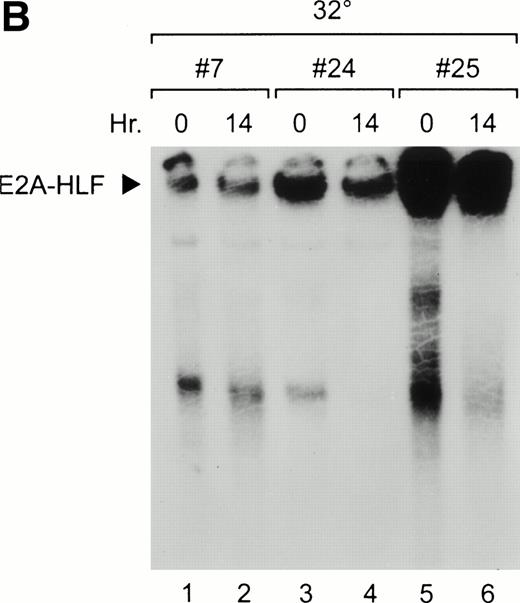
Three E2A-HLF-expressing M1tsp53val clones generated by retroviral infection were isolated and shown to express various levels of the chimeric protein by Western blot analysis (Fig 1A) and EMSA (Fig 1B). The E2A-HLF fusion protein expressed in these cells bound to the HLF consensus sequence probe, with the highest activity present in nuclear extracts from clone 25 and the lowest in those from clone 7 (Fig 1B, lanes 1, 2, 5, and 6). DNA-binding activity was unaffected 14 hours after a shift in temperature to 32°C, which induced a wild-type p53 conformation (lanes 2, 4, and 6).
Overexpression of E2A-HLF in M1p53tsvalcells. (A) Western blot analysis of 10 μg protein/lane blotted with HLF antisera showing variable levels of E2A-HLF expression in clones 7, 24, and 25 compared with a lack of expression in parental cells (P). (B) Nuclear lysates prepared from E2A-HLF clones 7, 24, and 25 and studied by EMSA using a 32P-labeled oligonucleotide probe containing the HLF consensus binding sequence and supershifted with an E2A antibody. Supershifted complexes containing E2A-HLF were stable at both 0 hours (mutant conformation of p53 at 37°C) and 14 hours after a shift in temperature (wild-type conformation of p53 at 32°C).
Overexpression of E2A-HLF in M1p53tsvalcells. (A) Western blot analysis of 10 μg protein/lane blotted with HLF antisera showing variable levels of E2A-HLF expression in clones 7, 24, and 25 compared with a lack of expression in parental cells (P). (B) Nuclear lysates prepared from E2A-HLF clones 7, 24, and 25 and studied by EMSA using a 32P-labeled oligonucleotide probe containing the HLF consensus binding sequence and supershifted with an E2A antibody. Supershifted complexes containing E2A-HLF were stable at both 0 hours (mutant conformation of p53 at 37°C) and 14 hours after a shift in temperature (wild-type conformation of p53 at 32°C).
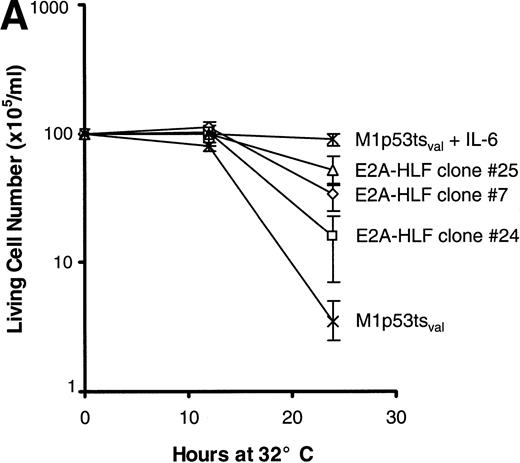
The survival of cells from each of the 3 clones was measured after the induction of wild-type p53 and compared with that of parental cells. The rate of cell death at 24 hours was significantly slower in clones expressing E2A-HLF than in the parental cell line (Fig 2A), but the E2A-HLF–induced block of p53-mediated apoptosis was not as pronounced as that observed when the M1tsp53val cells were grown in 12 ng/mL of IL-6 (top curve). The inhibitory effect of E2A-HLF on apoptotic cell death was also evident 46 hours after the temperature shift to 32°C (6.5%, 1.5%, and 5.7% cell survival for E2A-HLF–expressing clones 25, 7, and 24 v 0.83% for the parental M1 cells). However, by 72 hours, all cells from clones expressing E2A-HLF had undergone p53-induced apoptosis. A reduction in the proportion of cells dying by apoptosis was also apparent in E2A-HLF–expressing clones studied with a flow cytometric TUNEL assay, in which TdT was used to label the ends of fragmented DNA (Fig 2B). Eighty percent of the control (parental) cells were apoptotic 24 hours after induction of wild-type p53 at 32°C (lower left panel), which is 1.5- to 3.5-fold higher than the percentages of cells dying in clones that expressed E2A-HLF (38%, 29%, and 58% for clones 25, 24, and 7, respectively).
E2A-HLF suppresses p53-induced apoptosis in M1p53tsval cells. (A) Viable cell counts (means of triplicate determinations) performed at serial times after a shift in temperature to 32°C for 3 clones (7, 24, and 25) expressing E2A-HLF or for control cells, with or without the addition of IL-6. Comparisons of cell survival at 24 hours between controls and clones 7, 24, and 25 yielded P values of .004, .036, and .004, respectively. (B) Flow cytometric TUNEL assay of TdT-labeled DNA performed in cells grown at 37°C or 24 hours after a shift to 32°C. Respective differences in the percentages of cells undergoing apoptosis between the control and clones 7, 24, and 25 were statistically significant at the P < .0001 level.
E2A-HLF suppresses p53-induced apoptosis in M1p53tsval cells. (A) Viable cell counts (means of triplicate determinations) performed at serial times after a shift in temperature to 32°C for 3 clones (7, 24, and 25) expressing E2A-HLF or for control cells, with or without the addition of IL-6. Comparisons of cell survival at 24 hours between controls and clones 7, 24, and 25 yielded P values of .004, .036, and .004, respectively. (B) Flow cytometric TUNEL assay of TdT-labeled DNA performed in cells grown at 37°C or 24 hours after a shift to 32°C. Respective differences in the percentages of cells undergoing apoptosis between the control and clones 7, 24, and 25 were statistically significant at the P < .0001 level.
Both p53 and IL-6 regulate expression of Nfil3.
Nfil3 is a bZIP protein homologous to HLF within the basic DNA-binding region. It is regulated by IL-3 in pro-B lymphocytes and can function as either a transrepressor or transactivator of gene transcription through interaction with the HLF DNA-binding site.9 10Thus, we reasoned that Nfil3 might be regulated by IL-6 and could either block or accelerate apoptosis in M1p53tsval murine myeloid leukemia cells. Nfil3 mRNA was readily detectable in M1p53tsval cells grown in the absence of IL-6 at 37°C, a temperature at which these cells express a nonfunctional mutant p53 protein (Fig 3, lane 1). When the temperature was shifted to 32°C and p53 assumed its wild-type conformation, Nfil3 mRNA levels were progressively downregulated in cells growing without IL-6 (lanes 2 and 3). Decreased mRNA levels were observed within 1 hour of the change in temperature to 32°C (data not shown), reaching 10-fold lower levels by 12 hours (lane 3). Nfil3 expression was specifically downregulated in response to wild-type p53 activity, but not in response to a change in temperature alone. Indeed, the levels of Nfil3 transcripts were unaffected by a shift in culture temperature of parental M1 cells, which have homozygous loss of the p53 gene (data not shown). When IL-6 was added to the growth medium at 37°C, Nfil3 was modestly upregulated (∼1.5-fold, lanes 4 and 5). The cytokine also blocked Nfil3 downregulation in the presence of wild-type p53 at 32°C (lanes 6 and 7).
Regulation of Nfil3 expression by p53 and IL-6 in M1p53tsval cells. Northern blot analysis of total RNA (20 μg/lane) prepared from M1p53tsval cells grown at 37°C (lanes 1, 4, and 5) or 32°C (lanes 2, 3, 6, and 7) in the absence (lanes 1 through 3) or presence (lanes 4 through 7) of IL-6. Blots were hybridized with a mouse Nfil3 cDNA probe, stripped, and rehybridized with a mouse Gapdh control probe.
Regulation of Nfil3 expression by p53 and IL-6 in M1p53tsval cells. Northern blot analysis of total RNA (20 μg/lane) prepared from M1p53tsval cells grown at 37°C (lanes 1, 4, and 5) or 32°C (lanes 2, 3, 6, and 7) in the absence (lanes 1 through 3) or presence (lanes 4 through 7) of IL-6. Blots were hybridized with a mouse Nfil3 cDNA probe, stripped, and rehybridized with a mouse Gapdh control probe.
Cytokine-dependent regulation of Nfil3 expression was also apparent from the binding activity of the protein in nuclear lysates of M1p53tsval cells studied by EMSA (Fig 4). Nfil3 binding was maintained over the course of the experiment in cells grown in the absence of IL-6 at 37°C (lanes 1, 2, and 3), but decreased rapidly when the wild-type p53 was induced, reaching 10-fold lower levels by 14 hours after a temperature shift to 32°C (lane 6). By contrast, the addition of IL-6 to the growth medium immediately before induction of wild-type p53 led to a 3.2-fold increase in Nfil3 binding after 14 hours (lane 9). A similar result was obtained with lysates from cells grown at 37°C in IL-6 (lanes 10 and 11), indicating a dominant effect of this cytokine on Nfil3 RNA expression.
Regulation of Nfil3 binding activity by p53 and IL-6. Nuclear extracts prepared from M1p53tsval mouse myeloid leukemia cells were studied by EMSA with a 32P-labeled oligonucleotide probe containing the HLF consensus binding sequence. Levels of the Nfil3 complex are shown in the absence of IL-6 in cells maintained at either 37°C (lanes 1 through 3) or after a shift to 32°C (lanes 4 through 6), allowing p53 to assume a wild-type conformation. The effects of IL-6 addition on Nfil3 binding activity are shown at both 32°C (lanes 7 through 9) and 37°C (lanes 10 and 11).
Regulation of Nfil3 binding activity by p53 and IL-6. Nuclear extracts prepared from M1p53tsval mouse myeloid leukemia cells were studied by EMSA with a 32P-labeled oligonucleotide probe containing the HLF consensus binding sequence. Levels of the Nfil3 complex are shown in the absence of IL-6 in cells maintained at either 37°C (lanes 1 through 3) or after a shift to 32°C (lanes 4 through 6), allowing p53 to assume a wild-type conformation. The effects of IL-6 addition on Nfil3 binding activity are shown at both 32°C (lanes 7 through 9) and 37°C (lanes 10 and 11).
Enforced expression of NFIL3 blocks the effects of IL-6 on p53-mediated apoptosis.
M1p53tsval clones that overexpressed the human NFIL3cDNA were obtained by retroviral infection. Two representative clones had stable NFIL3 mRNA levels after p53 had assumed its wild-type conformation in cells cultured for 14 hours at 32°C (Fig 5A, top panel). By contrast, the same Northern blot hybridized with the murine Nfil3 probe showed downregulation of the endogenous mRNA (middle panel).
M1p53tsval cell clones expressing human NFIL3. (A) Northern analysis of total RNA (20 μg/lane) prepared from 2 NFIL3-expressing clones (top panel) and probed with a human NFIL3 cDNA probe. Note the stability of the LTR-driven NFIL3 mRNA (solid arrowhead) in the absence of IL-6 at 32°C, the permissive temperature for p53. Rehybridization of the Northern blot with a mouseNfil3 probe demonstrated downregulation of the endogenousNfil3 mRNA (middle panel, open arrowhead) due to p53. Results for the Gapdh RNA loading control are also shown (bottom panel; ○). (B) EMSA results showing NFIL3 binding activity of clones 2, 3, 4, 7, and 8 (lanes 3 through 12) versus control cells (lanes 1 and 2) at 37°C and 14 hours after shift to 32°C.
M1p53tsval cell clones expressing human NFIL3. (A) Northern analysis of total RNA (20 μg/lane) prepared from 2 NFIL3-expressing clones (top panel) and probed with a human NFIL3 cDNA probe. Note the stability of the LTR-driven NFIL3 mRNA (solid arrowhead) in the absence of IL-6 at 32°C, the permissive temperature for p53. Rehybridization of the Northern blot with a mouseNfil3 probe demonstrated downregulation of the endogenousNfil3 mRNA (middle panel, open arrowhead) due to p53. Results for the Gapdh RNA loading control are also shown (bottom panel; ○). (B) EMSA results showing NFIL3 binding activity of clones 2, 3, 4, 7, and 8 (lanes 3 through 12) versus control cells (lanes 1 and 2) at 37°C and 14 hours after shift to 32°C.
Antibody-perturbed DNA-protein complexes containing human and mouse Nfil3 have slightly different mobilities, allowing them to be discriminated from each other in EMSA assays (Fig 5B). In parental cells, levels of the murine Nfil3-DNA complex decreased dramatically in cells cultured for 14 hours at the permissive temperature (lanes 1 and 2). Two different patterns of DNA binding of retrovirally transduced human NFIL3 were observed after the cells were shifted to 32°C. NFIL3-DNA complexes were maintained at high levels in some clones, exemplified by clones 3 (lanes 3 and 4) and 7 (lanes 11 and 12); whereas in others, exemplified by clones 8 (lanes 5 and 6), 2 (lanes 7 and 8), and 4 (lanes 9 and 10), levels of the supershifted complex decreased with a concomitant increase in more rapidly migrating complexes, presumably due to the activation of the apoptotic program by 14 hours at 32°C.
The survival of each of 5 clones overexpressing NFIL3 in the presence of p53 was compared with that of parental cells and cells infected with empty vector by shifting the temperature to 32°C (Fig 6A). Similar cell death profiles were observed for each of the clones with high levels of enforcedNFIL3 expression and for the M1p53tsval controls in the absence of IL-6. After a temperature shift to 32°C, nearly all of the control cells survived in the presence of an IL-6 concentration of 12 ng/mL (Fig 6B), consistent with the ability of the cytokine to block p53-mediated apoptosis in this experimental system.13NFIL3 overexpression interfered with the ability of IL-6 to block p53-mediated cell death, as shown in Fig 6B for clones 3 and 8. Programmed cell death was confirmed by the TUNEL assay, which demonstrated DNA labeling indicative of apoptosis in 8% of the parental cells 24 hours after the shift in temperature in the presence of 12 ng/mL of IL-6, as compared with 29% of the cells overexpressing NFIL3 (Fig 6C, clone 8). Thus, although Nfil3 is upregulated by IL-6 in M1p53tsval cells, it does not appear to be involved in mediating the anti-apoptotic effects of this cytokine. Rather, constitutive expression of NFIL3 interferes with the ability of IL-6 to protect these cells from p53-induced cell death.
NFIL3 interferes with IL-6 rescue of M1p53tsval cells. (A) Viable cell counts (means of triplicate determinations) performed at serial times after a shift in temperature to 32°C for 5 NFIL3 clones and uninfected control. (B) Viable cell counts of NFIL3 clone 3 and 8 and control performed in the presence of 12 ng/mL IL-6. (C) Flow cytometric TUNEL assay of TdT-labeled DNA in NFIL3 clone 8 and control grown in the presence of IL-6 at 37°C and 24 hours after a temperature shift to 32°C.
NFIL3 interferes with IL-6 rescue of M1p53tsval cells. (A) Viable cell counts (means of triplicate determinations) performed at serial times after a shift in temperature to 32°C for 5 NFIL3 clones and uninfected control. (B) Viable cell counts of NFIL3 clone 3 and 8 and control performed in the presence of 12 ng/mL IL-6. (C) Flow cytometric TUNEL assay of TdT-labeled DNA in NFIL3 clone 8 and control grown in the presence of IL-6 at 37°C and 24 hours after a temperature shift to 32°C.
Regulation of p53-induced genes (PIGs) in M1p53tsvalcells expressing E2A-HLF.
When induced in response to DNA damage, p53 acts through both transcriptional and nontranscriptional pathways to promote cell cycle arrest and, in some cellular contexts, programmed cell death.19 Because E2A-HLF interferes with the ability of p53 to promote cell death, we were interested in determining whether it might interfere with upregulated expression of any of the known transcriptional targets of p53. As shown by Northern blot analysis (Fig 7), Bax, aBcl-2–related cell death gene induced by p53,20,21 increased progressively in control M1p53tsval cells induced to express wild-type p53 by a temperature shift to 32°C (Fig 7A). In all 3 clones expressing E2A-HLF, the increased levels of Bax expression resembled that of parental cells. p21WAF1/CIP1, a cdk inhibitor known to block cell cycle progression from the G1- to S-phase of the cell cycle, was induced to a similar degree by wild-type p53, both in the presence and absence of E2A-HLF (Fig 7B), as was EI24/PIG8 (Fig 7C), a previously described PIG of unknown function.22 23 Thus, the transcriptional activation of Bax and p21 by p53 occurs normally in cells expressing E2A-HLF, suggesting that the oncogenic fusion protein selectively interferes with other, as yet unidentified, downstream targets of the p53 protein.
E2A-HLF does not interfere with PIG expression in M1p53tsval cells. Northern blot analysis of total RNA (20 μg) prepared from parental M1p53tsval cells and M1p53tsval clones 7, 24, and 25 expressing E2A-HLF grown at 32°C for the times indicated. The blot was hybridized with Bax(A), p21WAF1/CIP1 (B), Ei24 (C), andGapdh (D) cDNA probes.
E2A-HLF does not interfere with PIG expression in M1p53tsval cells. Northern blot analysis of total RNA (20 μg) prepared from parental M1p53tsval cells and M1p53tsval clones 7, 24, and 25 expressing E2A-HLF grown at 32°C for the times indicated. The blot was hybridized with Bax(A), p21WAF1/CIP1 (B), Ei24 (C), andGapdh (D) cDNA probes.
To examine the regulation of additional apoptosis-associated genes, we used a multiprobe RNAse protection assay that detects mRNA transcripts expressed by genes in the Bcl-2 and Fas death receptor pathways (Fig 8). Results for theBcl-2 family of genes correlated with Northern blot results forBax and Bcl-2. Bax was upregulated twofold after activation of wild-type p53 in both E2A-HLF–expressing cells and control cells. Bcl-2 was downregulated twofold under conditions of wild-type p53 activation. mRNA levels of other members of this family were not significantly affected by activation of p53 or E2A-HLF. In the Fas-mediated death pathway, levels of the Fasreceptor increased 10-fold in both E2A-HLF–expressing clone 25 and in control cells when wild-type p53 was activated. mRNA levels of other members in this pathway were not dramatically altered by either wild-type p53 activation or E2A-HLF expression. Therefore, of the transcriptional targets of p53 examined, all were regulated independently of the expression of E2A-HLF.
RNAse protection assay using multiple probes detecting genes in the Bcl-2 and Fas receptor-mediated death pathways. Twenty micrograms of RNA isolated from parental M1p53tsval cells or M1p53tsval cells (clone 25) expressing E2A-HLF cultured at 37°C (0 hour) or at 32°C (14 hours) were hybridized to the mouse probes. RNA levels for each gene were compared with levels of expression of the housekeeping genes Gapdh and L32.
RNAse protection assay using multiple probes detecting genes in the Bcl-2 and Fas receptor-mediated death pathways. Twenty micrograms of RNA isolated from parental M1p53tsval cells or M1p53tsval cells (clone 25) expressing E2A-HLF cultured at 37°C (0 hour) or at 32°C (14 hours) were hybridized to the mouse probes. RNA levels for each gene were compared with levels of expression of the housekeeping genes Gapdh and L32.
DISCUSSION
We have previously shown that E2A-HLF protects pro-B cells from apoptosis due to IL-3 deprivation, presumably by activating an evolutionarily conserved survival program.3 The results reported here demonstrate that the fusion protein also protects against p53-mediated cell death. Apoptosis, as measured by cell viability assays and TdT labeling of nuclear DNA fragments, was clearly suppressed in cells constitutively expressing E2A-HLF, despite temperature-dependent activation of p53 in a wild-type conformation. This finding substantiates earlier observations of enhanced survival of E2A-HLF–positive murine pro-B cells after ionizing irradiation.3
NFIL3 is a growth factor-regulated member of the bZIP family of transcription factors that shares sequence identity with HLF in its basic DNA binding region. It was originally identified by its ability to recognize a transcription factor binding site of the adenovirusE4 promoter and independently shown to bind to a similar sequence in the human IL-3 promoter.9-11 Endogenous murine Nfil3 is upregulated by IL-3, suggesting that it acts as a downstream transcriptional effector of IL-3–initiated signals.12 In the present study, we show that Nfil3 is downregulated by p53 in a murine myeloid leukemia cell line (M1p53tsval), except when the cells are grown in the presence of IL-6. Although Nfil3 was upregulated in the presence of this cytokine, constitutively expressed NFIL3 did not replace the antiapoptotic function of IL-6 in myeloid cells, as it had in murine Baf-3 cells in the absence of IL-3. Rather, it enhanced programmed cell death in our M1p53tsval system and antagonized survival signals emanating from the IL-6 cytokine. We attribute this effect to the demonstrated ability of NFIL3 to function not only as an activator of gene expression but also as a repressor, depending on the cellular context.9-11
The bZIP domain shared by HLF and NFIL3 is highly conserved, bearing close similarity to sequences of the CES-2 (cell death specification) protein of Caenorhabditis elegans.24,25 CES-2 is thought to repress transcription of a downstream gene, ces-1, leading ultimately to specific activation of programmed cell death in the superfluous sister cells of serotonergic neurosecretory motor neurons during early nematode development.24 25 Thus, we postulated that E2A-HLF activates a pathway in pro-B cells that would normally be repressed by a ces-2–like protein, thereby promoting the aberrant survival of pro-B cells during development. Conceivably, E2A-HLF acts through a comparable pathway in myeloid cells, accounting for its ability to block p53-induced apoptosis. The failure of NFIL3 to rescue M1p53tsval cells from apoptotic death indicates that the respective regulatory cascades in lymphoid and myeloid cells may differ in certain key aspects, including the roles played by NFIL3.
Loss of the p53 tumor suppressor gene is a common event in multistep pathways leading to carcinogenesis. The mechanism of action of p53 involves either stimulation of an apoptotic pathway or induction of stable growth arrest, depending not only on cell lineage and the presence of specific cytokines, but on other, poorly understood factors as well.19,26 p53 can induce cell-cycle arrest at the G1/S-phase boundary, primarily through upregulation of the G1-cyclin dependent kinase inhibitor p21WAF1/CIP1. Stimulation of an apoptotic pathway can occur at the transcriptional level through induction of Bax,21 a member of the Bcl-2gene family.20 The mechanism of this activation appears to involve a shift in dimer equilibrium from Bax/Bcl-2 heterodimers to Bax/Bax homodimers.27-29 In M1 leukemia cells, we found that p53-induced upregulation of Bax is accompanied by downregulated expression of Bcl-2. However, E2A-HLF expression did not affect p53-induced changes in the levels of either Baxand Bcl-2 mRNAs or that of p21, indicating that E2A-HLF does not block apoptosis by altering the levels of expression of these genes. Recently, Polyak et al23 proposed a mechanism of p53-mediated apoptosis that is independent of Baxregulation. It involves the transcriptional activation of redox-related genes, several of which are induced at increased levels (≥10-fold) in response to enforced expression of p53.23 These findings suggest that p53 may activate a group of oxidoreductases that subsequently increase the content of reactive oxygen species (ROS) in cells, resulting in injury to mitochondria and stimulation of caspases, whose many substrates are thought to account for the terminal events of apoptosis.23
We suggest that E2A-HLF blocks p53-induced apoptosis through activation of a ces-1–like gene30 whose product then represses a genetic program required for apoptotic cell death.25,30 According to our model, genes repressed by aces-1 ortholog may include those involved in the p53-mediated death cascade before caspase activation. Because many of the genes that are specifically upregulated in p53-expressing cells encode proteins that could generate or respond to oxidative stress,23 they are likely candidates for the inhibitory effects of E2A-HLF. Our findings extend the concept that inhibition of p53-mediated cell death may be a general feature of cancers that respond poorly to conventional therapy.31 Because it is not expressed in normal tissues, E2A-HLF would afford an ideal therapeutic target in pro-B leukemia cells harboring the t(17;19) chromosomal translocation. A better understanding of the role of E2A-HLF in the p53-activated regulatory cascade could facilitate the design of targeted inhibitors of this novel pathway in pro-B leukemia cells.
ACKNOWLEDGMENT
The authors thank Richard J. Cross for assistance with flow cytometric analysis; Elizabeth Mann and Bart Jones for technical assistance; Deo Kumar Srivastava, Deepthi Jayawardene, and James Boyett for statistical analysis; and John Gilbert for scientific editing.
Supported in part by National Cancer Institute Grants No. CA 59571, CA 71907, CA 70089, and CA 21765 (Cancer Center Core); and by the American Lebanese Syrian Associated Charities (ALSAC), St Jude Children’s Research Hospital.
Address reprint requests to A. Thomas Look, MD, Department of Experimental Oncology, St Jude Children’s Research Hospital, 332 N Lauderdale, Memphis, TN 38105-2794; e-mail: thomas.look@stjude.org.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.












This feature is available to Subscribers Only
Sign In or Create an Account Close Modal