Abstract
Bcl-3 is a proto-oncogene involved in the chromosomal translocation t(14;19) found in some patients with chronic lymphocytic leukemia. It shares structural similarities with and is a member of the IκB family of proteins. In this report, involvement of Bcl-3in hematopoietic growth factor-stimulated erythroid proliferation and differentiation was examined. In TF-1 cells, an erythroleukemia cell line, granulocyte-macrophage colony-stimulating factor (GM-CSF) and erythropoietin (Epo) greatly enhanced Bcl-3 expression at both the protein and mRNA levels in association with stimulation of proliferation. Bcl-3 protein was also highly expressed in early burst-forming unit-erythroid (BFU-E)–derived erythroid precursors (day 7) and decreased during maturation (days 10 and 14), suggesting that Bcl-3 is involved in normal erythroid proliferation. In these hematopoietic cells, Bcl-3 was hyperphosphorylated. GM-CSF and Epo modulated the subcellular localization of Bcl-3. Upon stimulation of TF-1 cells with GM-CSF or Epo, the nuclear translocation ofBcl-3 was dramatically enhanced. Overexpression of Bcl-3 in TF-1 cells by transient transfection along with the NF-κB factors p50 or p52 resulted in significant induction of an human immunodeficiency virus–type 1 (HIV-1) κB-TATA-luceriferase reporter plasmid, demonstrating that Bcl-3 has a positive role in transactivation of κB-containing genes in erythroid cells. Stimulation with GM-CSF enhanced c-myb mRNA expression in these cells. Bcl-3 in nuclear extracts of TF-1 cells bound to a κB enhancer in the c-mybpromoter together with NF-κB2/p52 and this binding activity was enhanced by GM-CSF stimulation. Furthermore, cotransfection of Bcl-3 with p52 or p50 in TF-1 cells resulted in significant activation of ac-myb κB-TATA-luceriferase reporter plasmid. These findings suggest that Bcl-3 may participate in the transcriptional regulation of certain κB-containing genes involved in hematopoiesis, includingc-myb.
© 1998 by The American Society of Hematology.
Bcl-3 WAS FIRST IDENTIFIED from a chronic lymphocytic leukemia (CLL) patient with the chromosomal translocation t(14;19) (q32; q13.1).1 This breakpoint junction does not affect the structural integrity of Bcl-3, but results in the overexpression of Bcl-3 mRNA in leukemic cells.1Bcl-3 overexpression is proposed to contribute to the development of CLL through dysregulation of a gene(s) normally regulated by NF-κB transcription factors and important in cell proliferation and differentiation.1,2 The Bcl-3 protein has a distinct pattern of ankyrin-related repeats and is structurally similar to other members of the IκB family of proteins, including IκBα, IκBβ, and IκBγ.3-5
NF-κB/Rel designates a widely distributed family of transcription factors that modulate the expression of genes involved in immune and acute-phase responses as well as the response to signals for rapid gene expression.5 The DNA-binding forms of NF-κB transcription factors are heterodimers or homodimers composed of different combinations of 5 structurally related DNA-binding proteins, p50, p52, RelA (p65), RelB, and c-Rel. These proteins share a highly conserved amino-terminal sequence called the Rel homology region (RHR).5 Specific heterodimers or homodimers of these proteins bind to target enhancer elements (κB) present in the promoters of regulated genes. The activity of these proteins is regulated through cytoplasmic retention by physical interaction with cytoplasmic inhibitors termed IκB.5-7 IκB proteins interact with the RHR of NF-κB proteins through their ankyrin repeats.8 Stimulation of cells with NF-κB inducers leads to rapid phosphorylation and degradation of IκBs, allowing NF-κB to translocate to the nucleus to activate targets.5-8
Based on protein structural similarities and interaction with NF-κB/Rel proteins, Bcl-3 is considered to be a member of the IκB family. However, despite structural homology, Bcl-3 and IκBs appear to have different roles in the regulation of NF-κB/Rel proteins. Whereas IκBα is primarily cytoplasmic, transiently transfected Bcl-3 protein is predominantly located in the nucleus.2,3,9IκBα inhibits nuclear translocation of NF-κB/Rel, whereas Bcl-3 does not.2,5-8 Bcl-3 has been reported to form complexes with p5010 or p529-11 homodimers that serve as transcriptional coactivators of κB-specific gene expression. Under certain conditions, Bcl-3 may also function to antagonize the DNA binding of nuclear p50 homodimers, which can be κB-specific repressors of active forms of NF-κB/Rel.3,5 12 Thus, these transient transfection studies suggest that Bcl-3 plays a positive regulatory role in κB-specific gene transcription. The in vivo role of Bcl-3 in different cell types is less clear.
In this study, the involvement of Bcl-3 in erythroid proliferation and differentiation was investigated using both an erythroleukemia cell line, TF-1,13 and normal human progenitor-derived erythroblasts. Proliferation of the TF-1 cells is completely dependent on hematopoietic growth factors, including granulocyte-macrophage colony-stimulating factor (GM-CSF), erythropoietin (Epo), and interleukin-3 (IL-3).13 GM-CSF and Epo stimulation of TF-1 proliferation resulted in marked induction of Bcl-3 expression. Furthermore, Bcl-3 was highly expressed in day-7 burst-forming unit-erythroid (BFU-E)–derived erythroid precursors and decreased with maturation, suggesting that Bcl-3 is involved in normal erythroid proliferation. In contrast to previous transfection studies,9-11 endogenous Bcl-3 in TF-1 cells and normal erythroblasts was located in both nucleus and cytoplasm. After growth factor stimulation of TF-1 cells, a gradual but dramatic translocation of Bcl-3 to the nucleus was observed. Transient transfection studies with TF-1 cells showed that overexpression of Bcl-3 along with p50 or p52 resulted in significant transactivation of a luciferase reporter gene driven by a human immunodeficiency virus–type 1 (HIV-1) κB enhancer, suggesting that Bcl-3 can modulate expression of regulated genes in vivo in TF-1 cells. Induction ofc-myb mRNA was observed in GM-CSF–stimulated TF-1 cells. Electrophoretic mobility shift assay (EMSA) using the NF-κB binding site of the c-myb promoter suggested that a nuclear complex of Bcl-3 and p52 bound to the c-myb promoter and this complex was induced by GM-CSF stimulation of TF-1 cells. Overexpression of Bcl-3 along with p52 or p50 was also able to significantly induce expression of a luciferase reporter gene driven by a κB site present in thec-myb promoter. These data indicate that Bcl-3 may participate in the regulation of κB-containing genes involved in hematopoiesis including c-myb.
MATERIALS AND METHODS
Culture of BFU-E–derived erythroblasts and TF-1 cells.
Peripheral blood was obtained from normal volunteer donors at The Milton S. Hershey Medical Center (Hershey, PA) under protocols approved by the Institution’s Clinical Investigation Committee. BFU-E–derived erythroblasts were cultured as described previously.14Briefly, peripheral blood mononuclear cells were separated on Ficoll-Paque (Pharmacia, Piscataway, NJ) and cultured in 0.9% methylcellulose media containing 30% fetal calf serum, 9.0 mg/mL deionized bovine serum albumin (Cohn fraction V; Sigma Chemical Co, St Louis, MO), 1.4 × 10−4 mol/L β-mercaptoethanol, and 2 U/mL Epo (recombinant Epo >100,000 U/mg; Amgen, Thousand Oaks, CA). Single BFU-E, when cultured in methylcellulose, proliferate and differentiate over 14 days to form large colonies containing 1 to 5 × 104 mature erythroblasts. These cells can be removed from culture at different days to study a well-defined population of normal human cells at distinct stages of maturation.14 Day-7 cells are poorly hemoglobinized blasts with a large proliferative capacity, day-10 cells are partially hemoglobinized proerythroblasts with decreased proliferative capacity, and day-14 cells are terminally differentiating polychromatophilic and orthochromatic normoblasts. Cells from maturing BFU-E–derived colonies were plucked from culture on days 7, 10, and 14. Cytocentrifuge preparations of aliquots of BFU-E–derived cells routinely identified greater than 99% as erythroid precursors.
TF-1 cells, a human erythroleukemia cell line,13 were cultured in RPMI 1640 medium containing 10% fetal calf serum and 1 to 2 ng/mL human recombinant GM-CSF (R & D Systems, Minneapolis, MN) or 5 U/mL Epo. To examine Bcl-3 induction, TF-1 cells were removed from growth factor for 24 hours and then stimulated with 2 ng/mL GM-CSF or with 5 U/mL recombinant Epo. Samples were collected at intervals over 0 to 24 hours. Viability of TF-1 cells after growth factor stimulation was determined by trypan blue exclusion. The percentage in apoptosis was determined with the ApoAlert Annexin V Apoptosis Kit (Clontech, Palo Alto, CA) and analysis with a fluorescence-activated cell sorter. The cell cycle status of TF-1 cells was determined by propidium iodide staining.15
Cell lysate preparation and nuclear/cytoplasmic fractionation.
Whole cell lysates were prepared by suspending 1 × 106 TF-1 cells or BFU-E–derived erythroblasts in cell lysate buffer (50 mmol/L Tris HCl, pH 8.0, 150 mmol/L NaCl, 0.05% NP40, 100 mmol/L NaF, 1 mmol/L EDTA, 1 mmol/L EGTA, 0.08 mmol/L phenylmethyl sulfonyl fluoride [PMSF], 0.01 mg/mL of leupeptin, and 0.01 mg/mL aprotinin). The suspension was vortexed and centrifuged at 10,000 rpm for 10 minutes. The supernatant was saved for Western blotting. Nuclear and cytoplasmic fractions were prepared as previously described by Schreiber et al.16 TF-1 cells (1 × 107) or BFU-E–derived erythroblasts (1.5 × 107) harvested at day 10 were washed twice with cold phosphate-buffered saline (PBS). The cell pellet was resuspended in 100 μL of cold buffer (10 mmol/L HEPES, pH 7.9, 10 mmol/L KCl, 0.1 mmol/L EDTA, 0.4% NP40, 1 mmol/L dithiothreitol [DTT], 0.5 mmol/L PMSF, and 1% volume protease inhibitor cocktail) and pipetted several times. The lysates were spun, and the supernatant was used for the cytosol preparation. The nuclear pellet was extracted with 50 μL of ice-cold buffer (20 mmol/L HEPES, pH 7.9, 0.4 mol/L NaCl, 1 mmol/L EDTA, 1 mmol/L DTT, and 1 mmol/L PMSF) and vigorously shaken for 15 minutes at 4°C. After centrifugation, the nuclear extract was collected and kept at −70°C.
Human tissues frozen at −70°C were mixed 1:5 (wt/vol) with fresh lysis buffer (10 mmol/L Tris HCl, pH 7.6, 5 mmol/L EDTA, 5 mmol/L MgCl2, 0.8 mmol/L PMSF, 0.01 mg/mL leupeptin, and 0.01 mg/mL aprotinin), homogenized, and centrifuged at 10,000 rpm for 10 minutes. The supernatant was removed, quantitated, and used in Western blot analysis.
Immunoblotting.
The whole cell lysate, nuclear, or cytoplasmic preparations were boiled for 5 minutes in protein sample buffer and separated on 12% or 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. Proteins were electroblotted onto Hybond-ECL nitrocellulose membrane (Amersham Life Sciences, Bucks, UK) according to the recommended procedures of the manufacturer. After blocking in 5% dry milk in TBST buffer,14 membranes then were incubated with anti–Bcl-3 antibody (Santa Cruz Biotechnology, Santa Cruz, CA; diluted 1:200). Donkey antirabbit antibody (1:2,000 dilution) was used as the secondary antibody and membranes were detected with the ECL-Western blotting system (Amersham). Preincubation of anti–Bcl-3 antibody with Bcl-3 peptide (Santa Cruz Biotechnology) competitively removed recognition by the antibody of the Bcl-3 protein band on Western blots (data not shown). As another control, an aliquot of the protein sample was run on a 10% polyacrylamide gel and IκBα protein levels were examined with anti-IκBα antibody17(1:2,000 dilution) and ECL. In some experiments, anti-E47 antibody (Santa Cruz; diluted 1:250) was used to demonstrate subcellular localization of this basic helix-loop-helix transcription factor.
Northern blot analysis.
Total TF-1 RNA was isolated using TRI REAGENT-RNA/DNA/PROTEIN isolation reagent (Molecular Research Center, Inc, Cincinnati, OH). RNA samples (40 μg/lane) were separated on 1.2% agarose-formaldehyde gels and alkaline transferred onto Zeta-Probe GT Genomic Tested Blotting membrane (Bio-Rad, Hercules, CA). Membranes were prehybridized for 5 minutes at 42°C in 50% formamide, 120 mmol/L Na2HPO4, 250 mmol/L NaCl, and 7% SDS. The32P-dCTP–labeled Bcl-3 cDNA probe was added and the hybridization was continued for 18 hours, followed by standard washing steps. Bcl-3 cDNA clone (kindly provided by Dr Timothy McKeithan, University of Chicago, Chicago, IL) was digested with EcoRI and HindIII to obtain the insert, followed by Geno-Bind DNA purification (Clontech). The cDNA probe was labeled by the random primer labeling method (Promega, Madison, WI).
Phosphatase treatment.
Whole cell lysates and nuclear/cytoplasmic extracts were prepared in lysate buffer (50 mmol/L HEPES, 250 mmol/L NaCl, 0.1% NP40, 5 mmol/L EDTA, 1 mmol/L DTT, 1 mmol/L PMSF, and 1% volume protease inhibitor cocktail) without any phosphatase inhibitors. Thirty micrograms of protein from normal human heart and TF-1 cells or 50 μg of protein from BFU-E–derived erythroblasts was incubated with 26 U of calf intestine phosphatase (CIP; GIBCO-BRL, Gaithersburg, MD) for 40 minutes at 37°C and then separated on 10% SDS-PAGE gel for Western analysis.3
TF-1 transfection and luciferase assay.
The HIV-1 κB-TATA-luciferase reporter plasmid was generated by transferring the insert, containing the HIV-1 κB enhancer and TATA box, from the κB-TATA-CAT into the pGL2 plasmid 5′ of the luciferase gene (Promega).18,19 The pGL2 basic vector lacking the TATA box and the κB enhancer was used as a negative control. The cDNAs encoding Bcl-3, p50, and p52 have been described previously.2,20,21 These cDNAs were cloned into the expression plasmid pCMV4 as described.19 22 To construct the c-myb κB-luciferase reporter plasmid, 4 copies of a 14-nucleotide c-myb κB element and 1 copy of the Herpes simplex virus thymidine kinase (tk) minimal promoter were cloned in front of a luciferase gene. In brief, the κB oligonucleotide sequence present in the c-myb promoter (Harhaj and Sun, manuscript in preparation) was cloned into the Kpn I/MluI sites of the pGL2 basic vector (Promega). Subsequently, the tk promoter was inserted between the κB sites and luciferase gene at theBgl II/Nhe I sites. TF-1 cells were transfected at a density of 1 × 106 cells/mL with Tfx-20 reagent (Promega) at 3:1 ratio Tfx-20 to DNA in the presence of GM-CSF. The quantity of reporter gene and effector plasmids used is present in the Results. After 48 hours of culture, the transfectants were collected and suspended in a lysis buffer (Reporter lysis buffer; Promega). Cell extracts were normalized for protein recovery (Bio-Rad) and then subjected to luciferase assay (Promega). Luciferase activity was quantitated using a single photon channel of a scintillation counter (Beckman, Fullerton, CA).
Electrophoretic mobility shift assay (EMSA).
EMSA was performed by the method described previously.23 A double-stranded oligonucleotide covering a κB site present in the promoter of the human c-myb gene (Harhaj and Sun, manuscript in preparation) was labeled as described by Ganchi et al.20 Four microliters (6 μg) of nuclear extracts was incubated with 0.5 to 2 μL of NF-κB (anti-p50, anti-p52, anti-p65, anti-RelB, and anti–c-Rel; from Dr Warner Greene, University of California, San Francisco, CA and San Francisco General Hospital) or anti–Bcl-3 antibodies in 12 μL of the binding buffer (0.5 μL of 1 μg/μL polydI-dC, 1 μL of 0.1 mol/L DTT, 3 μL of KCl-Dialysis buffer lacking KCl,24 12 μL H2O) for 10 minutes at room temperature. Following this, 1 μL of 32P-radiolabeled c-myb-κB probe (1 × 105 cpm) was added and incubated for another 25 minutes. The DNA-protein complex was resolved on a 5% native polyacrylamide gel.
RESULTS
GM-CSF and Epo induce Bcl-3 expression in TF-1 cells.
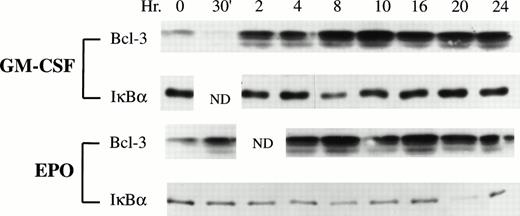
To explore the potential role of the proto-oncogene Bcl-3 in hematopoietic growth factor-stimulated proliferation, the expression ofBcl-3 was investigated by Western blotting analysis in GM-CSF–stimulated and Epo-stimulated TF-1 cells. The proliferation of TF-1 cells is dependent on growth factors, including GM-CSF, Epo, and IL-3, but the molecular mechanisms by which these hematopoietic growth factors induce the growth of TF-1 cells have only been partially identified. TF-1 cells were growth factor deprived for 24 hours. The viability of TF-1 cells 24 hours after growth factor deprivation was greater than 97%, and less than 5% of apoptotic cells were detected. After growth factor deprivation, 2 ng/mL of GM-CSF or 5 U/mL of Epo was added to the medium to induce cells to proliferate and the cells were subsequently continuously stimulated by growth factor. At different time points, stimulated TF-1 cells were collected for analysis ofBcl-3 expression. Bcl-3 protein was markedly induced after GM-CSF or Epo stimulation (Fig 1). At 2 hours (GM-CSF) or 4 hours (Epo) of stimulation, the level of Bcl-3 protein was approximately 3 times more than that in nonstimulated cells. The level of Bcl-3 protein remained elevated for at least 24 hours after stimulation. Both higher and lower molecular weight isoforms of Bcl-3 were induced. Because Bcl-3 has been proposed to be an IκB-like protein, the expression pattern of IκBα was also analyzed in these stimulated TF-1 cells. However, in contrast to Bcl-3, IκBα did not show increased expression in response to GM-CSF or Epo stimulation (Fig 1). These data suggest that hematopoietic growth factors participate in regulation of Bcl-3 expression in TF-1 cells.
Bcl-3 expression in GM-CSF–stimulated and Epo-stimulated TF-1 cells. Whole cell lysates were prepared from growth factor-induced TF-1 cells at the time points indicated. Thirty micrograms of protein was loaded on each lane of a 12% polyacrylamide gel (Bcl-3) or 15 μg of protein was loaded on each lane of a 10% polyacrylamide gel (IκB). The membranes were blotted with anti–Bcl-3 (1:200) or anti-IκB (1:2,000) antibody as a control and then detected with ECL. N.D. indicates time points that were not done. Three independent experiments showed induction of Bcl-3 expression.
Bcl-3 expression in GM-CSF–stimulated and Epo-stimulated TF-1 cells. Whole cell lysates were prepared from growth factor-induced TF-1 cells at the time points indicated. Thirty micrograms of protein was loaded on each lane of a 12% polyacrylamide gel (Bcl-3) or 15 μg of protein was loaded on each lane of a 10% polyacrylamide gel (IκB). The membranes were blotted with anti–Bcl-3 (1:200) or anti-IκB (1:2,000) antibody as a control and then detected with ECL. N.D. indicates time points that were not done. Three independent experiments showed induction of Bcl-3 expression.
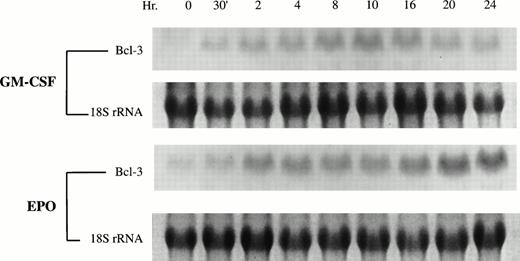
To examine the mechanism underlying the induction of Bcl-3 by hematopoietic growth factors, total RNA was extracted from TF-1 cells after stimulation by GM-CSF or Epo. Northern blotting showed thatBcl-3 transcripts increased significantly after 30 minutes of TF-1 stimulation by GM-CSF and continued to be elevated for 24 hours (Fig 2). This expression pattern is consistent with results of Western blotting. With Epo, a similar increase in Bcl-3 mRNA was observed. As a control, 18S rRNA showed no significant change (Fig 2). Taken together, these data suggest that growth factor stimulation of TF-1 proliferation results in enhanced Bcl-3 expression and that induced Bcl-3expression is mediated at the level of transcription.
GM-CSF and Epo induce Bcl-3 mRNA expression in TF-1 cells. Total RNA was isolated from GM-CSF–induced or Epo-induced TF-1 cells and Northern blotting analysis was performed using 40 μg RNA from each sample. 32P-dCTP–labeled Bcl-3 cDNA was used as a probe. 18s rRNA is shown as a control for equivalent loading.
GM-CSF and Epo induce Bcl-3 mRNA expression in TF-1 cells. Total RNA was isolated from GM-CSF–induced or Epo-induced TF-1 cells and Northern blotting analysis was performed using 40 μg RNA from each sample. 32P-dCTP–labeled Bcl-3 cDNA was used as a probe. 18s rRNA is shown as a control for equivalent loading.
After growth factor deprivation of TF-1 cells for 24 hours, in 3 experiments, 55% to 60% of cells were determined to be in G0/G1 of the cell cycle, and the rest of the cells were in S or G2/M phase. The percentage of cells in S phase increased significantly to a mean peak of 52% ± 2% at 4 to 10 hours after growth factor stimulation (P ≤ .02). The percentage of cells in G2 and M phase also increased significantly, and the increase peaked at 16 to 20 hours after growth factor stimulation (P < .002). The doubling time of TF-1 cells was 24 hours. No significant differences were noted in the cell cycle status of cells after GM-CSF compared with Epo stimulation. It is noteworthy that the increase in Bcl-3 protein and mRNA preceded the entrance of the majority of cells into S phase, but in TF-1 cells, Bcl-3 expression did not correlate with a specific phase of the cell cycle.
Nuclear expression of Bcl-3 is enhanced by hematopoietic growth factors.
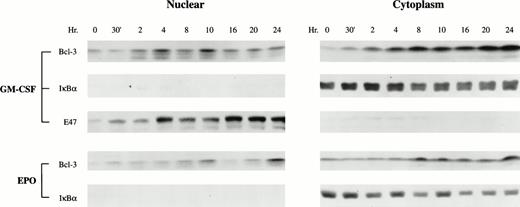
Previously, transiently transfected Bcl-3 protein has been predominantly localized in the nucleus.2,3,9 The N-terminal part of Bcl-3 resembles a nuclear localization signal.2Although this sequence is not perfectly conserved compared with other nuclear localization signals, deletion of this sequence abolished the nuclear localization of Bcl-3.2 The subcellular localization of endogenous Bcl-3 in a physiological setting, such as in hematopoietic growth factor-stimulated proliferation, remains unclear. To determine the subcellular localization of Bcl-3 in TF-1 cells, nuclear and cytoplasmic extracts were isolated from GM-CSF–induced or Epo-induced cells and Western blotting analysis was performed. Bcl-3 protein was weakly detectable in both cytoplasm and nucleus before growth factor stimulation (Fig 3). The effect of growth factor stimulation on the subcellular localization of Bcl-3 was determined. Growth factor stimulation greatly enhanced the level of cytoplasmic Bcl-3 (Fig 3). However, in addition, the nuclear level of Bcl-3 also was greatly increased, suggesting that growth factors enhance Bcl-3 nuclear translocation. To control for the quality of subcellular fractionation, nuclear and cytoplasmic extracts were examined for IκBα and E47 localization. IκBα, a cytoplasmic protein, was not detected in the nuclear fraction (Fig 3). E47, a basic helix-loop-helix transcription factor,25 was primarily detected in the nucleus (Fig 3).
GM-CSF and Epo enhance Bcl-3 nuclear translocation. Growth factor-induced TF-1 cells (1 × 107) were used for nuclear or cytoplasm separation. Twenty-five micrograms of nuclear or 20 μg of cytosolic protein extracts was loaded on each lane of a 12% polyacrylamide gel and subjected to Western blotting with anti–Bcl-3 antibody. As a control, 15 μg of nuclear or cytoplasmic extract was loaded on a 10% polyacrylamide gel to detect IκB. For GM-CSF–stimulated cells, 30 μg of nuclear or cytoplasmic extract was loaded on each lane of 12% gel and detection was with anti-E47 antibody as another control for quality of subcellular fractionation. Representative results are shown from 3 independent experiments.
GM-CSF and Epo enhance Bcl-3 nuclear translocation. Growth factor-induced TF-1 cells (1 × 107) were used for nuclear or cytoplasm separation. Twenty-five micrograms of nuclear or 20 μg of cytosolic protein extracts was loaded on each lane of a 12% polyacrylamide gel and subjected to Western blotting with anti–Bcl-3 antibody. As a control, 15 μg of nuclear or cytoplasmic extract was loaded on a 10% polyacrylamide gel to detect IκB. For GM-CSF–stimulated cells, 30 μg of nuclear or cytoplasmic extract was loaded on each lane of 12% gel and detection was with anti-E47 antibody as another control for quality of subcellular fractionation. Representative results are shown from 3 independent experiments.
Bcl-3 expression in normal human erythroblasts is correlated with proliferation.
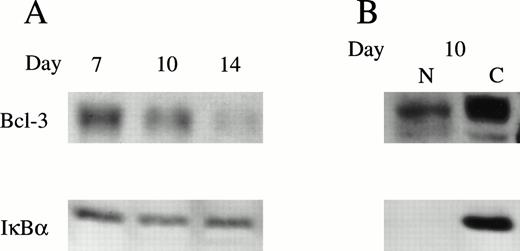
The expression of Bcl-3 protein was determined in normal erythroid proliferation and differentiation. Human BFU-E–derived erythroid precursors were removed from culture on days 7, 10, and 14 of maturation.14 Day-7 cells have a large proliferative capacity, day-10 cells are only partially hemoglobinized with decreased proliferative capacity, and day-14 cells are largely terminally differentiating normoblasts. Western blotting assay was performed using whole cell lysates from day-7, -10, and -14 cells (Fig 4A). In day-7 cells, which are rapidly proliferating, Bcl-3 had the highest expression level and decreased as erythroid precursors terminally differentiated. This dynamic pattern suggests that Bcl-3 expression is associated with normal erythroid proliferation rather than differentiation. Although Bcl-3 was minimally detectable in day-14 cells, IκBα showed little decline during differentiation and substantial quantities were still present at day 14. In day-10 BFU-E–derived cells, subcellular localization studies with nuclear and cytoplasmic extracts also showed that Bcl-3 was present in both nucleus and cytoplasm, whereas IκBα was primarily cytoplasmic (Fig 4B).
Bcl-3 expression in day-7, -10, and -14 BFU-E–derived erythroblasts. (A) Normal human BFU-E–derived erythroblasts were harvested and the whole cell lysates from 4 × 105 (Bcl-3) or 2 × 105 (IκB) cells were separated on a 10% polyacrylamide gel. Western analysis was performed with anti–Bcl-3 or anti-IκB antibodies and ECL. (B) Nuclear and cytoplasmic extracts were separated from day-10 cells. Fifty micrograms of nuclear (N) or cytoplasmic (C) extract was loaded onto each lane of a 10% gel and subjected to Western blotting. Two experiments were performed with anti–Bcl-3 or anti-IκB antibodies with identical results.
Bcl-3 expression in day-7, -10, and -14 BFU-E–derived erythroblasts. (A) Normal human BFU-E–derived erythroblasts were harvested and the whole cell lysates from 4 × 105 (Bcl-3) or 2 × 105 (IκB) cells were separated on a 10% polyacrylamide gel. Western analysis was performed with anti–Bcl-3 or anti-IκB antibodies and ECL. (B) Nuclear and cytoplasmic extracts were separated from day-10 cells. Fifty micrograms of nuclear (N) or cytoplasmic (C) extract was loaded onto each lane of a 10% gel and subjected to Western blotting. Two experiments were performed with anti–Bcl-3 or anti-IκB antibodies with identical results.
Bcl-3 is hyperphosphorylated in TF-1 and BFU-E–derived erythroblasts.
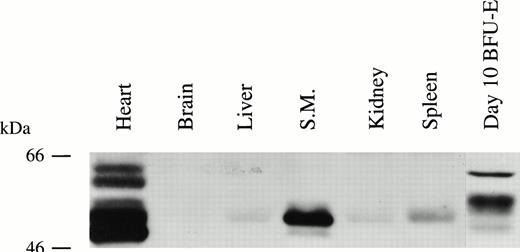
Bcl-3 mRNA has previously been shown to be expressed in several mammalian tissues.3 Expression of Bcl-3 protein was examined here in 7 human tissues. Bcl-3 was highly expressed in heart, skeletal muscle, and erythroid precursors; weakly detectable in liver, kidney, and spleen; and barely detectable in the brain (Fig 5). Parallel Northern blotting (Clontech) showed that the pattern of Bcl-3 protein correlated with mRNA expression (data not shown) and a single mRNA band was observed.
Bcl-3 is expressed in many human tissues. Protein extracts from normal human tissues were prepared as described in the Materials and Methods. Thirty micrograms of protein was loaded on each lane of a 10% polyacrylamide gel and detection was with anti–Bcl-3 antibody.
Bcl-3 is expressed in many human tissues. Protein extracts from normal human tissues were prepared as described in the Materials and Methods. Thirty micrograms of protein was loaded on each lane of a 10% polyacrylamide gel and detection was with anti–Bcl-3 antibody.
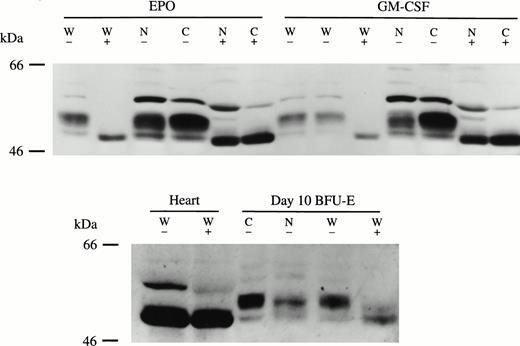
The molecular weight of Bcl-3 deduced from the protein sequence is 47 kD.1 Bcl-3 protein detected from normal human tissues such as heart, skeletal muscle, spleen, and liver showed a slightly higher molecular weight (Fig 5). Western blotting analysis showed multiple isoforms of Bcl-3 in GM-CSF– or Epo-stimulated TF-1 cells, and the same result was obtained with BFU-E–derived erythroblasts (Figs 1, 3, and 4). In BFU-E–derived erythroblasts (Fig 5) or TF-1 cells (Fig 6), Bcl-3 was larger than that in other human tissues. It is known that Bcl-3 is a proline- and serine-rich protein with high phosphorylation potential.3,4To test if the observed heterogeneity in size was due to protein phosphorylation, TF-1 and BFU-E–derived erythroblast cells and normal heart were treated with CIP. CIP treatment resulted in molecular weight reduction of Bcl-3 in both TF-1 (Fig 6, top) and BFU-E–derived (Fig 6, bottom) cells. However, CIP had no effect on the molecular weight of Bcl-3 from human heart tissue. In Fig 6, the highest molecular weight band was thought to represent a nonspecific protein, because it was inconsistently observed. These data demonstrate that the different size of Bcl-3 protein in tissue compared with TF-1 or BFU-E–derived cells is at least partially due to differential Bcl-3 phosphorylation. GM-CSF and Epo stimulated the appearance of higher and lower molecular weight Bcl-3 isoforms (Fig 1) and both are found in TF-1 and BFU-E nucleus and cytoplasm (Figs 4 and 6). The functional significance of Bcl-3 hyperphosphorylation in these hematopoietic-derived cells remains to be determined, but in other systems Bcl-3 dephosphorylation resulted in decreased activity.3 4
Bcl-3 is hyperphosphorylated in TF-1 cells and BFU-E–derived erythroblasts. (Top) Whole cell lysates (W) and nuclear (N) and cytoplasmic (C) extracts were prepared from Epo-induced or GM-CSF–induced TF-1 cells. Thirty micrograms of each extract was incubated with or without 26 U of CIP at 37°C for 40 minutes and then subjected to Western blotting assay with anti–Bcl-3 antibody and ECL. (Bottom) Thirty micrograms of whole cell lysate (W) from normal human heart tissue or 50 μg from day-10 BFU-E–derived erythroblasts was also incubated with or without 26 U CIP and subjected to Western blotting with anti–Bcl-3 as described in the Materials and Methods. Three experiments were performed with similar results. (+) with CIP; (−) without CIP.
Bcl-3 is hyperphosphorylated in TF-1 cells and BFU-E–derived erythroblasts. (Top) Whole cell lysates (W) and nuclear (N) and cytoplasmic (C) extracts were prepared from Epo-induced or GM-CSF–induced TF-1 cells. Thirty micrograms of each extract was incubated with or without 26 U of CIP at 37°C for 40 minutes and then subjected to Western blotting assay with anti–Bcl-3 antibody and ECL. (Bottom) Thirty micrograms of whole cell lysate (W) from normal human heart tissue or 50 μg from day-10 BFU-E–derived erythroblasts was also incubated with or without 26 U CIP and subjected to Western blotting with anti–Bcl-3 as described in the Materials and Methods. Three experiments were performed with similar results. (+) with CIP; (−) without CIP.
Overexpression of Bcl-3 in TF-1 cells activates an HIV-1 κB enhancer.
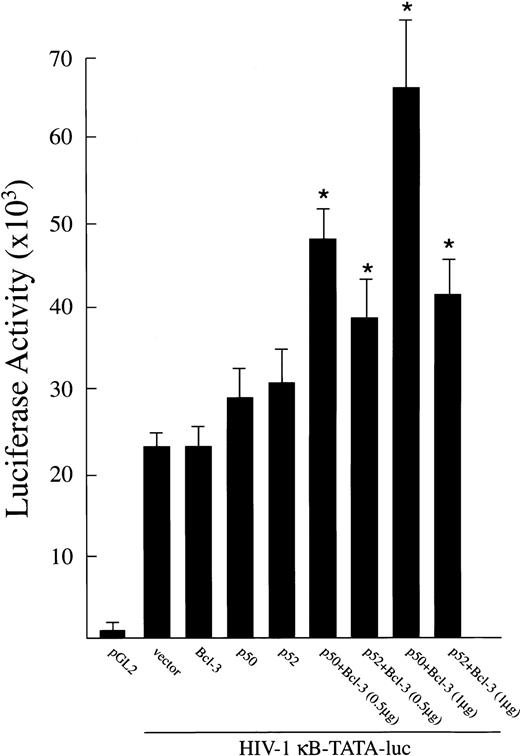
To determine whether Bcl-3 has a role in gene activation in TF-1 cells, functional reporter gene assays were performed with an HIV-1 κB-TATA-luciferase reporter plasmid.19,22 This plasmid was cotransfected into proliferating TF-1 cells (cultured in the presence of GM-CSF) along with cDNA for Bcl-3, p50, or p52. In these experiments, indicated amounts of plasmids expressing p50, 52, or Bcl-3 were transfected either separately or in combination along with 0.5 μg of HIV-1 κB-TATA-luciferase reporter plasmid into TF-1 cells. The total amount of transfected DNA (2.0 μg) was kept constant by adding appropriate amounts of expression vector without insert. The pGL2 vector was transfected as the negative control. As shown in Fig 7, a high level of κB-TATA-luc was expressed when this reporter plasmid was transfected alone. This result is consistent with our finding that endogenous Bcl-3 and other NF-κB factors are induced in TF-1 cells by the growth factor GM-CSF.26 Significant further induction of luciferase was observed in TF-1 cells cotransfected with the κB-TATA reporter plasmid and Bcl-3 together with p50 or p52 (P < .05). These results demonstrate that both endogenously expressed and transfected Bcl-3 and NF-κB factors p50 or p52 are capable of positively stimulating gene expression from the HIV-1 κB enhancer in TF-1 cells.
Activation of an HIV-1 κB-TATA-luciferase reporter plasmid after overexpression of Bcl-3 in TF-1 cells. A total of 0.5 μg of plasmids expressing Bcl-3, p50, or p52 was cotransfected with 0.5 μg of the HIV-1 κB-TATA-luciferase reporter plasmid into TF-1 cells separately or in combination. The pGL2 basic vector (0.5 μg) was used as negative control. Where noted, 1.0 μg of Bcl-3 was cotransfected. The total amount of transfected DNA was kept constant by adding appropriate amounts of expression vector without insert. At 48 hours after transfection, cells were collected for luciferase assay. Results are expressed as the mean ± SEM (×103 cpm). Five experiments were performed. *A significant increase above the κB-TATA reporter plasmid (P ≤ .05).
Activation of an HIV-1 κB-TATA-luciferase reporter plasmid after overexpression of Bcl-3 in TF-1 cells. A total of 0.5 μg of plasmids expressing Bcl-3, p50, or p52 was cotransfected with 0.5 μg of the HIV-1 κB-TATA-luciferase reporter plasmid into TF-1 cells separately or in combination. The pGL2 basic vector (0.5 μg) was used as negative control. Where noted, 1.0 μg of Bcl-3 was cotransfected. The total amount of transfected DNA was kept constant by adding appropriate amounts of expression vector without insert. At 48 hours after transfection, cells were collected for luciferase assay. Results are expressed as the mean ± SEM (×103 cpm). Five experiments were performed. *A significant increase above the κB-TATA reporter plasmid (P ≤ .05).
In control experiments (not shown), 1.5 μg of plasmid expressing NF-κB factors p50 or p52 was cotransfected with 0.5 μg of HIV-1 κB-TATA-luciferase reporter plasmid in TF-1 cells. Luciferase activity was not significantly different from cotransfection with the reporter plasmid alone.
Bcl-3 binds to and transactivates a κB site in thec-myb promoter.
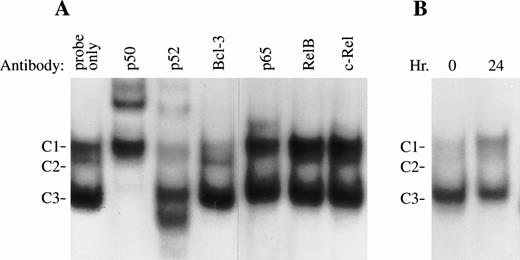
Previous studies have shown that transiently transfected Bcl-3 or purified Bcl-3 proteins interact with both p50 and p52 subunits of NF-κB/Rel proteins.2,3,4,9,10 Depending on experimental conditions, different functional results have been reported. Bcl-3 protein has been observed to either inhibit the DNA binding activity of NF-κB p50 and p52 in vitro3 or facilitate p52 or p50 activity by forming a complex with these NF-κB proteins on DNA.4,9,10 To examine the role of Bcl-3 in κB binding under physiological conditions, the interaction of endogeneous Bcl-3 with NF-κB transcription factors in growth factor-stimulated TF-1 cells was examined. EMSA was performed using an NF-κB binding site that recently was identified from the promoter region of the humanc-myb gene and nuclear extracts from TF-1 cells stimulated with GM-CSF for 24 hours. This site was chosen because induction ofc-myb mRNA in response to GM-CSF stimulation was observed in these TF-1 (data not shown) and c-myb has previously been shown to have an important role in erythropoiesis.27-30 Three major protein/DNA complexes were detected (Fig 8, C1, C2, and C3). Antibody supershift assays showed that all 3 complexes, C1, C2, and C3, immunoreacted with the anti-p52 antibody. The anti-p50 antibody supershifted both C2 and C3 complexes, whereas anti-p65 (RelA) supershifted the C2 complex. None of these complexes was immunoreactive with antibodies for RelB or c-Rel. These results suggest that p50 and p52 bind to the NF-κB site as either homodimers or heterodimers with each other or with p65. Formation of the C1 complex was reproducibly inhibited by anti–Bcl-3 antibody, suggesting that Bcl-3 is a part of this complex, which is also composed of p52. In nonstimulated TF-1 cells, the C1 complex containing Bcl-3 and p52 was much weaker than that detected in induced cells (Fig 8B). These results are consistent with the hypothesis that induction of Bcl-3 expression by GM-CSF contributes to enhancement of Bcl-3 DNA binding and increased formation of C1.
Bcl-3 is associated with NF-κB p52 in TF-1 cells (EMSA). (A) Six micrograms of nuclear extracts prepared from TF-1 cells stimulated with GM-CSF for 24 hours was incubated with different NF-κB antibodies for 10 minutes before adding a32P-labeled c-myb κB binding oligonuclear probe. Three DNA-protein complexes were generated. Complexes 2 and 3 (C2 and C3) were supershifted by anti-p50; C1, C2, and C3 were shifted by anti-p52. C1 was reproducibly inhibited by anti–Bcl-3 antibody (with a long exposure, a supershifted band was also visible). However, anti-RelB and anti–c-Rel had no effect on any of these complexes. Similar results were observed in 3 experiments. (B) EMSA was performed with growth factor-deprived TF-1 cells or TF-1 cells induced with GM-CSF for 24 hours. The C1 complex was greatly increased by GM-CSF stimulation, whereas other complexes had no significant change. This experiment was repeated 3 times with similar results.
Bcl-3 is associated with NF-κB p52 in TF-1 cells (EMSA). (A) Six micrograms of nuclear extracts prepared from TF-1 cells stimulated with GM-CSF for 24 hours was incubated with different NF-κB antibodies for 10 minutes before adding a32P-labeled c-myb κB binding oligonuclear probe. Three DNA-protein complexes were generated. Complexes 2 and 3 (C2 and C3) were supershifted by anti-p50; C1, C2, and C3 were shifted by anti-p52. C1 was reproducibly inhibited by anti–Bcl-3 antibody (with a long exposure, a supershifted band was also visible). However, anti-RelB and anti–c-Rel had no effect on any of these complexes. Similar results were observed in 3 experiments. (B) EMSA was performed with growth factor-deprived TF-1 cells or TF-1 cells induced with GM-CSF for 24 hours. The C1 complex was greatly increased by GM-CSF stimulation, whereas other complexes had no significant change. This experiment was repeated 3 times with similar results.
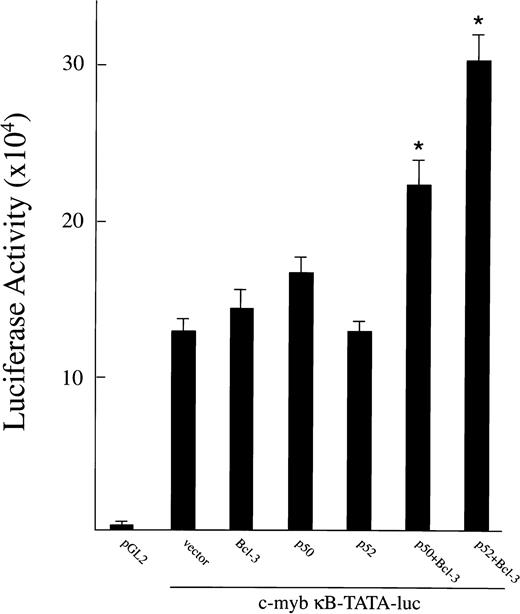
To further examine whether Bcl-3 has a role in regulating c-mybexpression, the c-myb κB-TATA-luciferase plasmid was cotransfected into proliferating TF-1 cells (cultured with GM-CSF) along with cDNA for Bcl-3, p50, or p52. Methods are as described for the HIV-1 κB-TATA-luciferase reporter plasmid. As shown in Fig 9, significant levels of c-mybκB-TATA-luciferase were expressed when this reporter plasmid was transfected alone. This is consistent with our finding that Bcl-3 and other NF-κB factors are induced by GM-CSF26 and that significant amounts of c-myb mRNA are present in these cells (data not shown). A significant induction of luciferase was observed when the TF-1 cells were transfected with the c-myb reporter plasmid along with Bcl-3 together with p52 or 50 (P ≤ .05). These results demonstrate that endogenously induced and transfected Bcl-3/NF-κB factors are capable of inducing gene expression from thec-myb NF-κB site in vivo in TF-1 cells.
Overexpression of Bcl-3 and p50 or p52 activates ac-myb κB-TATA-luciferase reporter plasmid. A total of 0.5 μg of plasmids expressing Bcl-3, p50, or p52 was cotransfected with 1.0 μg of c-myb κB-TATA-luciferase reporter plasmid into TF-1 cells separately or in combination. The pGL2 basic vector was used as negative control. The total amount of transfected DNA (2 μg) was kept constant by adding appropriate amounts of expression vector without insert. At 48 hours after transfection, cells were collected for luciferase assay. Results are expressed as the mean ± SEM (×103 cpm). Three experiments were performed. *A significant increase above the c-myb κB-TATA reporter plasmid (P < .05).
Overexpression of Bcl-3 and p50 or p52 activates ac-myb κB-TATA-luciferase reporter plasmid. A total of 0.5 μg of plasmids expressing Bcl-3, p50, or p52 was cotransfected with 1.0 μg of c-myb κB-TATA-luciferase reporter plasmid into TF-1 cells separately or in combination. The pGL2 basic vector was used as negative control. The total amount of transfected DNA (2 μg) was kept constant by adding appropriate amounts of expression vector without insert. At 48 hours after transfection, cells were collected for luciferase assay. Results are expressed as the mean ± SEM (×103 cpm). Three experiments were performed. *A significant increase above the c-myb κB-TATA reporter plasmid (P < .05).
DISCUSSION
GM-CSF and Epo induce erythroid proliferation by triggering a cascade of signal transduction events. Upon binding to their specific receptors, these growth factors induce activation of second messengers, including the protein tyrosine kinase JAK2, Stat5, ras, Raf-1, and MAP kinase.31-36 Induction of these signaling proteins results in activation of specific transcription factors, including GATA-1, SCL and other basic helix-loop-helix (bHLH) transcription factors, NF-E2, and RBTN2, which in turn control erythroid proliferation and differentiation.25 37-42 In this study, involvement of the proto-oncogene Bcl-3 in the signaling mechanisms of GM-CSF and Epo is demonstrated. The expression of Bcl-3 is greatly enhanced by hematopoietic growth factor stimulation at the level of both transcription and translation. A dynamic expression pattern ofBcl-3 in normal erythroid cells is demonstrated, suggesting that Bcl-3 is also involved in normal erythropoiesis.
The induction of Bcl-3 by both GM-CSF and Epo involves enhanced expression of Bcl-3 mRNA, suggesting that these growth factors induce transcription of the Bcl-3 gene. A number of potential regulatory sequences have been found in the 5′-flanking region of the Bcl-3 gene.1 These sequences include binding sites for Sp1, AP-1, AP-2, and NF-κB, indicating possible involvement of these transcriptional factors in regulation of Bcl-3 gene expression.1,3 In addition to these elements, we observed two other potential regulatory sequences, CAGCTG and CAACTG.1 These two DNA sequences are highly similar to the E-protein binding site CANNTG, termed E-box.43E-proteins belonging to the bHLH family of transcription factors form heterodimers with tissue-specific transcription factors such as SCL, which then bind to E-box motifs to participate in the transcriptional regulation of genes involved in cell growth.39,43,44 We and others have previously shown that SCL, E47, and HEB are involved in BFU-E–derived human erythroid proliferation and differentiation.39,40 44 GM-CSF stimulation of TF-1 cells induces E47 expresssion (Fig 3), and this was also observed for HEB and E2-2 (data not shown). Bcl-3transcription is likely controlled through recognition of binding sites in the promoter region of Bcl-3 by growth factor-regulated transcription factors involved in erythroid proliferation and differentiation, which may include SCL heterodimers.
The studies reported here demonstrate that endogenous Bcl-3 is located in both the nuclear and cytoplasmic compartments. Similar results have been obtained by immunofluorescent staining of TF-1 cells (not shown). These results are not in full agreement with previous reports in which the transiently transfected Bcl-3 is primarily expressed in the nucleus.2,3,9 Such discrepancy suggests that the nuclear expression of Bcl-3 may be regulated under physiological conditions. The cytoplasmic retention of endogenous Bcl-3 in TF-1 and BFU-E–derived cells may be due to physical association with other proteins or to posttranslational modifications. Bcl-3 has been shown to form nuclear complexes with NF-κB p50 and p52,9,10,11 but it is not known what factors form cytoplasmic complexes with Bcl-3. Our studies also demonstrate that Bcl-3 is hyperphosphorylated in both TF-1 cells and normal erythroblasts. High and low molecular weight isoforms are present in nucleus and cytoplasm; therefore, the phosphorylation state does not appear to determine subcellular localization. Higher and lower molecular weight isoforms are also induced by growth factor stimulation. The function of different phosphorylation states of Bcl-3 is not clear at this time. Phosphorylation of Bcl-3 has previously been shown to be important for full activity in other cell types.3,4 For example, dephosphorylation of Bcl-3 in thymocytes greatly decreased the ability of Bcl-3 to augment the DNA binding activity of endogenous p50 homodimers.4
GM-CSF and Epo not only induce the expression of Bcl-3 but also enhance nuclear translocation upon stimulation of TF-1 cells with these hematopoietic growth factors. Interaction of Bcl-3 with NF-κB/Rel transcription factors has been demonstrated in a number of previous studies, although the functional consequences of these interactions remains controversial.3,5,9-12 Although some studies demonstrated that Bcl-3 inhibits the DNA binding activity of NF-κB proteins, other studies showed that Bcl-3 binds to a κB enhancer together with p52 or p50 and serves as a transcriptional activator. Consistent with the latter finding, we demonstrated here that overexpression of Bcl-3 along with p50 or p52 in TF-1 cells results in induction of both HIV-1 and c-myb κB-TATA-luciferase reporter genes and that Bcl-3 has a function in gene activation in vivo in TF-1 cells. These results provide evidence that Bcl-3 may play a positive role in transactivation of κB-containing genes in erythroid cells. In TF-1 cells, GM-CSF stimulates increased c-myb mRNA expression. We also present data that suggest that Bcl-3 forms a complex with NF-κB p52 on a κB element present in the promoter of thec-myb gene and that formation of this complex is enhanced by stimulation with GM-CSF. Bcl-3 interaction with nuclear NF-κB proteins, including p52, may contribute to induction of expression of specific genes involved in erythroid proliferation, includingc-myb. The importance of c-myb in erythropoiesis has previously been demonstrated.27-30 The reported increase inBcl-3 in response to mitogenic signaling, associated with induction of immediate genes c-fos and c-myc, is consistent with our observations.1
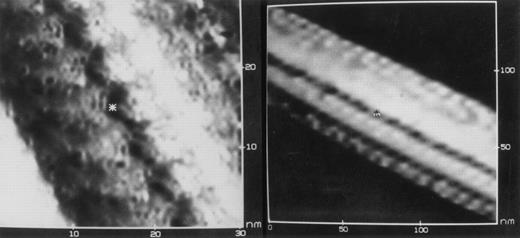
Deoxyhemoglobin S crystal. Chromatographically purified HbS (in 5 mmol/L KCl, 10 mmol/L Tris, pH 6.5) was deoxygenated, air-dried onto a graphite chip under a stream of nitrogen, and examined by scanning tunneling electron microscopy (STEM). Images at two levels of resolution are shown for a portion of a single crystalline bundle of HbS fibers that was 1,450 nm long and 65 nm in diameter. The size of the individual constituent subunits (6 × 5 nm) is consistent with their identity as hemoglobin tetramers. The vivid detail at the interface between tetramers (asterisk) suggests that STEM could be used to help define the fine structure of HbS polymers. (Courtesy of Mary M. Christopher, Yi Lin, Roy Matthew, D. Fennell Evans, and Robert P. Hebbel, Departments of Medicine and Chemical Engineering, University of Minnesota, Minneapolis, MN 55455.)
Deoxyhemoglobin S crystal. Chromatographically purified HbS (in 5 mmol/L KCl, 10 mmol/L Tris, pH 6.5) was deoxygenated, air-dried onto a graphite chip under a stream of nitrogen, and examined by scanning tunneling electron microscopy (STEM). Images at two levels of resolution are shown for a portion of a single crystalline bundle of HbS fibers that was 1,450 nm long and 65 nm in diameter. The size of the individual constituent subunits (6 × 5 nm) is consistent with their identity as hemoglobin tetramers. The vivid detail at the interface between tetramers (asterisk) suggests that STEM could be used to help define the fine structure of HbS polymers. (Courtesy of Mary M. Christopher, Yi Lin, Roy Matthew, D. Fennell Evans, and Robert P. Hebbel, Departments of Medicine and Chemical Engineering, University of Minnesota, Minneapolis, MN 55455.)
ACKNOWLEDGMENT
The authors thank Dr Timothy W. McKeithan for providing theBcl-3 cDNA and Dr Warner C. Greene for the antipeptide specific antisera for NF-κB/Rel proteins. We are grateful to Dr Toshio Kitamura for providing TF-1 cells. The authors thank Maxine Gerberich for careful preparation of the manuscript. We appreciate the technical assistance of Carol Stine.
Supported by National Institutes of Health Grants No. DK46778 (B.A.M.), CA 68471 (S.-C.S.), and MO1 RR10732 (GCRC grant) and by a grant from The Pennsylvania State University Cancer Center. S.-C.S. is a scholar of the American Society of Hematology. B.A.M. is the recipient of an American Cancer Society Faculty Award.
Address reprint requests to Barbara A. Miller, MD, Department of Pediatrics, The Milton S. Hershey Medical Center, PO Box 850, Hershey, PA 17033-0850; e-mail: bamll@psu.edu.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal