Abstract
Multicolor spectral karyotyping (SKY) was performed on bone marrow samples from 50 patients with multiple myeloma (MM) in anticipation of discovering new previously unidentified translocations. All samples showed complex karyotypes with chromosome aberrations which, in most cases, were not fully characterized by G-banding. Patients of special interest were those who showed add(14)(q32), add(8)(q24) and those whose G-banding karyotypes showed poor chromosome morphology. Three new recurring chromosome translocations not previously reported in MM were identified. Two of the translocations involve recurring aberrations at band 14q32.3, the site of the IgH locus, with different exchange partners. The most frequently recurring rearrangement was a subtle translocation at 14q32.3 designated as a t(14;16)(q32;q22∼23), which was identified in six patients. A second and larger translocation at 14q32, identified in two patients, was designated as a t(9;14)(p13;q32), previously associated with Waldenstrom’s macroglobulinemia and lymphoplasmacytoid lymphoma. A third translocation, identified in two patients, involved a whole-arm t(6;8)(p10;q10) translocation. The SKY technique was able to refine the designations of over 156 aberrations not fully characterized by G-banding in this study and resolved additional chromosome aberrations in every patient studied except two. The t(14;16)(q32;q22∼23) identified by SKY in this study suggests this may be a frequent translocation in MM associated with complex karyotypes and disease progression. Therefore, the SKY technique provides a useful adjunct to routine G-banding and fluorescence in situ hybridization studies in the cytogenetic analysis of MM.
MULTIPLE MYELOMA (MM) is a plasma cell disorder characterized, at the cytogenetic level, by complex karyotypes with frequent numerical and structural aberrations. The number of abnormal karyotypes reported in MM varies from 20% to 60%, but is about 40% in most published series.1,2 With conventional chromosome banding techniques the most consistent findings in MM have been the chromosomal aberrations add(14)(q32), t(11;14)(q13;q32), t(8;14)(q24;q32), and chromosome 1q aberrations.3,4Recently, several new recurring translocations in MM cell lines have been identified, including t(4;14)(p16.3;q32.3), t(6;14)(p25;q32), and t(14;16)(q32.3;q23).5-7 Several of these aberrations are similar to those seen in other B-cell disorders, involving a “promiscuous” array of exchange partners with the IgH locus8; therefore, it remains unclear if any of these aberrations are of primary importance in MM.
Molecular cytogenetic analysis by fluorescence in situ hybridization (FISH) has become a standard adjunct to routine banding methods in the clinical cytogenetics laboratory. The development of even newer molecular cytogenetic methods such as multicolor FISH (m-FISH), and multicolor spectral karyotyping (SKY), have also been shown to be powerful techniques in the expansion of FISH techniques available for the analysis of complex chromosomal rearrangements.9,10These techniques, in conjunction with FISH probes for single genes, hold the promise of helping bridge the gap between traditional banding techniques and molecular genetic analysis. SKY is a molecular cytogenetic technique that allows the simultaneous display of each chromosome in a different color.10 This technique makes possible the identification of chromosomal bands of unknown origin, including translocations, insertions, complex rearrangements, and small marker chromosomes. The SKY technique holds great promise but is limited, in some respects, by the inability to detect chromosomal inversions, very small deletions, insertions, or translocations.
Spectral karyotyping has recently been reported to identify hidden chromosome abnormalities in hematological malignancies.11In myeloma, evidence of new recurring translocations with the SKY technique has been found.12 Two cases with a novel 14;20 translocation and three regions involving recurring translocations including 3q27∼29,17q24∼25 and 20q11.2∼12 have been identified.12 These results indicate that MM shows even more karyotypic complexity than previously thought.
The application of the SKY technique as an adjunct to G-banding and FISH studies in the clinical cytogenetics laboratory could potentially help delineate the more complex chromosome aberrations seen in MM and provide new clinical insights. To test the feasibility of spectral karyotyping in a clinical setting, we have analyzed G-banded and spectral karyotypes from the same specimens on 50 patients with MM.
MATERIALS AND METHODS
Sample selection was based on showing complex karyotypes with chromosome aberrations which were not fully characterized by G-banding. Cases clearly showing the t(11;14)(q13;q32) were excluded from the study because the SKY technique is not necessary to identifiy this large rearrangement. Bone marrow of the 50 MM patients was processed for routine chromosome studies as previously described.13 An abnormal clone was identified as two or more metaphases displaying either the same structural abnormality or the same extra chromosome, or at least three cells with the same missing chromosome. Aberrations were designated according to ISCN (1995).14 Chromosome aberrations ascertained by SKY were assigned breakpoints if the aberrations were identified in two or more cells and if the comparison of DAPI banding of the same metaphase corresponded with the G-banding of other metaphase cells.
SKY methods.
The SKY probe mixture and hybridization reagents were prepared by Applied Spectral Imaging (Carlsbad, CA). Briefly, the chromosome painting probes were generated as described elsewhere, by flow sorting human chromosomes and DNA amplification using degenerate oligonucleotide primed polymerase chain reaction (DOP-PCR).15 A combinatorial labeling of five fluorochromes, including spectrum green, Texas red, spectrum orange, Cy5, and Cy5.5 were used to generate the 24 colors. Slides for spectral karyotyping were treated basically according to the manufacturer’s protocol with the probe cocktail hybridized to the slides for 2 days at 37°C. Chromosomes were counterstained with DAPI/antifade solution.
Image acquisition was performed using a SD200 Spectracube (Applied Spectral Imaging, Inc, Carlsbad, CA) mounted on a Zeiss Axioplan II microscope (Gottingen, Germany) using a custom designed optical filter (SKY-1; Chroma Technology, Brattleboro, VT) that allows for simultaneous excitation of all dyes and measurement of their emission spectra. Light travels through a Sagnac interferometer (Applied Spectral Imaging, Carlsbad, CA) in the optical head, and an interferogram is generated at all image points which is deduced from the optical path difference of the light that depends on the wavelength of the emitted fluorescence. The spectrum is recovered by Fourier transformation.16 The spectral data are displayed by assigning red, green, or blue colors to certain ranges of the spectrum. The red, green, blue (RGB) display renders a similar color to chromosomes that are labeled with spectrally overlapping fluorochromes. Based on the measurement of the discrete emission spectra at all pixels of the image, the hybridization colors are then converted to classification colors by applying an algorithm that results in the assignment of a discrete color to all pixels with an identical spectrum. DAPI banding images are acquired as part of the image acquisition process and analyzed using a DAPI specific optical filter. The DAPI images were used in conjunction with spectral classifications and G-banding for the identification of chromosome aberrations.
FISH.
Conventional dual-color whole-chromosome painting probes (Vysis, Downers Grove, IL) for chromosomes 9, 14, and 16 were used according to manufacturer’s protocol to confirm the translocations of material to the add(14)(q32) chromosome (not shown). Telomere probe 14q32.3 and alpha satellite 16 (Oncor, Gaithersburg, MD) were used in combination according to manufacturer’s protocol to confirm the reciprocal translocations t(14;16)(q32;q22∼23) (not shown).
RESULTS
We examined 50 bone marrow samples by routine G-banding and reanalyzed the same sample from each patient with spectral karyotyping for the presence of new previously unidentified translocations (Figs 1 through 3). Recurring nonrandom translocations identified from add(14)(q32), add(8)(q24), or refined designations are presented in Table 1, while the composite G-banding karyotypes and refinements of nonclonal aberrations by spectral karyotyping are presented in Table2. A total of 156 chromosome aberrations not identified by routine G-banding were at least partially resolved with the spectral karyotyping technique (Table 2). Among the 50 patients, numerous translocations were identified by SKY that occurred only in one patient; therefore, the description of results refers only to recurring translocations identified in more than one patient.
Spectral karyotyping of bone marrow chromosomes. Demonstration of simultaneous hybridization of 24 combinatorially labeled chromosome painting probes shown in display colors, aberrant chromosomes are highlighted by arrows (A). Spectra-based classification of the display colors shown in the same metaphase chromosomes, numbers beside chromosomes denote origin of translocated material (B). Karyotype of classification-colored chromosomes (C).
Spectral karyotyping of bone marrow chromosomes. Demonstration of simultaneous hybridization of 24 combinatorially labeled chromosome painting probes shown in display colors, aberrant chromosomes are highlighted by arrows (A). Spectra-based classification of the display colors shown in the same metaphase chromosomes, numbers beside chromosomes denote origin of translocated material (B). Karyotype of classification-colored chromosomes (C).
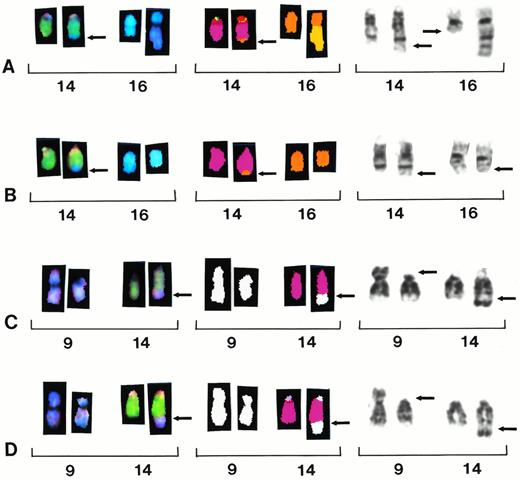
Partial karyotypes from four different patients showing recurring translocations t(14;16)(q32;q22∼q23) and t(9;14)(p13;q32). Each row represents a different patient sample with brackets within each row indicating a different representation of the same reciprocal translocation. Chromosomes are presented in SKY display colors (left brackets), SKY classification colors (center brackets), and G-banding (right brackets). Patient sample no. 8 shows translocation t(14;16)(q32;q22∼23) (A). Note the apparent breakpoint 16q22 versus smaller der 16 in patient below. Patient sample no. 3 shows translocation t(14;16)(q32;q22∼23) (B). Note that the breakpoint appears lower at band 16q23 in this patient. Patient samples no. 1 and 11 show the identification of a large segment designated add 14q32 by G-banding, refined to t(9;14)(p13;q32) (C and D).
Partial karyotypes from four different patients showing recurring translocations t(14;16)(q32;q22∼q23) and t(9;14)(p13;q32). Each row represents a different patient sample with brackets within each row indicating a different representation of the same reciprocal translocation. Chromosomes are presented in SKY display colors (left brackets), SKY classification colors (center brackets), and G-banding (right brackets). Patient sample no. 8 shows translocation t(14;16)(q32;q22∼23) (A). Note the apparent breakpoint 16q22 versus smaller der 16 in patient below. Patient sample no. 3 shows translocation t(14;16)(q32;q22∼23) (B). Note that the breakpoint appears lower at band 16q23 in this patient. Patient samples no. 1 and 11 show the identification of a large segment designated add 14q32 by G-banding, refined to t(9;14)(p13;q32) (C and D).
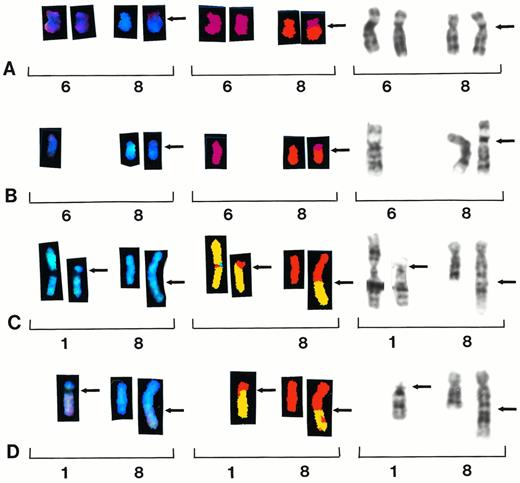
Partial karyotypes from four different patients showing recurring translocations t(6;8)(p10;q10) and t(1;8)(p11;q24). Each row represents a different patient sample, with brackets within each row indicating a different representation of the same reciprocal translocation. Chromosomes are presented in SKY display colors (left brackets), SKY classification colors (center brackets), and G-banding (right brackets). Patient samples no. 33 and 39 show whole-arm translocation t(6;8)(p10;q10) (A and B). Patient samples no. 7 and 28 show reciprocal translocation t(1;8)(p11;q24) (C and D).
Partial karyotypes from four different patients showing recurring translocations t(6;8)(p10;q10) and t(1;8)(p11;q24). Each row represents a different patient sample, with brackets within each row indicating a different representation of the same reciprocal translocation. Chromosomes are presented in SKY display colors (left brackets), SKY classification colors (center brackets), and G-banding (right brackets). Patient samples no. 33 and 39 show whole-arm translocation t(6;8)(p10;q10) (A and B). Patient samples no. 7 and 28 show reciprocal translocation t(1;8)(p11;q24) (C and D).
Recurring Chromosome Aberrations by G-Banding and Refined Designations by Spectral Karyotyping
| Patient No. . | G-Banding . | SKY Results . |
|---|---|---|
| 1 | add(14)(q32) | t(9;14)(p13;q32) |
| 3 | add(14)(q32) | t(14;16)(q32;q22 ∼ 23) |
| 6 | t(8;14)(q24;q32) | t(14;16)(q32;q22 ∼ 23) |
| 8 | add(14)(q32) | t(14;16)(q32;q22 ∼ 23) |
| 9 | add(14)(q32) | t(14;16)(q32;q22 ∼ 23) |
| 11 | add(14)(q32) | t(9;14)(p13;q32) |
| 12 | t(11;14)(q13;q32) | t(3;14)(q27;q32) |
| 14 | add(14)(q32) | t(8;14)(q24;q32) |
| 24 | add(8)(q24) | t(8;22)(q24;q12) |
| 42* | del(16)(q22) | t(14;16)(q32;q22 ∼ 23) |
| 44 | add(14)(q32) | t(8;14)(q24;q32) |
| 46 | t(8;14)(q24;q32) | t(14;16)(q32;q22 ∼ 23) |
| 48 | add(8)(q24) | t(6;8)(q21;q24) |
| 50 | add(14)(q32) | t(8;14)(q24;q32) |
| Patient No. . | G-Banding . | SKY Results . |
|---|---|---|
| 1 | add(14)(q32) | t(9;14)(p13;q32) |
| 3 | add(14)(q32) | t(14;16)(q32;q22 ∼ 23) |
| 6 | t(8;14)(q24;q32) | t(14;16)(q32;q22 ∼ 23) |
| 8 | add(14)(q32) | t(14;16)(q32;q22 ∼ 23) |
| 9 | add(14)(q32) | t(14;16)(q32;q22 ∼ 23) |
| 11 | add(14)(q32) | t(9;14)(p13;q32) |
| 12 | t(11;14)(q13;q32) | t(3;14)(q27;q32) |
| 14 | add(14)(q32) | t(8;14)(q24;q32) |
| 24 | add(8)(q24) | t(8;22)(q24;q12) |
| 42* | del(16)(q22) | t(14;16)(q32;q22 ∼ 23) |
| 44 | add(14)(q32) | t(8;14)(q24;q32) |
| 46 | t(8;14)(q24;q32) | t(14;16)(q32;q22 ∼ 23) |
| 48 | add(8)(q24) | t(6;8)(q21;q24) |
| 50 | add(14)(q32) | t(8;14)(q24;q32) |
This patient was identified by conventional FISH.
Refinement of Chromosome Aberrations by G-Banding With Spectral Karyotyping
| Patient No. . | G-Band Karyotype . | SKY Refinements . |
|---|---|---|
| 1. | 46 ∼ 47,X,−X,del(3)(p25),add(4)(p16),del(9)(p11),+11,add(14)(q32),+15,+18,−22[cp11] | add(4)(p16) = der(4)t(3;4)(q13;p16) add(14)(q32) = der(14)t(9;14)(p13;q32) |
| 2. | 51,XY,add(1)(q11),+der(1)del(1)(p13)?add(1)(q44),add(2)(p21),+3,add(6)(p25),+7,der(8;12)(q10;q10), dup(9)(q13q22),der(11)add(11)(p15)add(11)(q15),−14, der(14)t(1;14)(q10;q32),+19,+21,add(22)(q13)[cp7]/51,idem,del(3)(p21), −der(8;12)(q10;q10),+t(8;11)(p11.2;q13),−13,der(14;22)(q10;q10),−20)[cp4] | add(1)(q11) = der(15)t(1;15)(p21;p21) add(2)(p21) = der(2)t(2;15)(p21;?q21) add(6)(p25) = der(6)t(1;6)(q32;?p23) der(11)add(11)(q15) = der(11)t(11;14)(q13;q21) |
| 3. | 44,X,−X,i(1)(q10),del(2)(q27), der(5)del(5)(q22q32)t(5;1;6)(p14;p36;p21;p12),−6,i(8)(q10),del(12)(p11p12), −13,add(14)(q32),add(15)(q26),del(17)(q21q21),+19, der(19)t(1;19)(q12;q13),add(22)(p11)[cp14] | add(14)(q32) = der(14)t(14;16)(q32;q22 ∼ 23) add(15)(q26) = der(15)t(8;15)(q13;q26) add(22)(p11) = der(22)t(1;22)(q10;p11) |
| 4. | 49 ∼ 52,XX,t(1;15)(p13;q35),+der(1)t(1;5),del(2)(q31),+3,+6,t(7;11)(p?15;q23),t(8;14)(q24;q32),+9,+der(11)t(7;11),t(13;20)(q?22;q?11)[cp17] | t(7;11)(p?15;q23) = der(11)t(11;17)(q24;q22) t(13;20)(q?22;q?11) = t(2;20)(q31;q13) normal 16 = der(20)t(16;20) |
| 5. | 41,X,−X,der(1)t(1;19)(q10;p10),del(3)(p11p21),t(3;14)(q21;q32),−4, del(6)(q11),t(8;22)(q24;q11),add(9)(p22),−10,−14, der(15;21)(q10;q10),der(16)t(1;16)(q10;p10)[cp20] | add(9)(p22) = der(9)t(9;14)(p22;q31) |
| 6. | 40 ∼ 41,X,−Y,−1,ider(1)(p22)t(1;?12)(q21;q13),−2, der(2)add(2)(p23)add(2)(q33),der(3)add(3)(p11)t(1;3)(q21;q29),add(5)(q31), der(6)t(1;6)(p22;p21.3),−8,der(8)del(8)(p21),t(8;14)(q24;q32), der(10)t(?6;10)(p21.1;q26),−11,der(11)t(1;11)(p11;q25),−12, add(12)(q10),−13,der(14)t(8;14)(q24.1;q32), der(15)t(1;15)(q21;p10)ins(15;?2)(p10;q12q33),add(16)(q22) × 2,add(17)(p10),der(17)t(7;17)(q11;p11.1)ins(17;17)(p11.1;p11.1q25), der(18)t(8;18)(q22;q22),−19,der(19t(1;19)(q12;q13.4), der(21)t(3;21)(p11;p11.2),+add(22)(p11.1),+mar1,+mar2[cp8] | der(11)t(1;11) = der(11)t(1;3;11) der(14)t(8;14) = der(14)t(14;16)(q32;q22 ∼ 23) der(15) = der(15)t(15;2;1) add(17)(p10) = t(12;21) add(22)(p11.1) = t(11;19) +mar 1 = Y +mar 2 = der(2)t(2;17)(q10;p10) |
| 7. | 45 ∼ 49,XY,t(1;8)(p11;q24),add(3)(p25),add(6)(q27),+7,der(7)t(7;?14;1)(q11;q11q32;q11),der(10)t(1;10)(q11;p15),−13,t(14;14)(q11.2;q32),+15,+18,+21[cp20] | add(3)(p25) = der(3)t(3;8)(p25;q?24) add(6)(q27) = der(6)t(3;6)(q?21;q27) |
| 8. | 44 ∼ 45,XY,add(2)(q33),?del(3)(p21p24),der(4)t(1;4)(q21;p16), −8,add(9)(p12),−13,add(14)(q32),add(16)(q12.1), der(16)t(1;16)(q21;q11.2),del(17)(p12),−20,+21,+mar[cp7] | add (9)(p12) = der(9)t(9;20)(p10;q11) add(14)(q32) = t(14;16)(q32;q22 ∼ 23) +mar = 8 |
| 9. | 43,XY,add(1)(q12),ider(1)(q10)trp(1)(q10q?25),add(2)(q33),?add(4)(q10),inv(11)(p15q10),−12,−13,add(14)(q32),−16,del(16)(q22)[cp4] | add(1)(q12) = der(1)t(1;19)(q10;q10) ider(1)(q10)trp(1)(q10q?25) = t(1;16;1) ?add(4)(q10) = del(13)(q12q22) inv(11)(p15q10) = der(12)t(11;12) add(14)(q32) = t(14;16)(q32;q22 ∼ 23) |
| 10. | 42,XY,del(1)(p21p22),add(2)(p?25),add(4)(q?32),der(6)t(6;16)(q11;q12.1),add(9)(q34),add(10)(p11.2),t(14;17)(q24;p13),del(16)(q12.1),−18,−20,−21[cp 2] | add(2)(p?25) = der(2)t(2;8)(p25;?q21) add(4)(q?32) = der(4)t(4;15)(q32;?q26) add(9)(q34) = der(9)t(5;9) t(14;17) = der(17)t(16;17) |
| 11. | 44 ∼ 46,XX,add(2)(p23),der(4)t(1;4)(q21;q35), del(5)(q15q31),del(6)(q?23),dup(7)(q22q36),−8,del(9)(p11),add(10)(q26),del(11)(q?23),−13,del(13)(q12q14),add(14)(q32),−16,add(16)(q22),add(17)(p11.2),del(20)(q11 .2),+mar[cp13] | add(2)(p23) = der(2)t(2;8)(p23;q23) add(10)(q26) = der(10)t(10;20)(q26;q12) add(14)(q32) = der(14)t(9;14)(p13;q32) add(16)(q22) = der(16)t(13;16)(q10;q10) add(17)(p11.2) = der(17)t(3;17)(q13;p11.2) +mar = der(16)t(16;10) |
| 12. | 46 ∼ 49,XY,del(3)(q24),del(6)(q?23),add(8)(p11.2),+add(9)(p ?21),add(10)(q24),der(11)t(1;11)(q25;q23),add(12)(q21.2),−13,der(14)t(11;14)(q12 ∼ 13,1;q32),+add(15)(p11.2),−16,+19,+der(20)t(1;20)(q12;q13.3)[cp7] | add(8)(p11.2) = der(8)t(8;19) add(10)(q24) = der(10)t(10;12)(q24;q24) add(12)(q21;2) = der(12)t(12;15)(q15;q21) der(14)t(11;14) = t(3;14)(q21;q32) |
| 13. | 47 ∼ 48,X,−Y,trp(1)(q21q32),der(1)?t(1;10)(p22;p11.2)trp(1)(q21q32),+add(3)(p12),−6,del(7)(p12 p15),+del(7)(q11.1),add(8)(q?23),+del(9)(p13),add(12)(p11.2), +der(12)add(12)(q22),−13,−13,der(14)t(6;14)(p21.1;q32), +add(19)(?q13.2)[cp6] | der(1)?t(1;10) = der(1)t(1;7;10)(p10;p?;p?) add(8)(q?23) = t(8;13)(q24q22) |
| 14. | 44,XY,der(4)t(4;?21)(p16;?q11) × 2,−5,del(5)(p13), −6,del(6)(q21),add(8)(q13),t(9;15)(q34;q22),−11,−12,add(13)(q34),del(13)(q13q22),−14,add(14)(q32),add(15)(q15),−16,del(16)(p11.2),add(17)(q21),−20,−22,−22[cp11] | der(4)t(4;?21) = der(4)t(4;5)(p16;p14) del(6)(q21) = der(6)t(6;8)(q21;?) add(8)(q13) = der(8)t(8;22)(q13;?) add(13)(q34) = der(13)t(13;16) del(13)(q13q22) = der(22)t(8;22) add(14)(q32) = der(14)t(8;14)(q24;q32) add(17)(q21) = der(17)t(17;21)(p21;?) |
| 15. | 45 ∼ 46,XX,t(1;14)(q23;q21),add(6)(p25),i(11)(q10), −13,der(15)t(11;15)(q13;p11),+17,add(17)(p11) × 2, del(21)(q22)[cp15] | add(6)(p25) = t(6;12)(p25;?q) der(15)t(11;15) = der(15)t(13;15) add(17)(p11) = der(17)t(13;17) |
| 16. | 52 ∼ 54,X,−X,?inv(1)(p21p22),+3,der(3;8)(q10;q10),+del(5)(p15),+add(6)(q?15),add(7)(p22),+9,+11,der(12)add(12)(p12)add(12)(q24.3),+15,t(15;16)(p11.1;p11.1), del(17)(p11.2),+?add(18)(p11.2),+19,+21[cp4] | +del(5)(p15) = der(5)t(5;12) +add(6)(q?15) = der(6)t(6;17) add(7)(p22) = der(7)t(X;7)(?q22;p22) der(12)add(12)(p12) = der(12)t(8;12)(p24;?q21) +?add(18)(p11.2) = der(17)t(6;17) |
| 17. | 61 ∼ 64,XX,+1,der(1)t(1;19)(q11;q?13),t(1;9)(q11;q?13),+3,dup(3)(q11q24),der(3;7)(q10;p10),+4, der(4)t(4;17)(q35;q21),add(7)(p11),8,+9,+9,der(9)t(1;19)(q11,q?13), add(1)(q32),del(p11),add(12)(p12),add(13)(p11) × 2,+14,+14, add(14)(p11),+15,+15,+17,add(17)(p12),del(17)(p11),+18,+18,+19,+20, +20,+21, i(21)(q10q12),+22,add(22)(p11),+mar[cp16] | der(1)t(1;9)(q11;q?13) = der(8)t(1;8)(p31;p10) add(7)(p11) = der(7)t(7;7) add(13)(p11) = der(13)t(11;13) add(17)(p12) = der(17)t(17;21)(p11;q11) +mar = 21 |
| 18. | 56 ∼ 58,XY,add(X)(q28),del(1)( q42),add(1)(p36.1),del(2)(q35) × 2,−3,add(3)(q29),−4,add(4)(q32),−5,+6,add(6)(q?23),−7,−9,−9,add(10)(q24),−15, −16,−16,add(17)(p11.2),−18,−18,−20[cp14] | add(1)(p36.1) = t(1;10)(p36.1q24) add(3)(q29) = der(3)t(3;18)(q29;q11) add(6)(q?23) = t(6;12)(q23;q22) add(17)(p11.2) = der(17)t(17;19)(p11:?q) add(X)(q28) = der(X)t(X;9)(q28;?q13) |
| 19. | 45 ∼ 46,X,−X,+add(1)(p11),del(1)(p12p13.2) × 2, del(3)(q26.3),del(5)(q12q15),der(7)t(1;7)(q10;p22)del(1)(q32), der(12)add(12)(p11.2)del(12)(q24.?2), der(12)add(12)(p11.2)t(11;12)(q11;q24.?2),−13,−14,+15,+15,+add(16)(q24),add(19)(q13.4),tas(19;?)(p13.1;?),add(20)(p13) ,+i(21)(q10)[cp20] | +add(1)(p11) = der(1)t(1;3)(p11;q26) der(7)t(1;7) = der(7)t(1;7;20) +add(16)(q24) = der(16)t(16;18)(q24;?q22) tas(19;?)(p13.1;?) = tas(1;19)(p36;p13) add(20)(p13) = der(20)t(1;20)(?;p13) |
| 20. | 52 ∼ 52,X,−Y,+der(1;19)(q10;p10),+3,+add(4)(p12),+6, t(8;12)(q24;q13),+9,−13,+15,+18,+21,+21[cp20] | +add(4)(p12) = der(4)t(4;7)(p12;q22) |
| 21. | 54,XY,del(1)(p22p22),+3,+7,+8,+9,+11,+15,der(16)t(1;16)(q10;q22), +17,+19,add(19)(p13.3)[cp20] | add(19)(p13.3) = der(19)t(16;19)(q11;p13) |
| 22. | 55,XY,+3,add(3)(q27),+5,+7,t(8;22)(q24;q11),+9,+9,+11, +15,add(17)(q25),+19,+21[cp18) | add(3)(q27),t(8;22)(q24;q11), add(17)(q25) = t(3;17;22;8)(q27;q25;q11;q24) |
| 23. | 49 ∼ 50,X,−X,+add(1)(p11),+3,add(6)(q25),+add(7) (q36),−8,der(?8)add(?8)(p?21)add(?8)(q?24.1),+9,+add(9)(p?24),+11, add(12)(q?21),−13,+15,−16,+add(21)(q22),+mar[cp16] | +add(1)(p11) = der(16)t(1;16)(q10;q10) add(7)(q36) = der(7)t(7;8)(q36;q11) +add(21)(q22) = der(21)t(21;22)(q22;q13) +mar = 5 |
| 24. | 51 ∼ 52,XX,+1,del(1)(q11),i(1)(q10),+3,+7,add(8)(q24.1),+9,?11p,?t(12;22)(p13;q13),−13,−14,+15,+ 15,add(15)(p11.1),+21,+mar[cp13] | del(1)(q11) = der(14)t(1;14)(q22;q31) add(8)(q24.1) = der(8)t(8;22)(q24.1;q13) ?t(12;22)der(12)t(12;19)(p13;?) add(15)(p11.1) = der(15)t(14;15)(q23;p11) +mar = der(18)t(X;18)(q22;p11) |
| 25. | 53 ∼ 60,XY,−1,dup(1)(q11q32),+3,der(4)t(4;17)(q11;p12),+5,+5, del(5)(q15),del(5)(q22),+7,+9,der(10)t(10;11)(q26;q12),+11,+15, der(16)t(1;16)(q10;q24),der(17)t(4;17)(q22;p12),+18,+19,+21,add(21)(q22)[cp20] | del(5)(q22) = der(5)t(5;22) add(21)(q22) = der(21)t(19;21) |
| 26. | 42,X,del(X)(q27),del(1)(p21p22),der(6)t(6;21)(q11;q11.1),der(8)t(8;?12)(q22;q?22),der(12)r(12;8)(p13q24.1;q21.1q24.3),−13,der(15)t(15;16)(p?13;q11.2),−16,der(18)t(1;18)(q11;q23),−20,−21, del(22)(q13.2)[cp20] | der(8)t(8;?12)(q22;?22) = der(8)t(8;16)(q22;p11) der(12)r(12;8)(p13q24.1;q21.1q24.3) = r(12) |
| 27. | 43 ∼ 44,X,?X,add(1)(p36.3),add(1)(q32),add(2)(p21),del(4)(p12),add(5)(p15.3),del(6)(q13),add(7)(p13),der(8)t(1;8)(q10;p23),der(8;14)(q10;q10),−13,−14,add(15)(q26),add(17)(p12),add(18)(p11.2),+mar[cp7] | add(1)(p36.3) = t(1;1)(q36;q31) add(2)(p21) = der(2)t(2;4) add(5)(p15.3) = der(5)t(5;6)(p15;q23) add(7)(p13) = der(7)t(7;15)(p13;q25) add(15)(q26) = t(4;15)(p12;q25) |
| 28. | 47,XY,−1,t(1;8)(p11;q24),t(2;3)(q31;q27), der(3)del(3)(p21)?dup(3)(q21q25),add(4)(p12),del(5)(q22q31),?del(6)(q21q22), inv(7)(p13q32),+der(11)t(11;15)(p15;q15),add(12)(q24.1),del(13)(q12q14),der(16)t(1;16)(q21;q22),+add(19)(q13.3), add(21)(q22)[cp16] | add(12)(q24.1) = der(12)t(4;12)(p12;q24) +add(19)(q13.3) = der(19)t(1;19)(p13;q13) add(21)(q22) = der(21)t(21;21)(q22;?) |
| 29. | 50 ∼ 51,XY,+3,add(3)(p?26),+5,+6,del(6)(q23),+7,−8,+9,−13,+15,add(15)(p11.1),i(15)(q10),−16,+19, add(19)(q13.3),der(20)add(20)(p13)add(20)(q13.3),der(22)t(1;22)(q10;q10), del(22)(q11.3)[cp11] | add(3)(p?26) = der(3)t(3;8)(p10;q10) +6 = der(6)t(6;8)(p10;q10) add(15)(p11.1) = der(15)t(15;20)(p11;q10) |
| 30. | 52,X,−X,+7,+9,+11,der(11)t(7;11)(p14;p12), der(14)t(X;14)(p11.3;p13),+15,der(15)t(1;15)(q10;q26),del(16)(q22q22),+17,+19, der(19)t(1;19)(q10;q13),+21,add(21)(p22),der(22)t(3;22)(q12;q13)[cp10] | der(11)t(7;11) = der(11)t(5;11)(p14;p12) add(21)(p22) = der(21)t(9;21)(q34;q22) |
| 31. | 52 ∼ 59,XY,−Y,add(1)(q32),del(1)(p11),+1,inv(3)(p26q23), t(4;19)(q21;q13.3),add(5)(q22),der(5)t(1;5)(q11;p?14),der(5)t(1;5)(q21;q15), del(5)(p13),+5,add(6)(q11),+6,+7,+7,der(8)t(8;11)(q24;q13) × 2,+der(8)t(8;11)(q24;q13), t(1;8)(q11;p23),+15,der(16)t(1;16)(q11.1;p13.3),del(17)(p11),+19,+20,add(20)(p13),+dic(1;20)(p22;p13),+21,add(22)(q13), +add(22)(p11)[cp20] | add(6)(q11) = der(6)t(6;10)(q13;?q21 ∼ 22) der(8)t(8;11) = der(8)t(1;8)(q10;p23) add(20)(p13) = t(2;20) |
| 32. | 56 ∼ 68,XY,t(1;17),+der(1)t(1;17),?add(2) × 2,+3,−4,+5,−7,+8, der(8;21)(q10;q10) × 2,+9,+9,+der(11)t(1;11)ins(11;?) × 2, −12,−13,−14,+15,+15,−16,t(16;21)(q12;q22),−18,+21,−22[cp6] | +der(11)t(1;11)ins(11;?) = der(11)t(11;17;1) |
| 33. | 53 ∼ 54,XY,+3,+5,+6,add(6)(q23),+7,t(7;17)(p11.1;p12),−8,+9,+15,+19,+21,+mar[cp19] | add(6)(q23) = der(8)t(6;8)(p10;q10) |
| 34. | 58,−X,−Y,der(X;1)(p10;p10),del(1)(p32) × 2,−2,−4, der(6)t(3;6)(q10;p10) × 2,+7,−8,+9,−10,+11,−12,−13,−13,−14,−16,del(16)(q22q22),add(17)(p11.1),−21,−22[cp6] | del(1)(p32) = der(1)t(1;X)(p31;q22) del(16)(q22q22) = der(16)t(13;16)(q12;q22) add(17)(p11.1) = der(17)t(7;17)(p11;p11) |
| 35. | 43,XY,?t(1;4)(q?32;q?21),dup(1)(q11q44),inv(2)(p13q21),add(4)(q34),del(6)(q15),−10,−13,−14,del(22)(q12)[cp20] | add(4)(q34) = der(4)t(X;4)(?q22;q34) |
| 36. | 56,XX, + der(1;6)(q10;p10) × 2,+3,+5,add(7)(q10),+8, i(8)(q10) × 2,der(9)t(1;9)(q10;q12),add(9)(p10),+der(10)t(10;11)(q22;q22),add(10)(p12),−13,der(14;19)(q10;p10), +15,?add(16)(q24),+del(17)(p11.1),+19,−20,+21, +del(22)(q13)[cp11] | add(7)(q10) = der(7)t(7;16) +der(10)t(10;11)(q22;q22) = der(20)t(7;20) add(10)(p12) = der(10)t(10;11)(p12;q24) ?add(16)(q24) = der(16)t(7;16) +del(17)(p11.1) = der(15)t(15;17)(q15;q12) |
| 37. | 46 ∼ 47,XY,del(1)(p22p32),?t(2;8)(p13;q24),add(3)(p?22), t(4;9)(q21;q22),?7p,+9,?t(9;19)(p22;q?13.3),−10,t(10;17)(q21;p11.1), add(12)(q15),−13,der(16)t(1;16)(q21;q22),?add(18)(q21), +del(19)(q11),del(20)(q11.2),der(20)t(1;20)(q?12;p13),+mar[cp14] | ?t(2;8)(p13;q24) = der(8)t(8;15)(q24.1;p21) der(16)t(1;16) = der(16)t(6;16) +mar = del(15) |
| 38. | 51,X,?add(X)(q26),t(1;9)(q11;q13),+der(1;19)(q10;p10), der(2)t(2;?13)(q23;q12),t(2;3)(q23;p21),+3, der(8)add(8)(p21)t(8;22)(q24.1;q11),der(8)t(2;8)(q23;p23),+add(9)(p2?4),+11,−13,−14,+15,+21,der(22)t(8;22),+mar,1dmin[cp16] | der(8)t(2;8)(q23;p23) = der(8)t(4;8)(q10;p10) +mar = del(13) |
| 39. | 41 ∼ 43,X,−X,+1,+1,der(1;2)(q10p;10),der(1;5)(q10;p10)dup(1)(q21q25), der(1;13)(q10;q10),der(2)t(1;2)(p32;q10),t(4;7)(q11;q32),der(6;8)(p10;q10), −13,−14[cp19] | no change |
| 40. | 65,XX,−Y,−1,−2,+3,?del(3)(q27),add(3)(q12),−4,+6, del(6)(q23q23) × 2,+7,dup(7)(q22q31) × 2,+8,del(8)(q1?24.1), del(8)(q22),+9,add(9)(p24),−10,+11,−12,−13,−14,−16,−16,−17,+18,−20, +21,+mar[cp15] | dup(7)(q22q31) = der(7)t(4;7)(q?28;q?36) add(3)(q12) = der(3)t(3;12) |
| 41. | 45,XX,add(3)(q29),del(5)(q22),add(11)(q25),inv(12)(p11q11),−17,add(20)(q13.3)[cp15] | add(3)(q29) = der(3)t(3;3)(q29;?q) add(11)(q25) = der(11)t(3;11)(q?25;q25) add(20)(q13.3) = der(20)t(17;20)(q22;q13) |
| 42. | 41 ∼ 42,XX,del(1)(p32p13),t(1;6)(q21;q25),add(8)(p23),del(9)(p13),−11,−13,−14,del(16)(q21q22),−18,−20,−22,+mar[cp18] | add(8)(p23) = der(8)t(8;18)(p23;q?) del(16) = t(14;16)(q32;q22 ∼ 23) +mar = t(11;19) |
| 43. | 75 ∼ 78,XX,−Y,del(1)(p13),add(2)(q37),?add(2)(q11),+3,del(3)(q21), −4,add(4)(q35),+5,del(5)(q31q32),+6,del(6)(q?23),add(6)(q13),+7,add(7)(p?22), +8,+9,add(9)(p13),add(9)(q34) × 2,add(11)(q23) × 2,+12,+12,add(12)(p13) × 2,−13, del(13)(q14q32),−14,+15,−16,−16,−16,+17,add(17)(p13) × 2, +18,+18,+19,add(20)(q13),+21,add(21)(p11),+2mar[cp17] | add(9)(q34) = der(9)t(9;12)(q34;?q24) add(11)(q23) = der(11)t(11;16)(q22;?) add(12)(p13) = der(12)t(10;12)(?;p13) add(17)(p13) = der(17)t(13;17)(p11;p13) add(20)(q13) = der(20)t(20;20)(q13;?) |
| 44. | 58,XX,+3,+add(5)(q?33),der(6;8)(p10;q10),+7,+7,+9,+9, +11,+11,del(13)(q12q14),add(14)(q32),+15, +der(16)t(6;16)(p21.1;q12.1) × 2,+18,+19,+21,−22[cp9] | der(6)t(6;8)(p10;q10) = der(8)t(3;8)(p10;p21) add(14)(q32) = der(14)t(8;14)(q24;q32) |
| 45. | 45,XY,add(2)(q32),der(6)t(6;6)(q27;p11.2),t(11;13)(q?11;p?13), −13,der(14)t(1;14)(q21;q32),del(16)(q12.1),add(17)(q24), add(22)(q13)[cp13] | add(2)(q32) = t(2;22)(q32;q13) del(16)(q12.1) = der(16)t(16;22)(q11;q13) |
| 46. | 86 ∼ 90,XXYY,−1,−1,add(1)(p13),add(2)(q33) × 3,add(3)(p10), t(4;5)(q31.3;q?34),t(5;7)(q10;q10),del(6)(q23),der(6)t(3;6)(p21;q23), der(6)t(6;11)(q23;q13),t(8;14)(q24;q32),t(8;22)(q24.3;q13), add(9)(p21),+10,der(10)t(1;10)(p13;q26) × 2,−11,der(11)(q13), −12,−13,−14,+der(15;21)(q10;q10),del(16)(q22) × 2,?add(17)(p10),−19, add(20)(p?11.2),+add(21)(q22),der(22)t(1;22)(q?25;p11.2)[cp5] | t(8;14) = der(14)t(14;16)(q32;q22 ∼ 23) der(10)t(1;10) = der(8)t(1;8;1;8) der(22)t(1;22) = der(21)t(10;21)(p11;q21) +add(21)(q22) = der(10)t(Y;10)(?;q11) |
| 47. | 56 ∼ 57,X,−Y,+add(1)(p13),+add(2)(q37),+add(3)(p11),del(4)(p12p14),del(4)(q31),+del(5)(q31q33), add(5)(q22),+del(6)(q13),del(6)(q23),+7,der(8)t(8;15)(q10;q10), ?add(9)(q34),+del(11)(q23),+15,+15,+15,der(17)t(9;17)(q12;p11.1),+18,+19, +19[cp6] | +add(1)(p13) = der(1)t(1;16)(p10;p10) +add(3)(p11) = der(3)t(3;20)(p10;?q10) der(8)t(8;15) = der(8)t(2;8)(p10;p?11) |
| 48. | 51,XY,der(1)t(1;12)(p13;q13),del(1)(p13p32),add(4)(q28),+5,add(6)(q21),add(8)(q24.1),+9, add(10)(q24),+11,add(12)(q15),del(13)(q12q14),+15,+15,−16,+19[cp6] | der(1)t(1;12) = der(1)t(1;16)(q10;p10) add(4)(q28) = t(4;10)(q?25;q?25) add(8)(q24.1) = t(6;8)(q21;q25.1) add(12)(q15) = der(12)t(1;12)(p?32;q15) |
| 49. | 49 ∼ 51,X,der(Y)t(Y;2)(q12;p11.2),del(1)(p31p36.1),+5,inv(5)(q13q35), +7,+9,del(10)(p13),t(12;19)(q13;p13.3),−13,+15,der(16)t(1;16)(q21;q21), der(17)t(8;17)(q11.1;p11.2),+der(19)t(7;19)(q11.2;q13.4), del(20)(p11.2p11.2)[cp20] | no change |
| 50. | 42,X,−X,i(1)(q10),add(2)(q?31),der(3)t(3;9)(p11;q11), −4,add(6)(p?21.3),add(7)(q22),−8,+del(9)(p13),−13,−14,der(14)t(5;14)(p11.1;p11.1)add(14)(q32),−15,add(15)(q22),add(16)(q?11.2)[cp20] | add(2)(q?31) = t(2;8)(q33;q23) add(6)(p?21.3) = t(6;7)(p22;q32) der(14)t(5;14)add(14)(q32) = t(8;14)(q24;q32) add(15)(q22) = t(15;16)(q23;q12) |
| Patient No. . | G-Band Karyotype . | SKY Refinements . |
|---|---|---|
| 1. | 46 ∼ 47,X,−X,del(3)(p25),add(4)(p16),del(9)(p11),+11,add(14)(q32),+15,+18,−22[cp11] | add(4)(p16) = der(4)t(3;4)(q13;p16) add(14)(q32) = der(14)t(9;14)(p13;q32) |
| 2. | 51,XY,add(1)(q11),+der(1)del(1)(p13)?add(1)(q44),add(2)(p21),+3,add(6)(p25),+7,der(8;12)(q10;q10), dup(9)(q13q22),der(11)add(11)(p15)add(11)(q15),−14, der(14)t(1;14)(q10;q32),+19,+21,add(22)(q13)[cp7]/51,idem,del(3)(p21), −der(8;12)(q10;q10),+t(8;11)(p11.2;q13),−13,der(14;22)(q10;q10),−20)[cp4] | add(1)(q11) = der(15)t(1;15)(p21;p21) add(2)(p21) = der(2)t(2;15)(p21;?q21) add(6)(p25) = der(6)t(1;6)(q32;?p23) der(11)add(11)(q15) = der(11)t(11;14)(q13;q21) |
| 3. | 44,X,−X,i(1)(q10),del(2)(q27), der(5)del(5)(q22q32)t(5;1;6)(p14;p36;p21;p12),−6,i(8)(q10),del(12)(p11p12), −13,add(14)(q32),add(15)(q26),del(17)(q21q21),+19, der(19)t(1;19)(q12;q13),add(22)(p11)[cp14] | add(14)(q32) = der(14)t(14;16)(q32;q22 ∼ 23) add(15)(q26) = der(15)t(8;15)(q13;q26) add(22)(p11) = der(22)t(1;22)(q10;p11) |
| 4. | 49 ∼ 52,XX,t(1;15)(p13;q35),+der(1)t(1;5),del(2)(q31),+3,+6,t(7;11)(p?15;q23),t(8;14)(q24;q32),+9,+der(11)t(7;11),t(13;20)(q?22;q?11)[cp17] | t(7;11)(p?15;q23) = der(11)t(11;17)(q24;q22) t(13;20)(q?22;q?11) = t(2;20)(q31;q13) normal 16 = der(20)t(16;20) |
| 5. | 41,X,−X,der(1)t(1;19)(q10;p10),del(3)(p11p21),t(3;14)(q21;q32),−4, del(6)(q11),t(8;22)(q24;q11),add(9)(p22),−10,−14, der(15;21)(q10;q10),der(16)t(1;16)(q10;p10)[cp20] | add(9)(p22) = der(9)t(9;14)(p22;q31) |
| 6. | 40 ∼ 41,X,−Y,−1,ider(1)(p22)t(1;?12)(q21;q13),−2, der(2)add(2)(p23)add(2)(q33),der(3)add(3)(p11)t(1;3)(q21;q29),add(5)(q31), der(6)t(1;6)(p22;p21.3),−8,der(8)del(8)(p21),t(8;14)(q24;q32), der(10)t(?6;10)(p21.1;q26),−11,der(11)t(1;11)(p11;q25),−12, add(12)(q10),−13,der(14)t(8;14)(q24.1;q32), der(15)t(1;15)(q21;p10)ins(15;?2)(p10;q12q33),add(16)(q22) × 2,add(17)(p10),der(17)t(7;17)(q11;p11.1)ins(17;17)(p11.1;p11.1q25), der(18)t(8;18)(q22;q22),−19,der(19t(1;19)(q12;q13.4), der(21)t(3;21)(p11;p11.2),+add(22)(p11.1),+mar1,+mar2[cp8] | der(11)t(1;11) = der(11)t(1;3;11) der(14)t(8;14) = der(14)t(14;16)(q32;q22 ∼ 23) der(15) = der(15)t(15;2;1) add(17)(p10) = t(12;21) add(22)(p11.1) = t(11;19) +mar 1 = Y +mar 2 = der(2)t(2;17)(q10;p10) |
| 7. | 45 ∼ 49,XY,t(1;8)(p11;q24),add(3)(p25),add(6)(q27),+7,der(7)t(7;?14;1)(q11;q11q32;q11),der(10)t(1;10)(q11;p15),−13,t(14;14)(q11.2;q32),+15,+18,+21[cp20] | add(3)(p25) = der(3)t(3;8)(p25;q?24) add(6)(q27) = der(6)t(3;6)(q?21;q27) |
| 8. | 44 ∼ 45,XY,add(2)(q33),?del(3)(p21p24),der(4)t(1;4)(q21;p16), −8,add(9)(p12),−13,add(14)(q32),add(16)(q12.1), der(16)t(1;16)(q21;q11.2),del(17)(p12),−20,+21,+mar[cp7] | add (9)(p12) = der(9)t(9;20)(p10;q11) add(14)(q32) = t(14;16)(q32;q22 ∼ 23) +mar = 8 |
| 9. | 43,XY,add(1)(q12),ider(1)(q10)trp(1)(q10q?25),add(2)(q33),?add(4)(q10),inv(11)(p15q10),−12,−13,add(14)(q32),−16,del(16)(q22)[cp4] | add(1)(q12) = der(1)t(1;19)(q10;q10) ider(1)(q10)trp(1)(q10q?25) = t(1;16;1) ?add(4)(q10) = del(13)(q12q22) inv(11)(p15q10) = der(12)t(11;12) add(14)(q32) = t(14;16)(q32;q22 ∼ 23) |
| 10. | 42,XY,del(1)(p21p22),add(2)(p?25),add(4)(q?32),der(6)t(6;16)(q11;q12.1),add(9)(q34),add(10)(p11.2),t(14;17)(q24;p13),del(16)(q12.1),−18,−20,−21[cp 2] | add(2)(p?25) = der(2)t(2;8)(p25;?q21) add(4)(q?32) = der(4)t(4;15)(q32;?q26) add(9)(q34) = der(9)t(5;9) t(14;17) = der(17)t(16;17) |
| 11. | 44 ∼ 46,XX,add(2)(p23),der(4)t(1;4)(q21;q35), del(5)(q15q31),del(6)(q?23),dup(7)(q22q36),−8,del(9)(p11),add(10)(q26),del(11)(q?23),−13,del(13)(q12q14),add(14)(q32),−16,add(16)(q22),add(17)(p11.2),del(20)(q11 .2),+mar[cp13] | add(2)(p23) = der(2)t(2;8)(p23;q23) add(10)(q26) = der(10)t(10;20)(q26;q12) add(14)(q32) = der(14)t(9;14)(p13;q32) add(16)(q22) = der(16)t(13;16)(q10;q10) add(17)(p11.2) = der(17)t(3;17)(q13;p11.2) +mar = der(16)t(16;10) |
| 12. | 46 ∼ 49,XY,del(3)(q24),del(6)(q?23),add(8)(p11.2),+add(9)(p ?21),add(10)(q24),der(11)t(1;11)(q25;q23),add(12)(q21.2),−13,der(14)t(11;14)(q12 ∼ 13,1;q32),+add(15)(p11.2),−16,+19,+der(20)t(1;20)(q12;q13.3)[cp7] | add(8)(p11.2) = der(8)t(8;19) add(10)(q24) = der(10)t(10;12)(q24;q24) add(12)(q21;2) = der(12)t(12;15)(q15;q21) der(14)t(11;14) = t(3;14)(q21;q32) |
| 13. | 47 ∼ 48,X,−Y,trp(1)(q21q32),der(1)?t(1;10)(p22;p11.2)trp(1)(q21q32),+add(3)(p12),−6,del(7)(p12 p15),+del(7)(q11.1),add(8)(q?23),+del(9)(p13),add(12)(p11.2), +der(12)add(12)(q22),−13,−13,der(14)t(6;14)(p21.1;q32), +add(19)(?q13.2)[cp6] | der(1)?t(1;10) = der(1)t(1;7;10)(p10;p?;p?) add(8)(q?23) = t(8;13)(q24q22) |
| 14. | 44,XY,der(4)t(4;?21)(p16;?q11) × 2,−5,del(5)(p13), −6,del(6)(q21),add(8)(q13),t(9;15)(q34;q22),−11,−12,add(13)(q34),del(13)(q13q22),−14,add(14)(q32),add(15)(q15),−16,del(16)(p11.2),add(17)(q21),−20,−22,−22[cp11] | der(4)t(4;?21) = der(4)t(4;5)(p16;p14) del(6)(q21) = der(6)t(6;8)(q21;?) add(8)(q13) = der(8)t(8;22)(q13;?) add(13)(q34) = der(13)t(13;16) del(13)(q13q22) = der(22)t(8;22) add(14)(q32) = der(14)t(8;14)(q24;q32) add(17)(q21) = der(17)t(17;21)(p21;?) |
| 15. | 45 ∼ 46,XX,t(1;14)(q23;q21),add(6)(p25),i(11)(q10), −13,der(15)t(11;15)(q13;p11),+17,add(17)(p11) × 2, del(21)(q22)[cp15] | add(6)(p25) = t(6;12)(p25;?q) der(15)t(11;15) = der(15)t(13;15) add(17)(p11) = der(17)t(13;17) |
| 16. | 52 ∼ 54,X,−X,?inv(1)(p21p22),+3,der(3;8)(q10;q10),+del(5)(p15),+add(6)(q?15),add(7)(p22),+9,+11,der(12)add(12)(p12)add(12)(q24.3),+15,t(15;16)(p11.1;p11.1), del(17)(p11.2),+?add(18)(p11.2),+19,+21[cp4] | +del(5)(p15) = der(5)t(5;12) +add(6)(q?15) = der(6)t(6;17) add(7)(p22) = der(7)t(X;7)(?q22;p22) der(12)add(12)(p12) = der(12)t(8;12)(p24;?q21) +?add(18)(p11.2) = der(17)t(6;17) |
| 17. | 61 ∼ 64,XX,+1,der(1)t(1;19)(q11;q?13),t(1;9)(q11;q?13),+3,dup(3)(q11q24),der(3;7)(q10;p10),+4, der(4)t(4;17)(q35;q21),add(7)(p11),8,+9,+9,der(9)t(1;19)(q11,q?13), add(1)(q32),del(p11),add(12)(p12),add(13)(p11) × 2,+14,+14, add(14)(p11),+15,+15,+17,add(17)(p12),del(17)(p11),+18,+18,+19,+20, +20,+21, i(21)(q10q12),+22,add(22)(p11),+mar[cp16] | der(1)t(1;9)(q11;q?13) = der(8)t(1;8)(p31;p10) add(7)(p11) = der(7)t(7;7) add(13)(p11) = der(13)t(11;13) add(17)(p12) = der(17)t(17;21)(p11;q11) +mar = 21 |
| 18. | 56 ∼ 58,XY,add(X)(q28),del(1)( q42),add(1)(p36.1),del(2)(q35) × 2,−3,add(3)(q29),−4,add(4)(q32),−5,+6,add(6)(q?23),−7,−9,−9,add(10)(q24),−15, −16,−16,add(17)(p11.2),−18,−18,−20[cp14] | add(1)(p36.1) = t(1;10)(p36.1q24) add(3)(q29) = der(3)t(3;18)(q29;q11) add(6)(q?23) = t(6;12)(q23;q22) add(17)(p11.2) = der(17)t(17;19)(p11:?q) add(X)(q28) = der(X)t(X;9)(q28;?q13) |
| 19. | 45 ∼ 46,X,−X,+add(1)(p11),del(1)(p12p13.2) × 2, del(3)(q26.3),del(5)(q12q15),der(7)t(1;7)(q10;p22)del(1)(q32), der(12)add(12)(p11.2)del(12)(q24.?2), der(12)add(12)(p11.2)t(11;12)(q11;q24.?2),−13,−14,+15,+15,+add(16)(q24),add(19)(q13.4),tas(19;?)(p13.1;?),add(20)(p13) ,+i(21)(q10)[cp20] | +add(1)(p11) = der(1)t(1;3)(p11;q26) der(7)t(1;7) = der(7)t(1;7;20) +add(16)(q24) = der(16)t(16;18)(q24;?q22) tas(19;?)(p13.1;?) = tas(1;19)(p36;p13) add(20)(p13) = der(20)t(1;20)(?;p13) |
| 20. | 52 ∼ 52,X,−Y,+der(1;19)(q10;p10),+3,+add(4)(p12),+6, t(8;12)(q24;q13),+9,−13,+15,+18,+21,+21[cp20] | +add(4)(p12) = der(4)t(4;7)(p12;q22) |
| 21. | 54,XY,del(1)(p22p22),+3,+7,+8,+9,+11,+15,der(16)t(1;16)(q10;q22), +17,+19,add(19)(p13.3)[cp20] | add(19)(p13.3) = der(19)t(16;19)(q11;p13) |
| 22. | 55,XY,+3,add(3)(q27),+5,+7,t(8;22)(q24;q11),+9,+9,+11, +15,add(17)(q25),+19,+21[cp18) | add(3)(q27),t(8;22)(q24;q11), add(17)(q25) = t(3;17;22;8)(q27;q25;q11;q24) |
| 23. | 49 ∼ 50,X,−X,+add(1)(p11),+3,add(6)(q25),+add(7) (q36),−8,der(?8)add(?8)(p?21)add(?8)(q?24.1),+9,+add(9)(p?24),+11, add(12)(q?21),−13,+15,−16,+add(21)(q22),+mar[cp16] | +add(1)(p11) = der(16)t(1;16)(q10;q10) add(7)(q36) = der(7)t(7;8)(q36;q11) +add(21)(q22) = der(21)t(21;22)(q22;q13) +mar = 5 |
| 24. | 51 ∼ 52,XX,+1,del(1)(q11),i(1)(q10),+3,+7,add(8)(q24.1),+9,?11p,?t(12;22)(p13;q13),−13,−14,+15,+ 15,add(15)(p11.1),+21,+mar[cp13] | del(1)(q11) = der(14)t(1;14)(q22;q31) add(8)(q24.1) = der(8)t(8;22)(q24.1;q13) ?t(12;22)der(12)t(12;19)(p13;?) add(15)(p11.1) = der(15)t(14;15)(q23;p11) +mar = der(18)t(X;18)(q22;p11) |
| 25. | 53 ∼ 60,XY,−1,dup(1)(q11q32),+3,der(4)t(4;17)(q11;p12),+5,+5, del(5)(q15),del(5)(q22),+7,+9,der(10)t(10;11)(q26;q12),+11,+15, der(16)t(1;16)(q10;q24),der(17)t(4;17)(q22;p12),+18,+19,+21,add(21)(q22)[cp20] | del(5)(q22) = der(5)t(5;22) add(21)(q22) = der(21)t(19;21) |
| 26. | 42,X,del(X)(q27),del(1)(p21p22),der(6)t(6;21)(q11;q11.1),der(8)t(8;?12)(q22;q?22),der(12)r(12;8)(p13q24.1;q21.1q24.3),−13,der(15)t(15;16)(p?13;q11.2),−16,der(18)t(1;18)(q11;q23),−20,−21, del(22)(q13.2)[cp20] | der(8)t(8;?12)(q22;?22) = der(8)t(8;16)(q22;p11) der(12)r(12;8)(p13q24.1;q21.1q24.3) = r(12) |
| 27. | 43 ∼ 44,X,?X,add(1)(p36.3),add(1)(q32),add(2)(p21),del(4)(p12),add(5)(p15.3),del(6)(q13),add(7)(p13),der(8)t(1;8)(q10;p23),der(8;14)(q10;q10),−13,−14,add(15)(q26),add(17)(p12),add(18)(p11.2),+mar[cp7] | add(1)(p36.3) = t(1;1)(q36;q31) add(2)(p21) = der(2)t(2;4) add(5)(p15.3) = der(5)t(5;6)(p15;q23) add(7)(p13) = der(7)t(7;15)(p13;q25) add(15)(q26) = t(4;15)(p12;q25) |
| 28. | 47,XY,−1,t(1;8)(p11;q24),t(2;3)(q31;q27), der(3)del(3)(p21)?dup(3)(q21q25),add(4)(p12),del(5)(q22q31),?del(6)(q21q22), inv(7)(p13q32),+der(11)t(11;15)(p15;q15),add(12)(q24.1),del(13)(q12q14),der(16)t(1;16)(q21;q22),+add(19)(q13.3), add(21)(q22)[cp16] | add(12)(q24.1) = der(12)t(4;12)(p12;q24) +add(19)(q13.3) = der(19)t(1;19)(p13;q13) add(21)(q22) = der(21)t(21;21)(q22;?) |
| 29. | 50 ∼ 51,XY,+3,add(3)(p?26),+5,+6,del(6)(q23),+7,−8,+9,−13,+15,add(15)(p11.1),i(15)(q10),−16,+19, add(19)(q13.3),der(20)add(20)(p13)add(20)(q13.3),der(22)t(1;22)(q10;q10), del(22)(q11.3)[cp11] | add(3)(p?26) = der(3)t(3;8)(p10;q10) +6 = der(6)t(6;8)(p10;q10) add(15)(p11.1) = der(15)t(15;20)(p11;q10) |
| 30. | 52,X,−X,+7,+9,+11,der(11)t(7;11)(p14;p12), der(14)t(X;14)(p11.3;p13),+15,der(15)t(1;15)(q10;q26),del(16)(q22q22),+17,+19, der(19)t(1;19)(q10;q13),+21,add(21)(p22),der(22)t(3;22)(q12;q13)[cp10] | der(11)t(7;11) = der(11)t(5;11)(p14;p12) add(21)(p22) = der(21)t(9;21)(q34;q22) |
| 31. | 52 ∼ 59,XY,−Y,add(1)(q32),del(1)(p11),+1,inv(3)(p26q23), t(4;19)(q21;q13.3),add(5)(q22),der(5)t(1;5)(q11;p?14),der(5)t(1;5)(q21;q15), del(5)(p13),+5,add(6)(q11),+6,+7,+7,der(8)t(8;11)(q24;q13) × 2,+der(8)t(8;11)(q24;q13), t(1;8)(q11;p23),+15,der(16)t(1;16)(q11.1;p13.3),del(17)(p11),+19,+20,add(20)(p13),+dic(1;20)(p22;p13),+21,add(22)(q13), +add(22)(p11)[cp20] | add(6)(q11) = der(6)t(6;10)(q13;?q21 ∼ 22) der(8)t(8;11) = der(8)t(1;8)(q10;p23) add(20)(p13) = t(2;20) |
| 32. | 56 ∼ 68,XY,t(1;17),+der(1)t(1;17),?add(2) × 2,+3,−4,+5,−7,+8, der(8;21)(q10;q10) × 2,+9,+9,+der(11)t(1;11)ins(11;?) × 2, −12,−13,−14,+15,+15,−16,t(16;21)(q12;q22),−18,+21,−22[cp6] | +der(11)t(1;11)ins(11;?) = der(11)t(11;17;1) |
| 33. | 53 ∼ 54,XY,+3,+5,+6,add(6)(q23),+7,t(7;17)(p11.1;p12),−8,+9,+15,+19,+21,+mar[cp19] | add(6)(q23) = der(8)t(6;8)(p10;q10) |
| 34. | 58,−X,−Y,der(X;1)(p10;p10),del(1)(p32) × 2,−2,−4, der(6)t(3;6)(q10;p10) × 2,+7,−8,+9,−10,+11,−12,−13,−13,−14,−16,del(16)(q22q22),add(17)(p11.1),−21,−22[cp6] | del(1)(p32) = der(1)t(1;X)(p31;q22) del(16)(q22q22) = der(16)t(13;16)(q12;q22) add(17)(p11.1) = der(17)t(7;17)(p11;p11) |
| 35. | 43,XY,?t(1;4)(q?32;q?21),dup(1)(q11q44),inv(2)(p13q21),add(4)(q34),del(6)(q15),−10,−13,−14,del(22)(q12)[cp20] | add(4)(q34) = der(4)t(X;4)(?q22;q34) |
| 36. | 56,XX, + der(1;6)(q10;p10) × 2,+3,+5,add(7)(q10),+8, i(8)(q10) × 2,der(9)t(1;9)(q10;q12),add(9)(p10),+der(10)t(10;11)(q22;q22),add(10)(p12),−13,der(14;19)(q10;p10), +15,?add(16)(q24),+del(17)(p11.1),+19,−20,+21, +del(22)(q13)[cp11] | add(7)(q10) = der(7)t(7;16) +der(10)t(10;11)(q22;q22) = der(20)t(7;20) add(10)(p12) = der(10)t(10;11)(p12;q24) ?add(16)(q24) = der(16)t(7;16) +del(17)(p11.1) = der(15)t(15;17)(q15;q12) |
| 37. | 46 ∼ 47,XY,del(1)(p22p32),?t(2;8)(p13;q24),add(3)(p?22), t(4;9)(q21;q22),?7p,+9,?t(9;19)(p22;q?13.3),−10,t(10;17)(q21;p11.1), add(12)(q15),−13,der(16)t(1;16)(q21;q22),?add(18)(q21), +del(19)(q11),del(20)(q11.2),der(20)t(1;20)(q?12;p13),+mar[cp14] | ?t(2;8)(p13;q24) = der(8)t(8;15)(q24.1;p21) der(16)t(1;16) = der(16)t(6;16) +mar = del(15) |
| 38. | 51,X,?add(X)(q26),t(1;9)(q11;q13),+der(1;19)(q10;p10), der(2)t(2;?13)(q23;q12),t(2;3)(q23;p21),+3, der(8)add(8)(p21)t(8;22)(q24.1;q11),der(8)t(2;8)(q23;p23),+add(9)(p2?4),+11,−13,−14,+15,+21,der(22)t(8;22),+mar,1dmin[cp16] | der(8)t(2;8)(q23;p23) = der(8)t(4;8)(q10;p10) +mar = del(13) |
| 39. | 41 ∼ 43,X,−X,+1,+1,der(1;2)(q10p;10),der(1;5)(q10;p10)dup(1)(q21q25), der(1;13)(q10;q10),der(2)t(1;2)(p32;q10),t(4;7)(q11;q32),der(6;8)(p10;q10), −13,−14[cp19] | no change |
| 40. | 65,XX,−Y,−1,−2,+3,?del(3)(q27),add(3)(q12),−4,+6, del(6)(q23q23) × 2,+7,dup(7)(q22q31) × 2,+8,del(8)(q1?24.1), del(8)(q22),+9,add(9)(p24),−10,+11,−12,−13,−14,−16,−16,−17,+18,−20, +21,+mar[cp15] | dup(7)(q22q31) = der(7)t(4;7)(q?28;q?36) add(3)(q12) = der(3)t(3;12) |
| 41. | 45,XX,add(3)(q29),del(5)(q22),add(11)(q25),inv(12)(p11q11),−17,add(20)(q13.3)[cp15] | add(3)(q29) = der(3)t(3;3)(q29;?q) add(11)(q25) = der(11)t(3;11)(q?25;q25) add(20)(q13.3) = der(20)t(17;20)(q22;q13) |
| 42. | 41 ∼ 42,XX,del(1)(p32p13),t(1;6)(q21;q25),add(8)(p23),del(9)(p13),−11,−13,−14,del(16)(q21q22),−18,−20,−22,+mar[cp18] | add(8)(p23) = der(8)t(8;18)(p23;q?) del(16) = t(14;16)(q32;q22 ∼ 23) +mar = t(11;19) |
| 43. | 75 ∼ 78,XX,−Y,del(1)(p13),add(2)(q37),?add(2)(q11),+3,del(3)(q21), −4,add(4)(q35),+5,del(5)(q31q32),+6,del(6)(q?23),add(6)(q13),+7,add(7)(p?22), +8,+9,add(9)(p13),add(9)(q34) × 2,add(11)(q23) × 2,+12,+12,add(12)(p13) × 2,−13, del(13)(q14q32),−14,+15,−16,−16,−16,+17,add(17)(p13) × 2, +18,+18,+19,add(20)(q13),+21,add(21)(p11),+2mar[cp17] | add(9)(q34) = der(9)t(9;12)(q34;?q24) add(11)(q23) = der(11)t(11;16)(q22;?) add(12)(p13) = der(12)t(10;12)(?;p13) add(17)(p13) = der(17)t(13;17)(p11;p13) add(20)(q13) = der(20)t(20;20)(q13;?) |
| 44. | 58,XX,+3,+add(5)(q?33),der(6;8)(p10;q10),+7,+7,+9,+9, +11,+11,del(13)(q12q14),add(14)(q32),+15, +der(16)t(6;16)(p21.1;q12.1) × 2,+18,+19,+21,−22[cp9] | der(6)t(6;8)(p10;q10) = der(8)t(3;8)(p10;p21) add(14)(q32) = der(14)t(8;14)(q24;q32) |
| 45. | 45,XY,add(2)(q32),der(6)t(6;6)(q27;p11.2),t(11;13)(q?11;p?13), −13,der(14)t(1;14)(q21;q32),del(16)(q12.1),add(17)(q24), add(22)(q13)[cp13] | add(2)(q32) = t(2;22)(q32;q13) del(16)(q12.1) = der(16)t(16;22)(q11;q13) |
| 46. | 86 ∼ 90,XXYY,−1,−1,add(1)(p13),add(2)(q33) × 3,add(3)(p10), t(4;5)(q31.3;q?34),t(5;7)(q10;q10),del(6)(q23),der(6)t(3;6)(p21;q23), der(6)t(6;11)(q23;q13),t(8;14)(q24;q32),t(8;22)(q24.3;q13), add(9)(p21),+10,der(10)t(1;10)(p13;q26) × 2,−11,der(11)(q13), −12,−13,−14,+der(15;21)(q10;q10),del(16)(q22) × 2,?add(17)(p10),−19, add(20)(p?11.2),+add(21)(q22),der(22)t(1;22)(q?25;p11.2)[cp5] | t(8;14) = der(14)t(14;16)(q32;q22 ∼ 23) der(10)t(1;10) = der(8)t(1;8;1;8) der(22)t(1;22) = der(21)t(10;21)(p11;q21) +add(21)(q22) = der(10)t(Y;10)(?;q11) |
| 47. | 56 ∼ 57,X,−Y,+add(1)(p13),+add(2)(q37),+add(3)(p11),del(4)(p12p14),del(4)(q31),+del(5)(q31q33), add(5)(q22),+del(6)(q13),del(6)(q23),+7,der(8)t(8;15)(q10;q10), ?add(9)(q34),+del(11)(q23),+15,+15,+15,der(17)t(9;17)(q12;p11.1),+18,+19, +19[cp6] | +add(1)(p13) = der(1)t(1;16)(p10;p10) +add(3)(p11) = der(3)t(3;20)(p10;?q10) der(8)t(8;15) = der(8)t(2;8)(p10;p?11) |
| 48. | 51,XY,der(1)t(1;12)(p13;q13),del(1)(p13p32),add(4)(q28),+5,add(6)(q21),add(8)(q24.1),+9, add(10)(q24),+11,add(12)(q15),del(13)(q12q14),+15,+15,−16,+19[cp6] | der(1)t(1;12) = der(1)t(1;16)(q10;p10) add(4)(q28) = t(4;10)(q?25;q?25) add(8)(q24.1) = t(6;8)(q21;q25.1) add(12)(q15) = der(12)t(1;12)(p?32;q15) |
| 49. | 49 ∼ 51,X,der(Y)t(Y;2)(q12;p11.2),del(1)(p31p36.1),+5,inv(5)(q13q35), +7,+9,del(10)(p13),t(12;19)(q13;p13.3),−13,+15,der(16)t(1;16)(q21;q21), der(17)t(8;17)(q11.1;p11.2),+der(19)t(7;19)(q11.2;q13.4), del(20)(p11.2p11.2)[cp20] | no change |
| 50. | 42,X,−X,i(1)(q10),add(2)(q?31),der(3)t(3;9)(p11;q11), −4,add(6)(p?21.3),add(7)(q22),−8,+del(9)(p13),−13,−14,der(14)t(5;14)(p11.1;p11.1)add(14)(q32),−15,add(15)(q22),add(16)(q?11.2)[cp20] | add(2)(q?31) = t(2;8)(q33;q23) add(6)(p?21.3) = t(6;7)(p22;q32) der(14)t(5;14)add(14)(q32) = t(8;14)(q24;q32) add(15)(q22) = t(15;16)(q23;q12) |
Boldface indicates changes made to karyotyping designation by SKY.
Recurring aberrations of chromosome 14q32.
Seventeen patients showed aberrations of 14q32 by G-banding. Of these, four showed translocations correctly identified by G-banding (samples 2, 4 ,13, 45). However, 8 of 50 patients identified by G-banding with add(14)(q32) chromosomes were all refined by SKY and FISH to recurring translocations (Table 1). Three samples with add(14)(q32) chromosomes (samples 3, 8, 9) were identified as t(14;16)(q32;q22∼23) (Fig 2A and B) and three were designated t(8;14)(q24;q32) translocations (samples 14, 44, 50). Two patients showing larger add(14)(q32) translocations were identified as t(9;14)(p13;q32) (samples 1 and 11) (Fig 2C and D). One patient designated as t(11;14)(q13;q32) by G-banding (sample 12) was refined to a t(3;14)(q21;q32), and an additional patient (sample 5) was identified in this study by G-banding showing this same translocation, therefore identifying this as a recurring aberration.
Recurring aberrations involving chromosome 8.
Ten of the 50 patients showed recurring translocations involving 8q24 by G-banding, including three each with t(8;14)(q24;q32) and t(8;22)(q24;q12), and two each with t(1;8)(p11;q24), and add(8)(q24). Of the three designated t(8;14)(q24;q32) by G-banding, two were refined by SKY and FISH to t(14;16)(q32;q22∼q23) (samples 6 and 46). Of the two patients identified as add(8)(q24) by G-banding, one was resolved to t(8;22)(q24;q12) (sample 24) and the other to t(6;8)(q21;q24) (sample 48), respectively. Thus, following the SKY and FISH analysis, only one of three samples identified by G-banding showed a t(8;14)(q24;q32), while an additional sample showed the t(8;22)(q24;q12).
A new whole-arm t(6;8)(p10;q10) translocation was identified in two patients (samples 29 and 39) (Fig 3A and B). One patient was identified by G-banding (sample 39); however, the other one was identified by SKY (sample 29). Five additional patients showed whole-arm aberrations involving chromosome 8, including two patients with iso(8)(q), and one each with t(8;12)(q10;q10),t(8;21)(q10;q10) and t(8;22)(q10;q10) (Table 2).
Recurring aberrations of chromosome 1 identified by G-banding.
Two patients showed a t(1;8)(p11;q24) translocation (samples 7 and 28) by G-banding and both of these were confirmed by SKY analysis (Fig 3C and D). Four patients showed recurring translocations between chromosomes 1 and 16 designated t(1;16)(q21;q11∼22).
DISCUSSION
The chromosome aberrations found in clinical cytogenetic preparations of bone marrow samples in MM are unusual in their complexity. In many ways, these complex karyotypes resemble those found in solid tumors, because they combine both high numbers of numerical and structural chromosome aberrations. Although traditional cytogenetic methods have proven useful in providing correlations of clinical outcome, the current banding methods for bone marrow are incapable of resolving the more complex aberrations. The need for an adjunct technique for more precise karyotypic interpretation of MM is clear.
In the present study, we applied G-banding and spectral karyotyping to the same sample preparation in an attempt to extend the chromosomal aberrations previously reported in patients with MM and to further localize chromosomal regions that may be of importance in the etiology and progression of this disease. Seventeen patients showed aberrations of 14q32 by G-banding. Of these, 4 showed translocations correctly identified by G-banding. However, 8 of the 50 specimens showed an add(14)(q32) aberration by G-banding; of these, all were shown by SKY to involve recurring translocations. Three samples were refined to t(8;14)(q24;q32) and three samples were refined to t(14;16)(q32;q22∼23) (Fig 2A and B). The largest add(14)(q32) chromosome was resolved to a t(9;14)(p13;q32) translocation in two patients (Fig 2C and D). These results support and extend previous studies suggesting “promiscuous” translocations to the Ig locus at 14q32.8
The t(14;16)(q32;q22∼23) translocation identified in this study appears to involve a small range of breakpoints on chromosome 16 from q22∼q23. In two patients the breakpoint appears at 16q22 (Fig 2A), which makes the add(14)(q32) appear larger, while in two other patients the breakpoint appears more distal at q23 (Fig 2B) so that the add(14)(q32) segment looks smaller. In the other two samples, the breakpoints were unclear; therefore, we assigned the q22∼23 breakpoints to chromosome 16. The SKY probe cocktails were able to identify the add(14)(q32) material as chromosome 16 in all cases, suggesting a reciprocal translocation, but were unable to identify any reciprocal exchange of chromosome 14 material to the chromosome 16 (Fig2A and B). In these cases, a FISH probe for the 14q32.3 locus in conjunction with an alpha satellite probe to chromosome 16, was used to determine if 14q32 was translocated to 16. Indeed, 14q32 probe signal was found translocated to chromosome 16 by FISH, thus confirming the reciprocal translocation in all patients (not shown). Therefore, identification of chromosome 16 material at 14q32 by SKY ultimately led to the complete resolution of this subtle reciprocal translocation with the aid of standard FISH.
In one case (sample 42), a deletion of 16q22∼23, suggestive of the t(14;16)(q32;q22∼23), prompted us to use conventional FISH protocols to identify an additional patient with t(14;16)(q32;q22∼23) not detected by the SKY probes. We and others have previously reported add(14)(q32) chromosomes in the same G-band karyotypes with deletions of 16q22∼24, suggesting that this subtle translocation is missed by conventional techniques.13,17 It is conceivable that the analysis of more cases with add(14)(q32) and or del(16)(q22) may show hitherto unrecognized recurring translocations. Interestingly, a t(14;16)(q32.3;q23) translocation has recently been identified as a recurring aberration in MM cell lines and has been associated with c-maf overexpression.7 The 16q23 breakpoints in these cell lines appear to be dispersed over approximately 100 kb and appear to be separated from c-maf by less than 500 kb. The dispersion of breakpoints and distance from the oncogene appear to be compatible with dysregulation of c-maf by the strong 3′ IgH enhancer.7 We believe this translocation most likely represents the same translocation found in our patient samples.
A second recurring translocation to emerge from the SKY analysis of the add(14)(q32) chromosomes was refined to a t(9;14)(p13;q32) translocation (Fig 2C and D). This translocation was found in two patients. The SKY technique identified the add(14)(q32) as a t(9;14)(p13;q32); however, the 14q32 segment that was translocated to 9p13 was again too small to be resolved by SKY. In this case, the 14q32 translocation was identified at 9p13 by the FISH probe for 14q32.3 locus. The t(9;14)(p13;q32) translocation has been associated with lymphoplasmacytoid lymphoma, a subtype of B-cell non-Hodgkin’s lymphoma, and also associated with Waldenstrom’s macroglobulinemia. This rare lymphoma is characterized by an indolent clinical course and followed by transformation into large cell lymphoma.18 The translocation in this case juxtaposes the PAX-5 gene at 9p13 with the Ig regulatory elements at 14q32 apparently deregulating PAX-5, causing overexpression of PAX-5 mRNA.19
Translocations of 8q24 in this study were found in 13 patients by G-banding. Band 8q24 is the locus of the c-myc proto-oncogene that is involved in the pathogenesis of a variety of B-cell malignancies and has been reported previously in studies of MM as t(8;14)(q24;q32) and the variant t(8;22)(q24;q12). Four of the 13 designations by G-banding were changed as a result of the SKY analysis (Table 1). Of the two samples that could only be designated as add(8)(q24) (samples 24 and 48) by G-banding, one of these was resolved to be a t(8;22)(q24;q12) translocation while the other was refined to a t(6;8)(q21;q24) translocation. Two of three patients originally designated as t(8;14)(q24;q32) (samples 6 and 46) translocations were redesignated to t(14;16)(q32;q22∼23) translocations by SKY and FISH. Another sample was refined from a designation of ?t(2;8)(p13;q24.1) to t(8;15)(q24.1;p21). The redesignation of different translocations involving 8q24 in this study suggests that the t(8;14)(q24;q32) in MM may be incorrectly identified and in some cases may be the t(14;16)(q32;q22∼23). This concept is supported by the findings in MM cell lines that although IgH translocations in MM are nearly universal, they rarely involve c-myc.20
Whole-arm chromosome translocations are a special type of rearrangement in which chromosome arms exchange at the centromeric region. These translocations can result in derivative chromosomes, which are unbalanced for whole chromosome arms, resulting in partial trisomy for one arm and partial monosomy for the other arm. When one of these translocations occurs in poorly banded preparations with similar centromeric indexes, these rearrangements can be incorrectly identified. Two patients showed whole-arm t(6;8)(p10;q10) translocations. One of these patients was identified by G-banding; however, because of poor morphology and banding of chromosomes in the other patient, the translocation was incorrectly identified (Fig 3A and B). Five additional patients showed whole-arm aberrations involving chromosome 8, including two patients with iso(8)(q) and one each with t(8;12)(q10;q10),t(8;21)(q10;q10) and t(8;22)(q10;q10). Interestingly, in each of these eight cases the short arm of chromosome 8 was lost, suggesting the possibility of the loss of a putative tumor suppressor gene on 8p in the progression of chromosome aberrations in MM.
Chromosomal studies in MM point to the gradual accumulation of numerous chromosome aberrations, especially structural rearrangements of chromosome 1. Although large translocations involving chromosome 1 breakpoints at 1q21 have been identified in MM by routine banding, the recent cloning of a novel gene (BCL9) involving a t(1;14)(q21;q32) suggests it may play a role in the progression of B-cell malignancies.20 The function of the BCL9 gene is not yet known, yet some translocations of 1q21 result in overexpression of BCL9.21 We identified one patient in this study with t(1;14)(q21;q32) and also identified four additional patients with 1q21 breakpoints involving t(1;16)(q21;q11∼22), suggesting this recurring translocation may play a role in the progression of MM. In addition to 1q aberrations, a new recurring translocation t(1;8)(p11;q24) was identified by G-banding and confirmed by SKY. The 8q24 material translocated to 1p11 was the smallest reciprocal exchange of chromosomal material identified by SKY in this study (Fig 3C and D).
Although spectral karyotyping identified additional rearrangements in all cases except two, the technique does have limitations. The main drawback appears to be the limits of resolution of the painting probe cocktails, which are reported to be between 500 and 1,500 kb.10 Therefore, the technique is unable to completely resolve the very subtle translocations of less than 500 kb. The subtle translocations of 14q32 to the exchange partners in the t(14;16)(q32;q22∼23) and t(9;14)(p13;q32) translocations were both below the resolving power of SKY. To completely resolve these translocations, we applied a FISH probe for the telomere of 14q32. However, in most cases, the spectral karyotyping refined the G-band by identifying several of the derivative or add chromosomes. It appears clear that multiple adjunct techniques to G-banding, including both FISH and SKY, are necessary to resolve the complex karyotypes of MM.
The search for a primary cytogenetic event in MM has proven difficult because, by the very nature of the disease process, most karyotypes have already evolved by the time of diagnosis. As in other tumor types, numerous cytogenetic studies indicate that more advanced disease is associated with higher frequencies of chromosome aberrations.3,22,23 Recently, the association of poor prognosis with the presence of the specific chromosome aberrations of 11q and monosomy and/or deletion 13q has been reported.24 25 It is interesting to note that all patients in this study who showed t(14;16)(q32;q22∼23) also showed the poor prognostic indicator monosomy 13. Clearly a larger study is needed to correlate cytogenetic findings of patient samples with other prognostic indicators since this study of karyotypes had a selection bias. The bias in this study was toward complex karyotypes and aberrations of add(14)(q32). Therefore, the true frequency of the new or redesignated translocations and/or associations with poor prognostic indicators awaits to be ascertained in a larger study. However, because 6 of 50 patients with complex karyotypes showed t(14;16)(q32;q22∼23), this suggests that it may be a frequently occurring translocation overlooked by banding techniques. Multicolor spectral karyotyping appears to be an important adjunct to routine G-banding in the clinical cytogenetic analysis of MM.
Supported in part by CA55819 from the National Cancer Institute.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Jeffrey R. Sawyer, PhD, Cytogenetics Laboratory, Arkansas Children’s Hospital, 800 Marshall St, Little Rock, AR 72202.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal