Abstract
T-cell prolymphocytic leukemia (T-PLL) is a rare form of mature T-cell leukemia associated with chromosomal rearrangements implicatingMTCP1 or TCL1 genes. These genes encode two homologous proteins, p13MTCP1 and p14TCL1, which share no similarity with other known protein. To determine the oncogenic role of MTCP1, mice transgenic for MTCP1under the control of CD2 regulatory regions (CD2-p13 mice) were generated. No abnormality was detected during the first year after birth. A late effect of the transgene was searched for in a cohort of 48 CD2-p13 mice aged 15 to 20 months, issued from 3 independent founders. Lymphoid hemopathies, occurring in the three transgenic lines, were characterized by lymphoid cells with an irregular nucleus, a unique and prominent nucleolus, condensed chromatin, a basophilic cytoplasm devoid of granules, and an immunophenotype of mature T cells. The molecular characterization of Tcrb rearrangements demonstrated the monoclonal origin of these populations. Histopathological analysis of the cohort demonstrated early splenic and hepatic infiltrations, whereas lymphocytosis and medullar infiltrations were found infrequently. The engraftment of these proliferations in H2-matched animals demonstrated their malignant nature. Cumulative incidence of the disease at 20 months was 100%, 50%, and 21% in F3, F4, and F7 lines, respectively, and null in the control group. The level of expression of the transgene, as estimated by Western blotting in the transgenic lines correlated with the tumoral incidence, with the highest expression of p13MTCP1 being found in F3 mice. CD2-p13 transgenic mice developed an hemopathy similar to human T-PLL. These data demonstrate that p13MTCP1 is an oncoprotein and that CD2-p13 transgenic mice represent the first animal model for mature T-PLL.
T-CELL PROLYMPHOCYTIC leukemia (T-PLL) is a rare form of leukemia occurring in elderly people. It is characterized by an aggressive tumoral syndrome, associated with lymphocytosis, lymphadenopathy, hepatosplenomegaly, and skin lesions. Prolymphocytes are larger than lymphocytes and have a high nucleocytoplasmic ratio, a basophilic cytoplasm devoid of granules, moderately condensed chromatin, and a single prominent nucleolus. The immunophenotype is that of mature or premature T lymphocytes.1 Similar cells have been observed in some patients suffering from the genetic disease ataxia telangiectasia (AT). In these AT patients, the clonal lymphoid population (named ataxia telangiectasia clonal proliferation [ATCP]) is quiescent, but can transform in authentic T-PLL after several years.2 These observations suggest that ATCP is a preleukemic stage of T-PLL.3 Recurrent chromosomal aberrations associated with T-PLL and ATCP involve the 14q11 region, containing the TCRA/Dgene, and the Xq28 or 14q32.1 regions.1 Their molecular characterization led to the identification of the MTCP1 andTCL1 genes, respectively.4-6 Breakpoints of the seven t(X;14) translocations characterized at a molecular level are 5′ to or within the MTCP1 gene.4,5,7-9 This gene is organized into seven exons spread over approximately 10 kb.Alternative splicing generates A and B transcripts encoding two entirely different proteins, p8MTCP1 and p13MTCP1, an arrangement that is highly unusual in vertebrates.4 B transcripts contain the 7 exons of the gene and encode p13MTCP1, a 13-kD protein whose expression has only been detected so far in t(X;14)-associated T-cell proliferations.8,10 This protein shares significant homology with the product of TCL1, p14TCL1.11 Elucidation of the three-dimensional structure has confirmed that p13MTCP1 and p14TCL1 have a similar structure shared by no other known protein.12 13These indirect evidences suggest that p13MTCP1 is the oncogenic product of MTCP1. However, p13MTCP1 and p14TCL1 do not resemble other known oncoproteins, no functional role has been so far attributed to them, and in vitro transformation assays remained negative (unpublished data). The possibility thatMTCP1 is oncogenic was thus investigated by generating transgenic mice overexpressing p13MTCP1 in T cells. No phenotypic abnormality was observed before the age of 15 months, when clinically or biologically detectable hemopathies similar to human T-PLL started to appear. We present here the analysis of this transgenic cohort.
MATERIALS AND METHODS
CD2-p13 transgenic mice.
The CD2-p13 transgenic construct, was made by cloning in theEcoRI site of the CD2 vector,14 a fragment corresponding to the open reading frame of human MTCP1 coding for p13MTCP1, which shares 95% of identity with the murine p13MTCP1 protein.8Transgenic mice were generated using a Sal I-Xba I fragment encompassing the insert. Three founders were obtained (F3, F4, and F7) and bred. Transgenic heterozygote mice issued from these founders were studied and compared with nontransgenic siblings raised in identical conditions. Tumoral analysis was conducted on 77 mice aged from 15 to 20 months, including 48 transgenics (12 F3, 22 F4, and 14 F7) and 29 nontransgenic siblings.
Genotyping the mice.
Genotyping was performed on tail DNAs using the Nucleospin C&T kit (Macherey-Nagel, Hoerdt, France). Polymerase chain reaction (PCR) was performed on 5 μL DNA in a final mix containing 200 ng of CD2 primer (5′-GTGTGGACTCCACCAGTC-3′), 200 ng of III/IV primer (5′-CCCCTGACCATTAAA-3′), 0.2 mmol/L deoxyribonucleotides, 1× Taq buffer (Boehringer, Meylan, France), and 0.5 μL Taq polymerase (Boehringer). A 30-cycle amplification was performed (94°C for 15 seconds, 44°C for 15 seconds, and 72°C for 15 seconds) after an initial denaturation step of 3 minutes at 94°C. A specific band of 400 bp was shown on a 1.5% agarose gel. A final check of the transgenic status was performed by Southern blot on high molecular weight DNA extracted from splenic cells using the QIAamp Blood kit (Qiagen, Hilden, Germany). DNAs were digested byBamHI restriction enzyme and the resulting fragments were separated according to size by electrophoresis through a 0.8% agarose gel. After alkaline transfer on N+Hybond membranes (Amersham, Les Ulis, France), samples were hybridized to 32P-radiolabeled CD2 minigene 2-kb BamHI fragment and shown by autoradiography. A 2-kb band showed the transgenic status of the mice.
Western blotting.
Cell proteins were extracted with the Triple Detergent Lysis Buffer,15 quantified using the BCA kit (Pierce, Rockford, IL), sized-fractionated on 15% Tris-glycine sodium dodecyl sulfate-polyacrylamide gels (SDS-PAGE), and electrotransferred onto nitrocellulose. The membrane was blocked overnight in 10% nonfat dried milk in PBST (phosphate-buffered saline [PBS]: 7.6 g/L NaCl, 0.7 g/L Na2PO4, 0.2 g/L KPO4, and 0.1% Tween 20). The previously described antiserum anti-p13MTCP1 was diluted 1:1,000 in the blocking buffer and applied to the membrane.8 After 1 hour of incubation, the membranes were washed with PBST and incubated with goat antirabbit IgG-peroxidase conjugate (Boehringer) for 1 hour. After three washes in PBST, the membranes were overlaid with the chemiluminescent substrate solution and developed according to the manufacturer's instructions (ECL; Amersham). The same membrane was then probed using a monoclonal antibody against actin (AB-1; Oncogene Science, Paris, France) and a sheep antimouse peroxidase conjugate (Amersham) to control loading.
White blood cell (WBC) count.
Blood was collected from the cavernous sinus with a capillary tube in a tube coated with EDTA (Microtainer Brand; Becton Dickinson, Orders, France). Smears were immediately prepared and stained with May Grünwald Giemsa. Full counts were made on a cell counter (Coulter STKS; Coultronics, Margency, France).
Fluorescence-activated cell sorting (FACS) analysis of the splenic cells.
Spleens were weighed and dissociated in 10 mL RPMI 1640 (GIBCO BRL, Life Technologies, Cergy Pontoise, France) between two frosted slides. Cell suspensions were spun for 5 minutes at 1,500 rpm, the red blood cells were then lyzed in ACK (0.15 mol/L NH4Cl, 1 mmol/L KHCO3, 0.1 mmol/L EDTA), spun for 5 minutes at 1,500 rpm, and twice washed in 0.9% NaCl. Cells were counted and resuspended in PBS at 107 cells/mL. Immunophenotyping was performed with monoclonal antibodies anti-CD3, CD4, CD8, CD25, and B220 (PharMingen, San Diego, CA), directly coupled to fluorescein isothiocyanate (FITC; CD3, CD8) or to phycoerythrin (PE; CD8, CD25, B220). A 50-μL sample of cell suspension was incubated for 30 minutes at +4°C with 1 μL antibody at the appropriate dilution and washed in 2 mL of PBS-0.2% sodium azide. Cells were resuspended in 700 μL of the same buffer and analyzed on a fluorescent cell sorter (FACS Scan; Becton Dickinson).
Cell sorting.
Splenic cells of a 5-month-old F3 mouse were stained with monoclonal antibodies against CD4, CD8, CD3, or B220. Positive fractions were sorted on a FACS Scan cell sorter. The purity of the fractions was evaluated by analysis on a FACS Scan cytometer and was greater than 95% in all cases.
Analysis of the Tcrb gene configuration by Southern blot.
Tcrb rearrangements were analyzed by Southern blotting onHindIII-digested splenocyte DNAs. A serial dilution of tumoral DNA in normal splenic DNA (100%, 25%, and 10%) was included on each blot to quantify the intensity of clonal rearrangements. Clonal rearrangements accounting for less than 10% of the splenocytes were considered as not significant. The probes used were the 650-bpEcoRI fragment of 86T5 cDNA,16 which hybridizes to murine Tcrb Cβ1 and Cβ2 regions, and the 286-bp Pst I-Cla I fragment containing the Jβ2.6 segment of the murine Tcrb.
Histopathology.
Organs were fixed in AFA (80% ethanol, 15% formaldehyde, 5% acetic acid) for 2 to 4 hours and further processed for paraffin embedding. Three-micrometer sections were stained with hematoxylin-eosin and analyzed by two different pathologists.
Transplantation of leukemic cells.
Ten million leukemic cells resuspended in PBS were intravenously injected in 4- to 6-week-old C57BL/6 × DBA/2 mice. Neither speed nor efficiency of engraftment was enhanced in 2.5 Gy irradiated animals.
RESULTS AND DISCUSSION
Characterization of CD2-p13 transgenic mice.
We generated transgenic mice in which expression of p13MTCP1 was controlled by the CD2 regulatory sequences (CD2-p13 mice).14 Three transgenic mice (F3, F4, and F7) were obtained and bred. The expression of the transgene was evaluated on each lineage by Western blot on proteins extracted from thymus, spleen, and nonlymphoid organs. The three transgenic lines expressed p13MTCP1 in thymuses at different levels: high for F3, intermediate for F4, and weak for F7 (Fig 1A). Expression was weaker in spleens, but the gradation between the lineages was maintained. No expression was detected in the nonlymphoid organs (data not shown). The transgene expression was then analyzed in lymphoid subpopulations. The p13MTCP1 protein was detected in CD4+and CD8+ splenocytes at comparable levels, but not in B lymphocytes (Fig 1B). No difference in the size or distribution of the T-cell subpopulations was detected in a series of transgenic mice aged from 6 weeks to 1 year compared with nontransgenic siblings (data not shown).
Transgene expression in CD2-p13 mice. Detergent lysates were subjected to SDS-PAGE through a 15% gel. Immunoblot analysis was performed using the anti-p13 rabbit antiserum (upper panel) and loading was estimated using anti-actin monoclonal antibody (lower panel). (A) Expression analysis in the three transgenic lines. Thymocytes (Thymus) and splenocytes (Spleen) were analyzed in transgenics (F3, F4, and F7) and nontransgenic control mouse (C). (B) Expression analysis in the lymphoid subpopulations. CD3+ cells (CD3), CD4+ cells (CD4), CD8+ cells (CD8), and B220+ B cells (B) from a F3 transgenic mouse spleen were purified by cell sorting (purity >95%) and compared with total splenocytes of this mouse (total). Arrows indicate the p13-and actin-specific signals.
Transgene expression in CD2-p13 mice. Detergent lysates were subjected to SDS-PAGE through a 15% gel. Immunoblot analysis was performed using the anti-p13 rabbit antiserum (upper panel) and loading was estimated using anti-actin monoclonal antibody (lower panel). (A) Expression analysis in the three transgenic lines. Thymocytes (Thymus) and splenocytes (Spleen) were analyzed in transgenics (F3, F4, and F7) and nontransgenic control mouse (C). (B) Expression analysis in the lymphoid subpopulations. CD3+ cells (CD3), CD4+ cells (CD4), CD8+ cells (CD8), and B220+ B cells (B) from a F3 transgenic mouse spleen were purified by cell sorting (purity >95%) and compared with total splenocytes of this mouse (total). Arrows indicate the p13-and actin-specific signals.
Elderly CD2-p13 transgenic mice develop T-PLL.
The prolonged survey of the animal cohort resulted in the detection of mice suffering from leukemic syndromes. Seven mice from the cohort aged from 15 to 18 months died of various causes (2 urogenital tumors and 1 polyclonal lymphadenitis in transgenic and control animals) and of 4 cases of leukemia in F4 transgenics. Because of the short delay between the clinically detectable stage of the disease and the distress of the animals, the remainder of the cohort (44 transgenics) was killed at 18 to 20 months of age, as well as 29 nontransgenic age-matched siblings, and studied.
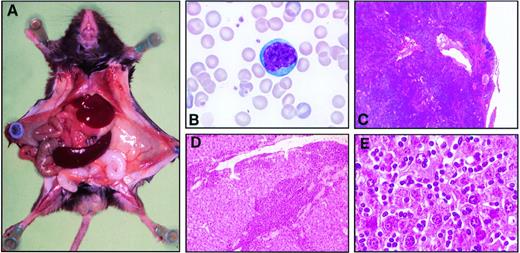
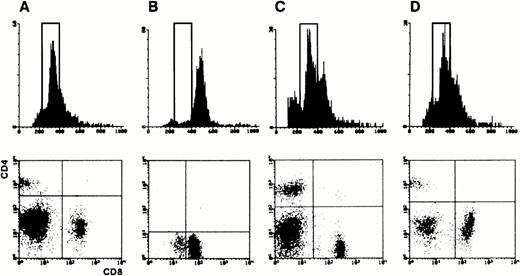
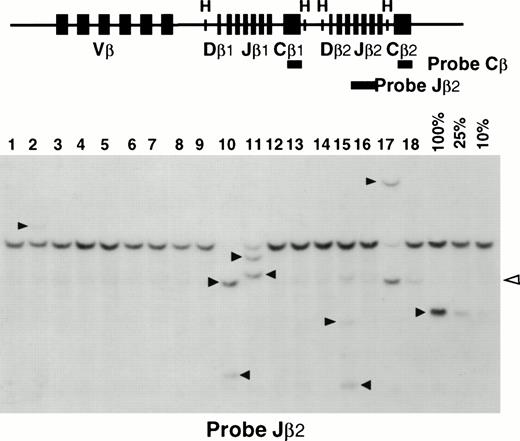
Cytological examination of blood smears and spleen appositions in transgenic mice showed frequent invasion of lymphoid cells characterized by an irregular nucleus with condensed chromatin containing a unique and prominent nucleolus and by a basophilic cytoplasm devoid of granules (Fig 2B). Histopathological examination demonstrated constant infiltrations of spleens and livers by these abnormal lymphoid cells in presence or absence of organomegaly (Fig 2A, C, D, and E). The immunophenotype of the leukemic cells, when tested, was CD3+CD8+CD4−CD25−B220−in 23 cases, and was CD3+CD4+CD8−CD25−B220−in 1 case (Fig 3). The rarity of CD4+ T-cell leukemia was not due to a lower level of expression of p13MTCP1 in the CD4+subset (Fig 1B). In humans, MTCP1 aberrations are associated with CD4+CD8+, CD4−CD8+, and CD4+CD8− T-PLLs.1 Subtle differences in T-cell differentiation and physiology in the two species may thus account for the preferential transformation of CD8+ cells in the transgenics. In all but 3 cases, these cells were larger than normal peripheral blood lymphocytes based on the forward scatter (FSC) histogram in FACS analysis (Fig 3B and C). The three normally sized clonal populations were reminiscent of the small cell variant of the human T-PLL, alternatively named T-cell chronic lymphocytic leukemia (Fig 3D).1,17 Clonality of T-cell proliferations was demonstrated by Southern blot analysis of Tcrbgene rearrangements (Fig 4). This leukemic disease fitted all criteria defining the human T-PLL.18
Histopathological analysis of the CD2-p13 mice. (A) Macroscopic view of a F4 CD2-p13 transgenic mouse showing an enlarged spleen of 1,100 mg. (B) Photomicrography of a typical T-cell prolymphocyte from a F4 CD2-p13 transgenic mouse. The blood smear was stained with May-Grünwaltd-Giemsa. (C through E) Histology of spleen (C) and liver (D and E) after hematoxylin-eosin staining. (C) The spleen has lost its normal architecture and is infiltrated by prolymphocytes beyond its capsule. (D) Low magnification of the liver showing large amounts of prolymphocytes surrounding portal tracts. (E) High magnification of the liver showing numerous prolymphocytes within sinusoids.
Histopathological analysis of the CD2-p13 mice. (A) Macroscopic view of a F4 CD2-p13 transgenic mouse showing an enlarged spleen of 1,100 mg. (B) Photomicrography of a typical T-cell prolymphocyte from a F4 CD2-p13 transgenic mouse. The blood smear was stained with May-Grünwaltd-Giemsa. (C through E) Histology of spleen (C) and liver (D and E) after hematoxylin-eosin staining. (C) The spleen has lost its normal architecture and is infiltrated by prolymphocytes beyond its capsule. (D) Low magnification of the liver showing large amounts of prolymphocytes surrounding portal tracts. (E) High magnification of the liver showing numerous prolymphocytes within sinusoids.
FACS analysis of splenic populations. (Upper panel) FSC histogram analyzing the cell size. A rectangle defines the normal sized splenocytes. (Lower panel) CD4 and CD8 double fluorescence patterns. (A) Analysis of normal control splenocytes. (B) Analysis of a transgenic mouse with a major tumoral load. Nearly all splenic cells are large sized CD8+ prolymphocytes. (C) Analysis of a transgenic mouse with minor tumoral load. FCS histogram shows a larger than normal sized cell population with a CD8+ phenotype (not shown). (D) Analysis of a small cell variant. An excess of CD8+ cells is clearly visible, but no larger than normal cells are detected on the FSC histogram.
FACS analysis of splenic populations. (Upper panel) FSC histogram analyzing the cell size. A rectangle defines the normal sized splenocytes. (Lower panel) CD4 and CD8 double fluorescence patterns. (A) Analysis of normal control splenocytes. (B) Analysis of a transgenic mouse with a major tumoral load. Nearly all splenic cells are large sized CD8+ prolymphocytes. (C) Analysis of a transgenic mouse with minor tumoral load. FCS histogram shows a larger than normal sized cell population with a CD8+ phenotype (not shown). (D) Analysis of a small cell variant. An excess of CD8+ cells is clearly visible, but no larger than normal cells are detected on the FSC histogram.
Analysis of Tcrb rearrangements. (Upper panel) A schematic representation of the murine Tcrb gene is shown with its Vβ, Dβ, Jβ, and Cβ segments. The relevant HindIII sites (H) are shown. The Jβ2 and Cβ probes are shown as thick lines. (Lower panel) An example of Southern blot analysis is shown. Mouse splenic DNAs are analyzed on lanes 1 through 18. Quantification of the clonal rearrangements is made by comparison to a serial dilution of tumoral DNA into normal spleen DNA (100%, 25%, and 10%) included on each membrane. Black arrows indicate the clonalTcrb rearrangements. The open arrow shows incomplete DJ rearrangements that were not taken into account. Samples in lanes 9, 10, and 17 demonstrated major tumor loads. Samples in lanes 2 and 15 demonstrated minor tumoral loads.
Analysis of Tcrb rearrangements. (Upper panel) A schematic representation of the murine Tcrb gene is shown with its Vβ, Dβ, Jβ, and Cβ segments. The relevant HindIII sites (H) are shown. The Jβ2 and Cβ probes are shown as thick lines. (Lower panel) An example of Southern blot analysis is shown. Mouse splenic DNAs are analyzed on lanes 1 through 18. Quantification of the clonal rearrangements is made by comparison to a serial dilution of tumoral DNA into normal spleen DNA (100%, 25%, and 10%) included on each membrane. Black arrows indicate the clonalTcrb rearrangements. The open arrow shows incomplete DJ rearrangements that were not taken into account. Samples in lanes 9, 10, and 17 demonstrated major tumor loads. Samples in lanes 2 and 15 demonstrated minor tumoral loads.
Invasion staging and incidence of the disease.
The analysis of the transgenic mice given above defined two stages of the disease. Major tumoral load was defined by clonal T-cell rearrangements in more than 25% of the splenocytes (Fig 4) and more than 60% of monomorphic CD3+CD8+ or CD3+CD4+ splenocytes in FACS analysis (Fig 3B). This had occurred in 10 cases (Table 1). In these cases, lymphocytosis and splenomegaly were frequent but not constant (mean WBC, 226 × 109 cells/L; range, 13 to 1,470 × 109 cells/L; mean spleen weight, 909 mg; range, 87 to 2,700 mg). Major tumoral load (splenomegaly of 30 times the normal weight [as shown on Fig 2A] and lymphocytosis as high as 2,000 × 109 cells/L) was occasionally associated with near normal behavior of the animals. A complete histological analysis of 3 florid cases showed an infiltration of all lymphoid organs, livers, and lungs. Infiltration of other organs was not constant: bone marrow was invaded in only 1 case and testes were normal in all cases. Tumor cells from these 10 major T-cell clonal hemopathies were intravenously injected into H2-compatible immunocompetent mice. In 9 cases, the tumors engrafted and lymphocytosis was detectable in 1 to 4 months, demonstrating the malignant nature of these proliferations.
T-PLL Incidence in 18 to 20 Months CD2-p13 Mice
| Transgenic Line . | Major Tumoral Load* . | Minor Tumoral Load-151 . | Small Cell Variants-152 . | T-PLL Free Mice . |
|---|---|---|---|---|
| F3 | 2/12 (17%) | 9/12 (75%) | 1/12 (8%) | 0/12 (0%) |
| F4 | 8/22 (36%) | 3/22 (14%) | 0/22 (0%) | 11/22 (50%) |
| F7 | 0/14 (0%) | 1/14 (7%) | 2/14 (14%) | 11/14 (79%) |
| Control | 0/29 (0%) | 0/29 (0%) | 0/29 (0%) | 29/29 (100%) |
| Transgenic Line . | Major Tumoral Load* . | Minor Tumoral Load-151 . | Small Cell Variants-152 . | T-PLL Free Mice . |
|---|---|---|---|---|
| F3 | 2/12 (17%) | 9/12 (75%) | 1/12 (8%) | 0/12 (0%) |
| F4 | 8/22 (36%) | 3/22 (14%) | 0/22 (0%) | 11/22 (50%) |
| F7 | 0/14 (0%) | 1/14 (7%) | 2/14 (14%) | 11/14 (79%) |
| Control | 0/29 (0%) | 0/29 (0%) | 0/29 (0%) | 29/29 (100%) |
*Major tumoral load as defined by more than 25% of clonal splenocytes estimated by Southern blotting and more than 60% of monomorphic cells in FACS analysis.
Minor tumoral load as defined by 10% to 25% of clonal splenocytes estimated by Southern blotting and no lymphocytosis or organomegaly.
Small cell variant as defined by 10% to 25% of clonal splenocytes estimated by Southern blotting, but no larger than normal CD8 or CD4 lymphocytes.
In 16 cases, the presence of a clonal T-cell population in the spleen was only detected by the Southern blot analysis (Fig 4) and was not linked to organomegaly or lymphocytosis. Intensity of these clonal rearrangements correlated with the extent of the invasion evaluated by FACS analysis and histological studies, except in one tumor in which the proportion of CD8+ large cells exceeded the intensity of the Tcrb clonal rearrangement, suggesting in this case the presence of a polyclonal prolympho-cytic cell population. Prolymphocytic infiltration of the liver was constantly demonstrated in these 16 cases by histological analyses. In all but 3 cases, clonal rearrangements were associated with the presence of larger than normal CD8+ T cells (Fig 3C). The cases of small cell variant occurred in F4 (1 case) and F7 (2 cases) lines.
Altogether, a T-cell hemopathy was detected in all 12 transgenic mice (100%) issued from founder F3, 11 of 22 transgenics (50 %) issued from founder F4, 3 of 14 transgenics (21%) issued from founder F7, and none of the 29 sibling controls (Table 1). The incidence of the disease correlated with the level of expression of p13MTCP1 in the transgenic lines (Fig 1A), suggesting a dose-response effect of p13MTCP1. However, other genetic factors are probably involved, because hemopathies appeared earlier and were more severe in the F4 transgenic line, whereas the incidence was higher in the F3 line.
MTCP1 is an oncogene.
The presence of T-cell leukemia in the three transgenic lines and in none of the nontransgenic siblings clearly demonstrated thatMTCP1 is an oncogene coding for a 13-kD oncoprotein. Conversely, no in vitro or in vivo transformation assays demonstrated any oncogenic activity for the alternative and more abundant product ofMTCP1, an ubiquitous 8-kD mitochondrial protein (Stern et al, unpublished data). The products of the MTCP1 andTCL1 genes, p13MTCP1 and p14TCL1, now constitute a new oncogene family of yet unknown biological function.
T-PLL in CD2-p13 transgenic mice.
The murine hemopathy arising in CD2-p13 transgenics is, to our knowledge, previously undescribed in mice and shares most of the clinical and biological features of the human T-PLL. Its late onset (after 15 months) in transgenics also mimics the human T-PLL that arises generally in the elderly. The only noticeable differences are that human T-cell prolymphocytic cells have a more diverse immunophenotype and frequently infiltrate the skin when compared with the CD2-p13 mice. The natural history of the human T-PLL was inferred from patients with AT. These studies showed that the emergence of clonal T-cell population with all the biological characteristics of T-cell prolymphocytes precede lymphocytosis by many years.2,3 The hemopathy observed in CD2-p13 transgenics fits this model. Furthermore, the early and latent hepatosplenic prolymphocytic invasion in transgenics, which precedes the leukemic syndrome, suggests a similar preclinical stage for the human disease. The latency period before emergence of the tumors, which is longer than in most murine models of oncogenesis, is in agreement with the necessity of several genetic events in addition to the activation ofMTCP1 (or TCL1) in the development of the T-PLL. To date, studies of the human disease have demonstrated the importance of the inactivation of the ATM gene19-21 and of the duplication of the long arm of chromosome 8.1 22 The striking similarity of the human and murine T-PLLs makes CD2-p13 mice a valuable model to investigate the biological functions of the family of oncoproteins formed by p13MTCP1 and p14TCL1, to identify the secondary events necessary for the malignant phenotype, and to test therapeutics.
NOTE ADDED IN PROOF
Interestingly, transgenic mice with TCL1, a gene with high similarity to MTCP1, under the control of the 1ck promoter developed similar mature CD8+ T-cell leukemias (Virgilio et al, Proc Natl Acad Sci USA 95:3885, 1998).
ACKNOWLEDGMENT
The authors thank P. Blanchet, M. Chopin, and M. Pla for their help in generating and raising the transgenic animals; D. Kioussis for the gift of the CD2 minigene construct; C. Guyonnet for reviewing liver sections; M. Schmid for cell sorting; C. Green for critical reading; and J. Boisse and R. Nancel for artwork.
Supported by l'INSERM and la Ligue Nationale Contre le Cancer.
Address reprint requests to Marc-Henri Stern, MD, PhD, UnitéINSERM U462, Centre Hayem, Hopital Saint Louis, 75475 Paris Cedex 10, France; e-mail: mh.stern@chu-stlouis.fr.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.
© 1998 by the American Society of Hematology.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal