Abstract
Clonal expansion of B-cell precursor acute lymphoblastic leukemia (ALL) is potentially regulated by survival, growth, and death signals transduced by the bone marrow (BM) microenvironment. Using a human BM stromal cell culture that supports the growth of normal human B-cell precursors, we established a pre-B ALL cell line designated BLIN-2. BLIN-2 has a clonal rearrangement of the Ig heavy chain locus, a dic(9;20) chromosomal abnormality, and a bi-allelic deletion of thep16INK4a and p19ARF genes. The most interesting feature of BLIN-2 is an absolute dependence on adherent human BM stromal cells for sustained survival and growth. BLIN-2 cultured in the absence of BM stromal cells undergo apoptosis, and direct contact with viable BM stromal cells is essential for optimal growth. BLIN-2 cells also grow on vascular cell adhesion molecule-1 (VCAM-1)–negative human skin fibroblasts, making it unlikely that a very late antigen-4 (VLA-4)/VCAM-1 interaction is required for BLIN-2 growth. Western blot analysis of BLIN-2 cells cultured in the presence or absence of BM stromal cells demonstrates that contact of BLIN-2 with BM stromal cells induces hyperphosphorylation of Rb. In contrast, the pre-B ALL cell line BLIN-1, which has a bi-allelic deletion of p16INK4ap19ARF but does not require BM stromal cells for growth, does not undergo Rb phosphorylation after BM stromal cell contact. The BLIN-2 cell line will facilitate identification of ligand/receptor interactions at the B-cell precursor/BM stromal cell interface and may provide new insight into microenvironmental regulation of leukemic cell survival and growth.
ACUTE LYMPHOBLASTIC leukemia (ALL) is a malignancy characterized by the clonal expansion of T- or B-lineage lymphoid progenitors, and approximately 70% of newly diagnosed cases involve CD19+ cells at various stages of B-cell precursor development.1-3 The cytogenetic and molecular genetic abnormalities in B-cell precursor ALL frequently consist of chromosomal translocations involving genes that encode transcription factors, and many of these transcription factor genes are members of the homeobox-containing HOX gene family.4,5Look5 has recently discussed how aberrant regulation ofHOX gene expression, subsequent to the aforementioned translocations, may contribute to the transformation process in ALL. For example, alterations in HOX gene expression could subvert the normal program of apoptotic fate characteristic of most lymphoid progenitors.5 However, much less understood is the potential contribution of bone marrow (BM) microenvironmental survival/growth stimuli to the clonal expansion of leukemic progenitors, potentially in the background of disrupted apoptotic programs.
Analyses of various cytokines, interleukins, and colony-stimulating factors for their capacity to support the survival and growth of B-cell precursor ALL in vitro have been reported.6-15 Importantly, several of these studies surveyed a range of cytokines and concluded that no single cytokine or combination of cytokines could support the clonogenic growth of leukemic cells from a significant number of cases.11,14 15 A potential common limitation in all these studies was a failure to adequately evaluate the clonogenic growth potential of leukemic blasts using a culture system that recapitulates the BM microenvironment. B-cell precursor ALL is a malignancy of BM origin. It follows that at least the earliest stages of B-cell precursor ALL exhibit a requirement on BM microenvironment-derived survival or growth factor signals for expansion of the leukemic cell clone. How these BM microenvironment-derived signals influence the initial stages of transformation or subsequent expansion of the dominant leukemic subclone is unknown.
A series of studies from Campana et al have described the development and use of a short-term (ie, 7 days) BM stromal cell-based in vitro assay for examining the contribution of BM stromal cells to the survival and programmed cell death of B-cell precursor ALL.16-18 Interestingly, a strong inverse correlation was observed between the inherent propensity of leukemic blasts from individual patients to survive on BM stromal cells and the probability that individual patients would achieve long-term, event-free survival.18 The probability of event-free survival at 4-year follow-up was lower among patients whose leukemic blasts survived for up to 7 days on BM stromal cells in vitro compared with patients whose leukemic blasts underwent apoptosis within 7 days on stromal cells.
Our laboratory has developed a BM stromal cell culture system that supports the survival and growth of normal human B-cell precursors.19 The stromal cells in this culture are predominantly fibroblast-like adventitial reticular cells that express vascular cell adhesion molecule-1 (VCAM-1).20 Optimal growth occurs in serum-free medium supplemented with interleukin-7 (IL-7), and direct contact with BM stromal cells is essential.19 21 In the current study, we describe the application of this BM stromal cell culture to evaluate the capacity of BM stromal cells to support the long-term growth of B-cell precursor ALL. We report the development and characterization of a new B-cell precursor ALL cell line, designated BLIN-2, that exhibits an absolute dependency on BM stromal cells for survival and growth. BLIN-2 is a novel cellular resource for elucidating the ligand/receptor interactions essential for the development of normal and leukemic human B-cell precursors.
MATERIALS AND METHODS
Establishment of BLIN-2.
Cryopreserved BM from a 3-year-old girl with newly diagnosed B-cell precursor ALL (based on immunophenotype and histopathology) was thawed from liquid nitrogen and centrifuged over a Ficoll-Hypaque gradient (Histopaque; Sigma Chemical Co, St Louis, MO) to remove dead cells. Approximately 2.0 × 106 recovered interface cells were washed three times in RPMI-1640 (Life Technologies, Grand Island, NY), supplemented with 2% fetal bovine serum (FBS; Hyclone, Ogden, UT), and plated onto a pre-established adherent layer of adult BM stromal cells in 96-well flat-bottom microtiter plates (Costar, Cambridge, MA). One objective at this stage was to determine whether leukemic B-cell precursors would exhibit optimal growth when cultured on BM stromal cells supplemented with IL-7, as we had previously shown for normal human pro-B cells.19 21 Leukemic cells were therefore cultured in the absence or presence of IL-7 (10 ng/mL; PeproTech, Rocky Hill, NJ) at an initial density of 3.5 × 104 cells/well. After 3 weeks in culture, approximately 50% of the wells had viable leukemic cells as judged by inverted light microscopy, independent of whether the leukemic cells were cultured in the presence or absence of IL-7. The cellular content of wells with viable cells was diluted 1:2 and passaged onto fresh adult BM stromal cells. After an additional 3 weeks, the leukemic cells were pooled from multiple wells and phenotyped. The leukemic cells established in culture at this stage were designated B lineage-2 (BLIN-2). Because IL-7 had no additional effect on BLIN-2 survival or growth beyond the supportive effect of the stromal cells, IL-7 supplementation was discontinued.
Other cells.
BLIN-1 is a surface μ+/ψ light chain+ pre-B ALL cell line originally established in this laboratory.22RAMOS is an Epstein-Barr virus (EBV)-negative, surface μ+/surface λ light chain+ Burkitt lymphoma cell line obtained from the American Type Culture Collection (ATCC; Rockville, MD). BLIN-1 and RAMOS were maintained in RPMI-1640 supplemented with 10% FBS, 100 U penicillin/mL, and 100 μg streptomycin/mL. Fluorescence-activated cell sorting (FACS)-purified fetal BM pro-B cells were isolated as previously described.21
Adherent cells.
Fetal BM was obtained from 19- to 21-week gestational age fetuses, in accordance with guidelines set forth by the University of Minnesota Committee on the Use of Human Subjects in Research. A detailed description of the methods we use for the establishment of human BM stromal cells has been published.23 24 Briefly, total BM mononuclear cells were isolated by Ficoll-Hypaque centrifugation and seeded into 75-cm2 tissue culture flasks (Falcon, Lincoln Park, NJ) in RPMI-1640 containing 10% FBS. Nonadherent cells were washed off after 2 hours at 37°C, and the adherent cells were cultured in EX-CELL 610 (JRH Biosciences, Lenexa, KS) supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL of streptomycin. An adherent layer of BM stromal cells was generally established within 1 week. The cells were then detached with 0.05% trypsin, 0.53 mmol/L EDTA (Life Technologies, Grand Island, NY) and transferred into new 75-cm2 flasks containing fresh EX-CELL 610/10% FBS. The adherent layer reached confluence within 1 week, and the cells were passaged a second time. Adult BM stromal cells were established and passaged in the same manner as fetal BM stromal cells, except that adult BM stromal cells required 2 to 3 more days to reach confluence. Upon reaching 90% confluence after second passage, fetal and adult BM stromal cells could be maintained in EX-CELL 610 serum-free medium.
Foreskin fibroblasts were originally obtained at first passage from Dr Elizabeth Wayner (formerly of the Department of Laboratory Medicine/Pathology, University of Minnesota, Minneapolis, MN). These cells were maintained and passaged as described above for BM stromal cells and were used between passages 8 and 11 in the experiments described herein.
Antibodies.
Monoclonal antibodies (MoAbs) recognizing specific Ig molecules or membrane proteins and their conjugation to fluorescein isothiocyanate (FITC) or biotin have been described.19-21,23 24 Streptavidin-phycoerythrin (PE) was purchased from Caltag Laboratories (South San Francisco, CA). Goat antimouse FITC was purchased from Southern Biotechnology Associates, Inc (Birmingham, AL). Rabbit antibody recognizing the human retinoblastoma (Rb) protein was purchased from Santa Cruz Biotechnology (Santa Cruz, CA) as a horseradish peroxidase conjugate (catalog no. SC-050-HRP) and was used in Western blotting. Rabbit anti-Rb was made against a peptide corresponding to amino acids 914-928 at the carboxy terminus of p110 Rb and recognizes phosphorylated and nonphosphorylated forms of Rb.
Immunofluorescent staining and flow cytometry.
Cytogenetics and fluorescence in situ hybridization (FISH).
Metaphase cells were harvested from short-term unstimulated cultures of the patient’s BM or from the actively growing BLIN-2 cell line and were subsequently G-banded using Wright stain according to standard cytogenetic procedures.25 In the initial analysis of BLIN-2, 15 metaphase cells were analyzed microscopically for definition of the clonal abnormalities. FISH was performed using chromosome 9 and chromosome 20 whole chromosome paint probes (VYSIS Inc, Downersgrove, IL) directly labeled in Spectrum orange and Spectrum green, respectively. FISH was performed with modification of the manufacturer’s instructions. Briefly, target DNA was denatured, hybridized with the probes for 16 hours, and washed in 2× SSC at 72°C. Chromosomes were counterstained with 4′, 6′-diamidino-2′-phenylindole (DAPI), and metaphase cells and hybridization signals were visualized under an Olympus microscope outfitted for fluorescence with a triple band pass (B-MAX) filter (Olympus Optical Co Ltd, Tokyo, Japan).
Growth assay.
Adherent cells were seeded into 96-well flat-bottom microtiter plates at 0.6 to 1.0 × 104 cells/well in EX-CELL 610/10% FBS. After 3 to 5 days, the adherent cells were washed with X-VIVO 10/0% FBS and BLIN-2 cells were seeded at 1 to 3 × 104 cells/well in a final volume of 200 μL of X-VIVO 10/0% FBS. X-VIVO 10 is a serum-free medium purchased from BioWhittaker, Inc (Walkersville, MD). It contains human serum albumin as a carrier protein and insulin and transferrin as growth-promoting supplements. The capacity of BLIN-2 cells to grow in the absence of BM stromal cell contact was tested by plating BLIN-2 cells into 6.5-mm transwells (Costar) containing a 0.4-μm polycarbonate membrane suspended above the adherent cells. The cultures were fed every 3 to 4 days by replacing 50% of the culture volume with fresh X-VIVO 10/0% FBS, unless otherwise stated. BLIN-2 growth was quantified using PE-conjugated anti-CD19 in a microsphere flow cytometric-based quantitation assay.21
Apoptosis and cell cycle analysis.
Cells were cultured at 1 × 106 cells/60-mm petri plate in 4 mL of X-VIVO 10 serum-free medium in the presence or absence of fetal BM stromal cells. At each time point, BLIN-2 cells were gently removed from culture without disrupting the adherent layer and simultaneously analyzed for subdiploid events and cell cycle using the method of Nicoletti et al.26 Briefly, the cell pellet was gently resuspended in 1 mL of hypotonic solution (50 μg/mL propidium iodide in 0.1% sodium citrate plus 0.1% Triton-X-100) and incubated overnight at 4°C in the dark. Analysis of intact and fragmented nuclei was achieved using a FACSCalibur flow cytometer (Becton Dickinson and Co, Mountain View, CA) and CELLQuest software. Chromatin degradation, a characteristic of apoptosis, was detected as a heterogeneous subdiploid population to the left of the peak corresponding to intact nuclei in G0/G1 phases of the cell cycle.
FITC-conjugated Annexin V was purchased from Pharmingen (San Diego, CA) and used according to the manufacturer’s instructions. Briefly, cells were washed once in ice-cold phosphate-buffered saline (PBS) and then washed once in prewarmed binding buffer (10 mmol/L HEPES, pH 7.4, 140 mmol/L NaCl, 2.5 mmol/L CaCl2). Cells were resuspended in 100 μL binding buffer containing 5 μL of Annexin V-FITC and 10 μL of propidium iodide (50 μg/mL of stock solution) and then incubated at room temperature for 15 minutes. An additional 400 μL of binding buffer was added to the cells before analysis using a FACSCalibur flow cytometer and CELLQuest software (Becton Dickinson, San Jose, CA).
Polymerase chain reaction (PCR) and Southern blotting.
TRI Reagent (Molecular Research Center, Cincinnati, OH) was used to extract DNA from BLIN-1, RAMOS, and BLIN-2 cells. Approximately 0.5 μg of DNA was amplified using 0.4 mmol/L of each specific primer, 0.4 mmol/L of dNTP, 1.5 mmol/L MgCl2, and 2.5 U AmpliTaq (Perkin Elmer, Branchburg, NJ) in a final volume of 50 μL. The mixture was overlaid with 100 μL mineral oil. The PCR reaction was performed in an automated DNA Thermal Cycler (Hybaid Ltd, Middlesex, UK) with the following parameters: denaturation at 95°C for 4 minutes, 30 cycles of 94°C for 1 minute, annealing at 55°C for 1 minute, 1 minute of extension at 72°C, and a final 10 minutes of extension at 70°C. Amplified products were electrophoresed on 1.5% agarose gels. The gels were washed in a denaturing solution containing 0.5N NaOH and 1.5 mol/L NaCl for 20 minutes at room temperature, followed by one wash in a neutralization solution containing 1.5 mol/L NaCl and 0.5 mol/L Tris-HCl, pH 7.5, for 30 minutes at room temperature. The DNA was then transferred onto a nylon membrane (Nytran; Schleicher & Schuell, Keene, NH) using vacuum blotting with a PosiBlot (Stratagene, La Jolla, CA) and the membrane was UV cross-linked with 120 mJ/cm2 (UV Crosslinker; Fisher Scientific, Pittsburgh, PA) and allowed to air-dry. Probes were end-labeled with ATP[γ-32P] using T4 Polynucleotide Kinase (Life Technologies) according to the manufacturer’s recommendations. Blots were prehybridized in 1 mol/L NaCl, 0.2 mol/L Tris-HCl, pH 7.5, 0.1% sodium dodecyl sulfate (SDS), containing 200 μg/mL denatured salmon sperm for 2 hours at 49°C to 51°C. Hybridization was conducted in the same solution and at the same temperature as the prehybridization, with the addition of ATP[γ-32P]-labeled probes for 18 hours. The membranes were washed sequentially with 1× SSC, 0.1% SDS twice at room temperature for 10 minutes each and then at 54°C to 58°C for 10 minutes, followed by a final wash at room temperature for 5 minutes. The membrane was developed by autoradiography using X-Omat AR film (Eastman Kodak, Rochester, NY) at −70°C. Exposure times ranged from 3 to 6 hours.
The primer pairs used were as follows: p15INK4bexon 2, 5′-CGA-GGA-GAA-CAA-GGG-CAT-3′ and 5′-GAA-TGC-ACA-CCT-CGC-CAA-CG-3′;p19ARF exon 1β, 5′-AGT-CTG-CAG-TTA-AGG-GGG-CAG-3′ and 5′-GGC-TAG-AGA-CGA-ATT-ATC-TGT-3′;p16INK4a exon 1α, 5′-GAG-GCG-GCG-AGA-ACA-TGG-TG-3′ and 5′-CTT-CTA-GGA-AGC-GGC-TGC-TG-3′;p16INK4a-p19ARF exon 2, 5′-GCT-TCC-TTT-CCG-TCA-TGC-CG-3′ and 5′-CAA-ATT-CTC-AGA-TCA-TCA-GTC-C-3′; actin, 5′-ATC-ATG-TTT-GAG-ACC-TTC-AA-3′ and 5′-CAT-CTC-TTG-CTC-GAA-GTC-CA-3′. The sequence of the internal probes used to detect specific amplified products by Southern blotting were as follows: p15INK4b exon 2, 5′-CAA-ATC-TAC-ATC-GCG-ATC-TAG-G-3′;p19ARF exon 1β, 5′-CAC-CAA-ACA-AAA-CAA-GTG-CG-3′;p16INK4a exon 1α, 5′-GAC-GCT-GGC-TCC-TCA-GTA-GC-3′;p16INK4a-p19ARF exon 2, 5′-CTG-TTC-TCT-CTG-GCA-GGT-CAT-G-3′; and actin, 3′-ATG-TCA-CGC-ACG-ATT-TCC-CG-5′.
Western blotting.
BLIN-1 or BLIN-2 cells were cultured on BM stromal cells or in medium alone for 18 hours. The leukemic cells were gently poured off the BM stromal cell adherent layer, lysed in 0.05 mol/L Tris, 0.25 mol/L NaCl, 0.5% NP-40, 1 mmol/L phenylmethylsulfonyl fluoride (PMSF), 1% aprotinin, 10 μg/mL leupeptin, 0.1 mol/L NaF, and 0.002 mol/L sodium orthovanadate and then vortexed for 30 minutes at 4°C. The lysates were centrifuged for 30 minutes at 13,500g at 4°C. The supernatant was removed and protein quantitation was conducted using the Coomassie Plus Protein Assay Reagent (Pierce, Rockford, IL). Forty to 50 μg of protein per sample was electrophoresed on a 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) gel and then transferred onto a nitrocellulose membrane (Bio-Rad Laboratories, Hercules, CA). The membrane was blocked in Tris-buffered saline-Tween (TBST; 50 mmol/L Tris, pH 7.6, 150 mmol/L NaCl, 0.1% Tween 20) containing 5% milk for 2 hours at room temperature. Horseradish peroxidase-conjugated rabbit anti-Rb was incubated with the blot at a final concentration of 2 μg/mL for 30 minutes in TBST containing 5% milk. The membrane was then washed 4 times at 10-minute intervals in TBST and developed by enhanced chemiluminescence according to the manufacturer’s instructions (Amersham Life Science, Arlington Heights, IL).
Quantitation of chemiluminescent Western blots was conducted by scanning densitometry using a model GS-700 Imaging Densitometer (Bio-Rad, Hercules, CA). Data analysis was conducted using Molecular Analyst ver 2.0 software. The intensity of individual bands was converted to a histogram profile of the sum of the pixel density, and the profiles were adjusted to remove background. The area under the curve was calculated in square millimeters and calibrated to an internal machine constant. Band intensity values were expressed as ratios of hyperphosphorylated Rb to the total Rb in each lane. Scanning densitometry was conducted using the facilities of the Biomedical Imaging and Processing Laboratory, University of Minnesota Medical School.
RESULTS
Establishment of the BLIN-2 cell line.
This study was initiated to establish BM stromal cell-dependent pre-B ALL cell lines as part of a long-term project to elucidate how BM stromal cells influence the development of this malignancy. Twenty-three consecutive BM specimens from patients with newly diagnosed ALL were plated on allogeneic BM stromal cell feeders in X-VIVO 10 serum-free medium in the presence or absence of IL-7. Of these 23 specimens, 17 showed a less than 10-fold increase in CD19+ cell number after 4 weeks in culture, whether IL-7 was present or not. Six specimens showed a greater than 10-fold expansion in CD19+ cell number. One of these cultures, designated BLIN-2, is the subject of this report.
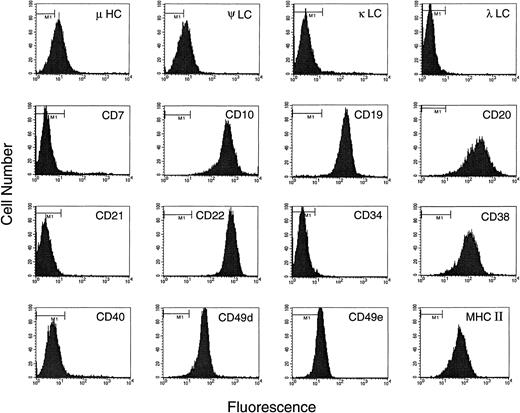
The BLIN-2 cell line was initiated in April 1993 using cryopreserved BM from a pediatric patient diagnosed with ALL. The cryopreserved leukemic BM specimen was 100% CD19+ and 50% CD34+ at the time of initial plating onto adult BM stromal cells, and a stable population of leukemic cells emerged after approximately 6 weeks. As shown in Fig 1, the established cell line expressed a typical pre-B phenotype (ie, CD19+, CD20+, CD22+, CD34−, weakly surface μ+, ψ light chain+, and κ/λ light chain−). Southern blot analysis of DNA isolated from the original cryopreserved leukemic BM specimen and BLIN-2 cells in culture for 6 months showed identical heavy and light chain Ig gene rearrangements (data not shown). DNA from the original cryopreserved leukemic BM specimen and BLIN-2 was amplified by PCR using consensus VH family primers and consensus primers that would amplify JH1, JH3, and JH4 genes. The amplified products were sequenced, and the original cryopreserved leukemic BM specimen and the established BLIN-2 cell line were shown to harbor identical VH3(DN4)JH4 rearrangements across 80 nucleotides of sequence (including identical N region insertions). This confirmed the clonal identity of the original BM leukemic cells and the established BLIN-2 cell line using a unique V(D)J rearrangement as a fingerprint.
Immunophenotype of BLIN-2. Background staining using isotype-matched myeloma proteins as negative controls is shown by the horizontal bar in the upper left part of each histogram.
Immunophenotype of BLIN-2. Background staining using isotype-matched myeloma proteins as negative controls is shown by the horizontal bar in the upper left part of each histogram.
G-banded chromosomal analysis of the patient’s BM at diagnosis in June 1993 showed a hypodiploid clone with a 45,XX complement, including an abnormal chromosome 9 with a deletion of its distal short arm and loss of one chromosome 20 as the sole karyotypic abnormalities. G-banded chromosomal analysis of the BLIN-2 cell line in June 1994 showed these same abnormalities and also a gain of a chromosome 8. The origin of the trisomy 8 in BLIN-2 cells is unknown. It may have been present as a rare subclone in the original BM specimen that expanded in culture, or it could have arisen during evolution of the cell line in culture.
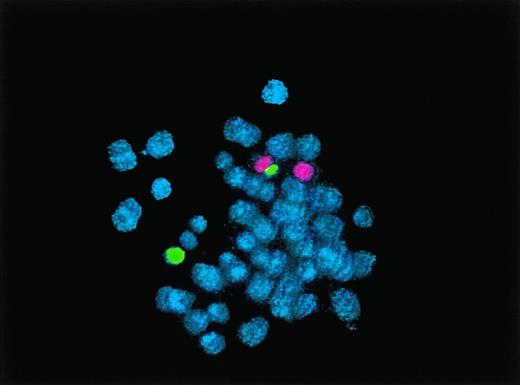
In February 1998, G-banded chromosomal analysis of the BLIN-2 cell line was repeated and supplemented by FISH using chromosome 9 and chromosome 20 paint probes (Fig 2). FISH showed that the abnormal chromosome 9 was actually a dicentric chromosome containing the short arm of a chromosome 20 joined to the long arm of a chromosome 9, with centromeric regions of both chromosomes included in the rearrangement. Thus, the karyotype of this cell line has been stable in culture and can be definitively designated as 46,XX,+8,dic(9;20)(p11;q11.1). This karyotype results in net losses of one copy each of the short arm of chromosome 9 and the long arm of chromosome 20 and a net gain of one copy of chromosome 8.
FISH analysis of BLIN-2 cells from February 1998. Normal chromosome 9 is shown in red, normal chromosome 20 in green, and dic(9;20) as red/green. Chromosomes were counterstained with DAPI (blue).
FISH analysis of BLIN-2 cells from February 1998. Normal chromosome 9 is shown in red, normal chromosome 20 in green, and dic(9;20) as red/green. Chromosomes were counterstained with DAPI (blue).
Stromal cell dependence of BLIN-2.
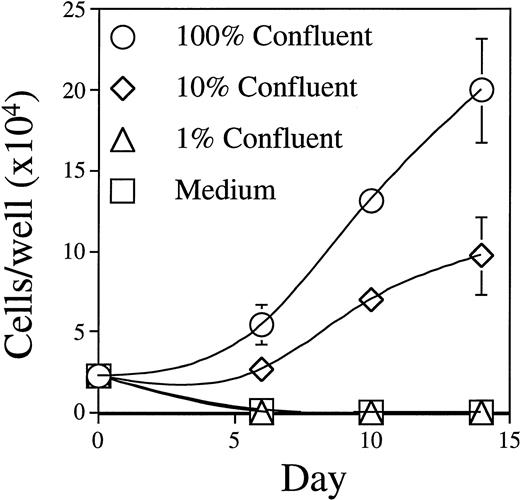
The BLIN-2 cell line was established on an adherent monolayer of BM stromal cells and has maintained a strict BM stromal cell requirement for survival and growth. BLIN-2 growth showed a good correlation with the number of BM stromal cells on which BLIN-2 was initially plated. As shown in Fig 3, when BLIN-2 cells were plated on 100% confluent BM stromal cells, there was an eightfold increase in BLIN-2 by day 14. In contrast, 10% confluent BM stromal cells only supported a 3.5-fold increase in BLIN-2 by day 14, and 1% confluent BM stromal cells or serum-free medium alone failed to support survival or growth. We emphasize that this BM stromal cell culture system does not require serum supplementation. Addition of up to 20% vol/vol FBS does not enhance growth of BLIN-2 on BM stromal cells. Furthermore, BLIN-2 cells cultured in the absence of BM stromal cells do not grow in X-VIVO 10 serum-free medium supplemented with FBS. Growth of BLIN-2 is also not contingent upon a specific source of human BM stromal cells, because we have continuously maintained BLIN-2 cells on BM stromal cells established from greater than 20 different donors.
Growth of BLIN-2 on BM stromal cells. BLIN-2 cells (2.5 × 104/well) were cultured on various concentrations of BM stromal cells, and the number of BLIN-2 cells was quantified on days 7, 10, and 14. Each symbol represents the mean ± SD of triplicate wells. One hundred percent confluency corresponds to approximately 6 × 103 BM stromal cells per 200-μL flat-bottom microtiter well. This experiment is representative of five similar experiments.
Growth of BLIN-2 on BM stromal cells. BLIN-2 cells (2.5 × 104/well) were cultured on various concentrations of BM stromal cells, and the number of BLIN-2 cells was quantified on days 7, 10, and 14. Each symbol represents the mean ± SD of triplicate wells. One hundred percent confluency corresponds to approximately 6 × 103 BM stromal cells per 200-μL flat-bottom microtiter well. This experiment is representative of five similar experiments.
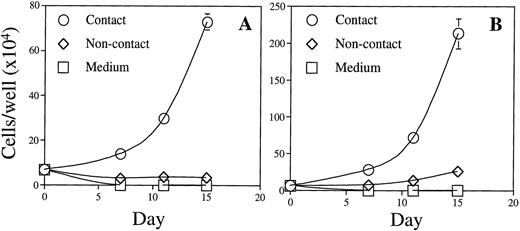
The role of direct BM stromal cell contact in supporting BLIN-2 growth was evaluated using a transwell system. Figure 4 compared the growth characteristics of an early passage of BLIN-2 cryopreserved in February 1995 and thawed in January 1998 for use in this experiment versus BLIN-2 cells that had been passaged continuously. Figure 4A and B shows that direct contact with BM stromal cells was essential for optimal BLIN-2 growth. The February, 1995 BLIN-2 cells cultured in transwell inserts slowly died over the 15-day culture period and increased 10-fold in contact with BM stromal cells (Fig 4A). In contrast, the continuously passaged BLIN-2 cells underwent a very modest fourfold increase by day 15; this compared with the 33-fold increase under contact conditions by day 15. When BLIN-2 cells were cultured in direct contact with BM stromal cells, and a transwell containing BLIN-2 cells was inserted above the BLIN-2/BM stromal cell contact culture, BLIN-2 cells in the transwell failed to grow, whereas BLIN-2 cells in direct contact with BM stromal cells grew normally (data not shown). These results make it unlikely that BLIN-2 cell contact induced the BM stromal cells to secrete a product that supported BLIN-2 growth.
The role of BM stromal cell contact in the growth of BLIN-2. BLIN-2 cells cryopreserved in February 1995 and thawed for use in this experiment in January 1998 (A) or maintained continuously on BM stromal cells (B) were cultured in direct contact with BM stromal cells, in transwell inserts (noncontact) suspended above the BM stromal cells, or in medium alone. BLIN-2 growth was quantified on days 7, 11, or 15. Each symbol represents the mean ± SD of triplicate wells. This experiment is representative of six similar experiments.
The role of BM stromal cell contact in the growth of BLIN-2. BLIN-2 cells cryopreserved in February 1995 and thawed for use in this experiment in January 1998 (A) or maintained continuously on BM stromal cells (B) were cultured in direct contact with BM stromal cells, in transwell inserts (noncontact) suspended above the BM stromal cells, or in medium alone. BLIN-2 growth was quantified on days 7, 11, or 15. Each symbol represents the mean ± SD of triplicate wells. This experiment is representative of six similar experiments.
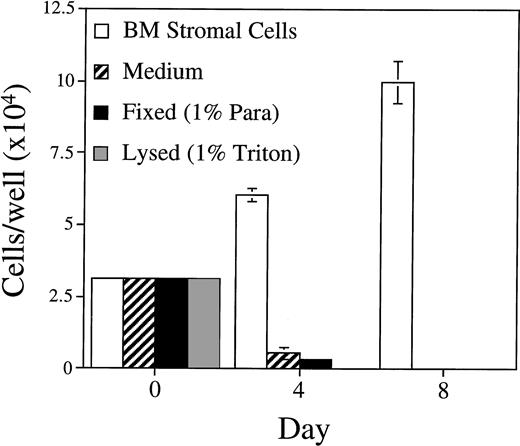
The importance of BM stromal cell integrity was tested by assaying the capacity of BLIN-2 cells to grow on Triton X-100–lysed or paraformaldehyde-fixed BM stromal cells. As shown in Fig 5, fixed or lysed BM stromal cells were unable to support BLIN-2 survival and growth. Thus, intact, metabolically active BM stromal cells are essential for BLIN-2 growth.
Growth of BLIN-2 on fixed or lysed BM stromal cells. BM stromal cells were fixed in 1% paraformaldehyde or lysed in 1% Triton X-100 and then washed five times in medium before the addition of BLIN-2 cells. BLIN-2 growth was quantified on days 4 and 8. Each bar represents the mean ± SD of triplicate wells. This experiment is representative of four similar experiments.
Growth of BLIN-2 on fixed or lysed BM stromal cells. BM stromal cells were fixed in 1% paraformaldehyde or lysed in 1% Triton X-100 and then washed five times in medium before the addition of BLIN-2 cells. BLIN-2 growth was quantified on days 4 and 8. Each bar represents the mean ± SD of triplicate wells. This experiment is representative of four similar experiments.
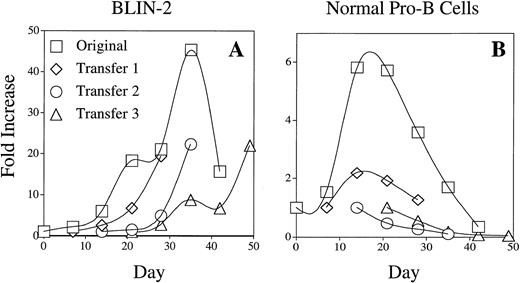
The growth kinetics of BLIN-2 on BM stromal cells (Figs 3, 4, and 5 and numerous other experiments) indicated that the population doubling time of BLIN-2 is relatively slow. BLIN-2 would typically undergo a twofold to threefold increase in cell number between days 0 and 7 and a fivefold to sevenfold increase in cell number between days 7 and 14. As presented in Fig 6A, BLIN-2 cells repetitively passaged on BM stromal cells showed a consistent sixfold to eightfold increase in cell number during the initial 14 days after transfer onto fresh stromal cells. This rate of population increase was similar to IL-7–stimulated normal human pro-B cells cultured on BM stromal cells (Fig 6B).
Comparison of BLIN-2 and normal pro-B cell growth on BM stromal cells. BLIN-2 and FACS-purified normal pro-B cells were plated at 2.5 to 5.0 × 104/well on BM stromal cells and cultured in X-VIVO 10/0% FBS. The pro-B cells were supplemented with 10 ng/mL of IL-7. BLIN-2 and pro-B cell numbers were quantified on day 7 and replated onto fresh BM stromal cells (transfer 1) at the initial cell concentration of 2.5 to 5.0 × 104/well. Quantitation of cell numbers and replating onto fresh BM stromal cells was repeated on day 14 (transfer 2) and day 21 (transfer 3).
Comparison of BLIN-2 and normal pro-B cell growth on BM stromal cells. BLIN-2 and FACS-purified normal pro-B cells were plated at 2.5 to 5.0 × 104/well on BM stromal cells and cultured in X-VIVO 10/0% FBS. The pro-B cells were supplemented with 10 ng/mL of IL-7. BLIN-2 and pro-B cell numbers were quantified on day 7 and replated onto fresh BM stromal cells (transfer 1) at the initial cell concentration of 2.5 to 5.0 × 104/well. Quantitation of cell numbers and replating onto fresh BM stromal cells was repeated on day 14 (transfer 2) and day 21 (transfer 3).
Apoptosis of BLIN-2 cells in the absence of BM stromal cells.
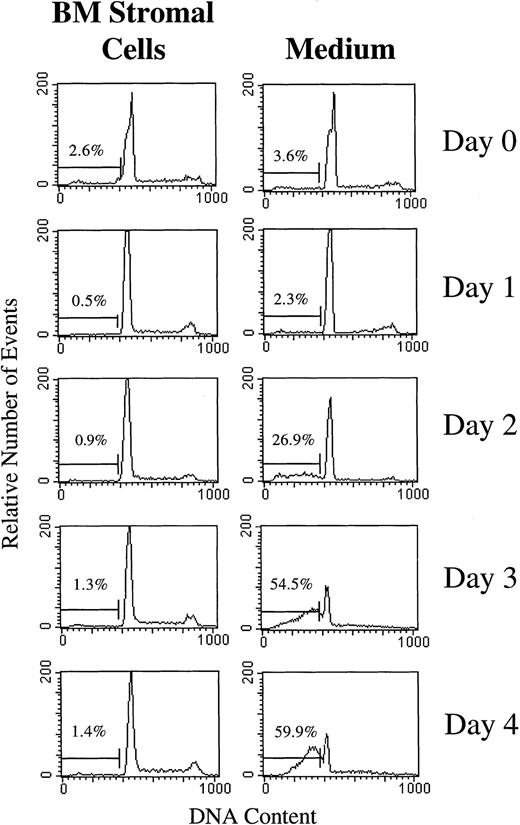
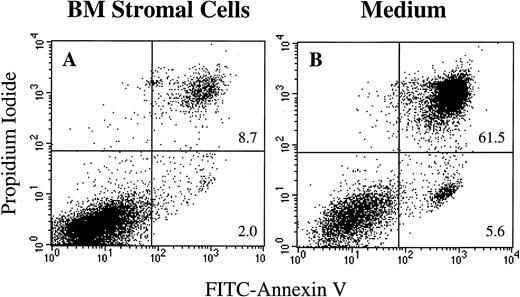
BLIN-2 cells were cultured on BM stromal cells or in serum-free medium alone, and propidium iodide-stained nuclei and nuclear fragments were analyzed by flow cytometry at various time points. Figure 7 shows that BLIN-2 cells cultured on BM stromal cells did not exhibit an increase in subdiploid events during 4 days of culture. In contrast, BLIN-2 cells cultured in serum-free medium alone showed no accumulation of subdiploid events at day 1, but underwent an increase to approximately 27% subdiploid events at day 2, that reached approximately 60% by day 4. In a separate experiment shown in Fig 8, Annexin V binding was coupled with propidium iodide staining to quantify early stage apoptotic events (ie, Annexin V+, propidium iodide−). BLIN-2 cells cultured on BM stromal cells for 4 days (Fig 8A) had 2% Annexin V+/propidium iodide− apoptotic events and less than 10% Annexin V+/propidium iodide+ events that represent dead cells. In contrast, BLIN-2 cells cultured in medium alone for 4 days (Fig 8B) had 5.7% Annexin V+/propidium iodide− apoptotic events and greater than 60% Annexin V+/propidium iodide+ dead cells.
Flow cytometric analysis of subdiploid events in BLIN-2 cells cultured in the presence or absence of BM stromal cells. BLIN-2/stromal cell cultures were gently shaken to displace BLIN-2 from the stromal cells. These stromal cell-displaced BLIN-2 cells and BLIN-2 cells in medium alone were then lysed in 0.1% Triton X-100, and the nuclei were isolated and stained with propidium iodide. The percentage of subdiploid events is listed in each histogram. This experiment is representative of six similar experiments.
Flow cytometric analysis of subdiploid events in BLIN-2 cells cultured in the presence or absence of BM stromal cells. BLIN-2/stromal cell cultures were gently shaken to displace BLIN-2 from the stromal cells. These stromal cell-displaced BLIN-2 cells and BLIN-2 cells in medium alone were then lysed in 0.1% Triton X-100, and the nuclei were isolated and stained with propidium iodide. The percentage of subdiploid events is listed in each histogram. This experiment is representative of six similar experiments.
Annexin V/propidium iodide dual staining of BLIN-2 cells cultured in the presence (A) or absence (B) of BM stromal cells for 4 days. BLIN-2/stromal cell cultures were gently shaken to displace BLIN-2 from the stromal cells. These stromal cell-displaced BLIN-2 cells and BLIN-2 cells in medium alone were then stained with propidium iodide and FITC-conjugated Annexin V. The numbers represent the percentage of propidium iodide+/Annexin V+or propidium iodide−/Annexin V+ cells as a function of total cells analyzed.
Annexin V/propidium iodide dual staining of BLIN-2 cells cultured in the presence (A) or absence (B) of BM stromal cells for 4 days. BLIN-2/stromal cell cultures were gently shaken to displace BLIN-2 from the stromal cells. These stromal cell-displaced BLIN-2 cells and BLIN-2 cells in medium alone were then stained with propidium iodide and FITC-conjugated Annexin V. The numbers represent the percentage of propidium iodide+/Annexin V+or propidium iodide−/Annexin V+ cells as a function of total cells analyzed.
BLIN-2 growth does not depend on very late antigen-4 (VLA-4)/VCAM-1 interaction.
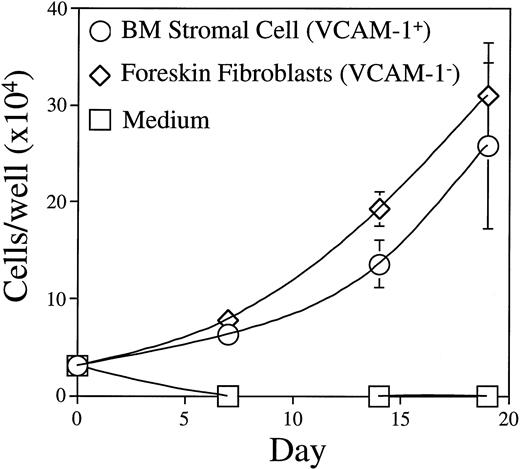
BM stromal cells used in our culture system are fibroblast-like adventitial reticular cells that express VCAM-1.20 23Figure 9 shows that human foreskin fibroblasts were as effective as BM stromal cells in supporting BLIN-2 growth, even though we could not detect VCAM-1 on the surface of foreskin fibroblasts by flow cytometry (data not shown). Furthermore, inclusion of saturating concentrations of MoAb recognizing VCAM-1, or the α4 or β1 subunits of VLA-4, had no effect on BM stromal cell-dependent growth when used individually or in combination (data not shown). Thus, VCAM-1 binding to VLA-4 and signaling pathways activated after VLA-4 cross-linking are probably not essential for BLIN-2 growth. BM stromal cells and foreskin fibroblasts were comparable in their capacity to support BLIN-2 growth. However, human umbilical vein endothelial cells, the murine S17 BM stromal cell line, and NIH-3T3 fibroblasts did not support BLIN-2 growth (data not shown).
Growth of BLIN-2 on VCAM-1+ BM stromal cells or VCAM-1− foreskin fibroblasts. BLIN-2 growth was quantified on days 7, 14, and 19. Each symbol represents the mean ± SD of triplicate wells. This experiment is representative of eight similar experiments.
Growth of BLIN-2 on VCAM-1+ BM stromal cells or VCAM-1− foreskin fibroblasts. BLIN-2 growth was quantified on days 7, 14, and 19. Each symbol represents the mean ± SD of triplicate wells. This experiment is representative of eight similar experiments.
Potential role of the retinoblastoma (Rb) pathway in BLIN-2 growth.
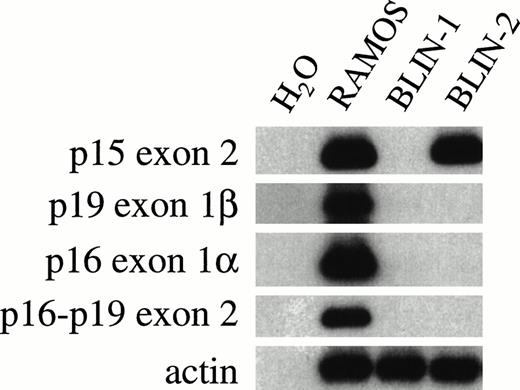
Homozygous deletions or point mutations in thep16INK4a cyclin-dependent kinase (CDK) inhibitor gene on chromosome 9p are found in many freshly isolated human tumors and tumor cell lines, including ALL.27,28 Furthermore, thep16INK4a locus in mouse29 and human30 31 includes a novel gene, designatedp19ARF (for alternative reading frame), that uses an exon (exon 1β) that is 13 to 20 kb centromeric to exon 1α used by p16INK4a. Because BLIN-2 has a dic(9;20), we evaluated the status of the p16INK4a,p19ARF, and p15INK4b genes by PCR. To reduce the possibility that analysis of BLIN-2 DNA would be confounded by contaminating BM stromal cell DNA, we FACS-purified BLIN-2 before conducting the PCR. As shown in Fig 10, BLIN-2 has deleted both alleles of p19ARF exon 1β, p16INK4aexon 1α, and p16INK4a-p19ARFexon 2. However, PCR amplification using p15INK4bexon 2-specific primers indicated that at least onep15INK4b allele was present. These results contrast with RAMOS cells that contain an intact INK4a locus and the BLIN-1 pre-B ALL cell line that has the three genes deleted.
Status of INK4 locus genes in BLIN-2. DNA was isolated from RAMOS, BLIN-1, and BLIN-2 using TRI reagent and amplified with primers specific for various INK4 locus exons, as described in Materials and Methods. BLIN-2 was separated from BM stromal cells by FACS sorting before DNA isolation. The RAMOS Burkitt lymphoma cell line served as a positive control for amplification of all INK4 locus genes.
Status of INK4 locus genes in BLIN-2. DNA was isolated from RAMOS, BLIN-1, and BLIN-2 using TRI reagent and amplified with primers specific for various INK4 locus exons, as described in Materials and Methods. BLIN-2 was separated from BM stromal cells by FACS sorting before DNA isolation. The RAMOS Burkitt lymphoma cell line served as a positive control for amplification of all INK4 locus genes.
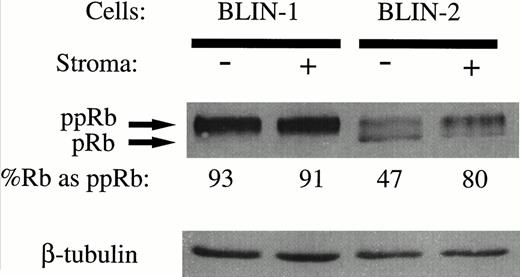
With the results in Fig 10 showing that bothp16INK4a alleles are deleted in BLIN-2, we hypothesized that growth of BLIN-2 on BM stromal cells may occur by virtue of continuous activation of the cyclin D/CDK/Rb pathway. As an initial approach in testing this hypothesis, we examined the status of Rb phosphorylation in BLIN-2 and BLIN-1 cells in the presence or absence of BM stromal cell contact. The pre-B ALL cell line BLIN-1 was used as a control, because it has both p16INK4aalleles deleted (Fig 10) but is not dependent on BM stromal cells for growth. As shown in Fig 11, two isoforms of Rb were present in BLIN-2 cells cultured in the presence or absence of BM stromal cells for 18 hours, but a difference in the relative ratio of hypo-phosphorylated (pRb) to hyper-phosphorylated (ppRb) Rb was detected. Scanning densitometry indicated that 80% of Rb was hyperphosphorylated in BLIN-2 cells cultured on BM stromal cells (Fig11, lane 4), whereas only 47% of Rb was hyperphosphorylated in BLIN-2 cells maintained in medium alone (Fig 11, lane 3). In contrast, the control BLIN-1 cell line exhibited only one predominant hyper-phosphorylated Rb isoform, independent of whether the cells were maintained on BM stromal cells or in medium alone.
Phosphorylation of Rb in BLIN-2 cells cultured in the presence or absence of BM stromal cells. BLIN-1 or BLIN-2 cells were cultured in X-VIVO 10 serum-free medium for 18 hours in the absence (−) or presence (+) of BM stromal cells. The cells were then harvested and lysed in 0.5% NP-40, and approximately 50 μg of protein per lane was electrophoresed on a 10% SDS-PAGE gel. The separated proteins were then transferred to nitrocellulose and Western blotted with rabbit antihuman Rb. The blot was restained with mouse antihuman β tubulin to control for equal protein loading. The numbers under each lane represent the percentage of Rb detected as the hyper-phosphorylated isoform (ppRb ÷ pRb + ppRb) by scanning densitometry.
Phosphorylation of Rb in BLIN-2 cells cultured in the presence or absence of BM stromal cells. BLIN-1 or BLIN-2 cells were cultured in X-VIVO 10 serum-free medium for 18 hours in the absence (−) or presence (+) of BM stromal cells. The cells were then harvested and lysed in 0.5% NP-40, and approximately 50 μg of protein per lane was electrophoresed on a 10% SDS-PAGE gel. The separated proteins were then transferred to nitrocellulose and Western blotted with rabbit antihuman Rb. The blot was restained with mouse antihuman β tubulin to control for equal protein loading. The numbers under each lane represent the percentage of Rb detected as the hyper-phosphorylated isoform (ppRb ÷ pRb + ppRb) by scanning densitometry.
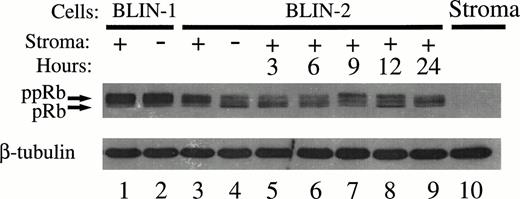
The kinetics of Rb phosphorylation in BLIN-2 was evaluated by time-course analysis. BLIN-2 cells were cultured in medium alone for 18 hours, replated onto fresh BM stromal cells, and assessed for subsequent changes in Rb phosphorylation. As expected, BLIN-2 cells cultured in medium alone for 18 hours expressed more hypo-phosphorylated Rb (Fig 12, lane 4), compared with BLIN-2 cells maintained on BM stromal cells for 18 hours (Fig 12, lane 3). The pattern of Rb phosphorylation in BLIN-1 was the same whether the cells were cultured on BM stromal cells (Fig 12, lane 1) or in medium alone (Fig 12, lane 2). These results are consistent with the results in Fig 11, except that, in the experiment shown in Fig12, three distinct phosphorylated Rb isoforms are visible. When BLIN-2 cells cultured in medium alone for 18 hours (Fig 12, lane 4) were transferred to fresh BM stromal cells, a shift toward hyper-phosphorylated Rb was detected between 6 and 9 hours. The absence of detectable Rb in BM stromal cells provided a useful control to confirm that BLIN-2 cell protein lysates were not contaminated with BM stromal cell proteins.
Time course of Rb phosphorylation in BLIN-2. Lanes 1 through 4, BLIN-1 and BLIN-2 were cultured for 18 hours and analyzed for Rb expression/phosphorylation as described in the Fig 10 legend. Lanes 5 through 9, BLIN-2 cells from lane 4 were replated on fresh BM stromal cells and reanalyzed at the times indicated for Rb expression/phosphorylation. The blot was restained with mouse antihuman β tubulin to control for equal protein loading.
Time course of Rb phosphorylation in BLIN-2. Lanes 1 through 4, BLIN-1 and BLIN-2 were cultured for 18 hours and analyzed for Rb expression/phosphorylation as described in the Fig 10 legend. Lanes 5 through 9, BLIN-2 cells from lane 4 were replated on fresh BM stromal cells and reanalyzed at the times indicated for Rb expression/phosphorylation. The blot was restained with mouse antihuman β tubulin to control for equal protein loading.
DISCUSSION
This report describes the establishment and characterization of a novel human pre-B ALL cell line with an absolute requirement on BM stromal cells or skin fibroblasts for optimal growth. A large number of ALL cell lines have been generated from patients with B-cell precursor or T-lineage ALL. The vast majority of these cell lines proliferate in FBS-supplemented tissue culture medium in the absence of any adherent feeder layer (Zhang et al32 and references therein), although one study did present a brief description of a B-cell precursor ALL cell line that required BM stromal cells for growth.33 We have previously reported that BM stromal cell-dependent human B-cell development can occur in the absence of IL-7,34 consistent with the fact that SCID patients with mutations in the γc subunit of the IL-7 receptor or Jak-3 tyrosine kinase have normal (or even elevated) numbers of B cells.35-38 Although some B-cell precursor ALL respond to IL-7, IL-7 by itself is not a potent growth stimulus for the majority of B-cell precursor ALL.11-15 Thus, BM stromal cell products in addition to IL-7 must be essential for survival and/or growth of normal and leukemic human B-cell precursors. We therefore sought to establish ALL cell lines that would preserve a dependence on BM stromal cells for long-term survival and growth in vitro to establish a model to evaluate the role of the BM microenvironment in the development of B-cell precursor ALL.
BLIN-2 has a typical pre-B cell phenotype, including the expression of cell surface μ heavy chains, ψ light chains, CD19, CD20, and CD22 (Fig 1). Data in Figs 3, 4, and 5 demonstrate that (1) viable, intact BM stromal cells are essential for BLIN-2 growth and (2) optimal growth requires direct contact. Although removal of BLIN-2 from BM stromal cells leads to inevitable apoptotic cell death, more recent passages of BLIN-2 exhibit slightly greater survival in noncontact (transwell) conditions (Fig 4A and B). Furthermore, earlier passages of BLIN-2 doubled every 4 to 5 days, whereas recent passages double approximately every 2 days (Fig 4A and B). These data suggest that selection of a subclone (or subclones) with greater proliferative capacity has occurred during continuous passage of BLIN-2 on BM stromal cells. The genetic differences that underlie this modest change in survival and proliferation status are unknown. The population doubling time of BLIN-2 on BM stromal cells is comparable to the population doubling time of normal pro-B cells stimulated with IL-7 (Fig 6). One interpretation of these data would argue that BLIN-2 and normal pro-B cells are dependent on similar BM stromal cell products for survival and growth. The difference in long-term growth characteristics (ie, permanent growth of BLIN-2 vis-à-vis limited growth of normal pro-B cells) could then be at least partially attributed to the absence of p16INK4a and/or p19ARF proteins in BLIN-2 cells, leading to dysregulation in G1 to S-phase entry and/or entry into apoptosis (see below). However, normal pro-B cells plated on BM stromal cells supplemented with IL-7 undergo partial differentiation to the pre-B and immature B stages of B-cell development.21 Thus, the ability of normal pro-B cells to differentiate and the inability of BLIN-2 to differentiate may also explain the results.
BLIN-2 cells removed from BM stromal cells gradually undergo apoptosis (Figs 7 and 8), consistent with the relatively slow growth rate of this cell line. BLIN-2 cells cultured in the absence of BM stromal cells exhibited a sharp increase in subdiploid events between days 1 and 2 that reached 60% by day 4 (Fig 7). However, analysis of the early stages of apoptosis by Annexin V binding to exposed phosphatidylserine residues indicated only a minor percentage (5.6%; Fig 8B) of Annexin V+/propidium iodide− events 4 days after the removal of BLIN-2 cells from BM stromal cells. The difference between the results in Figs 7 and 8 probably reflects the difference in the two assays. The Annexin V binding assay detects early stages of apoptosis in intact cells.39 In contrast, propidium iodide analysis of subdiploid events essentially detects nuclear fragments, which do not necessarily correlate with the number of apoptotic cells.40
A major question not resolved in the current study is the identity of the BM stromal cell-derived ligand (or ligands), and their cognate receptors on BLIN-2, essential for the survival and growth of BLIN-2. The data in Fig 4 indicate that direct contact with BM stromal cells is essential for optimal growth, but minimal survival/growth also occurs in noncontact conditions. We have tested a variety of cytokines and combinations of cytokines for their capacity to support survival/growth of BLIN-2 in the absence of BM stromal cells. Log range concentrations of IL-3, IL-6, IL-7, IL-9, IL-10, IL-11, stem cell factor, Flt3 ligand, basic fibroblast growth factor, or various combinations failed to enhance the survival of BLIN-2 cells beyond that which occurred in medium alone (N. Shah and T.W. LeBien, unpublished observations). A potential candidate in the contact equation would be VLA-4/VCAM-1, because BLIN-2 cells express the CD49d/α4 subunit of VLA-4 (Fig 1) and the BM stromal cells used in our culture system express VCAM-1.20 Studies using mouse and human early B-lineage cells have suggested a role for VLA-4/VCAM-1 in the growth and development of B-cell precursors.41,42Furthermore, chimeric mice derived by injecting embryonic stem cells containing a targeted deletion of the α4 gene into RAG-1– or RAG-2–deficient blastocysts exhibited a dramatic deficiency of α4−/− B cells in adult BM, blood, and spleen.43 However, other signaling interactions between B-lineage cells and BM stromal cells are not dependent on a VLA-4/VCAM-1 adhesion event.44 45 Independent of the precise role of VLA-4/VCAM-1 in other in vitro or in vivo experimental systems, VLA-4/VCAM-1 interaction plays little or no role in the survival and growth of BLIN-2. This conclusion is supported by two separate results. First, BLIN-2 cells show essentially identical growth rates on VCAM-1+ BM stromal cells and VCAM-1−foreskin fibroblasts (Fig 9). Second, inclusion of saturating concentrations of MoAb against the α4 or β1 subunits of VLA-4 and/or MoAb against VCAM-1 has no effect on the growth of BLIN-2 on BM stromal cells (N. Shah and T.W. LeBien, unpublished observations).
As shown in Fig 2, BLIN-2 harbors a dic(9;20), a recurring chromosomal abnormality previously identified in a small subset of children46,47 and adults48 with pre-B ALL. Whether a direct relationship exists between the presence of a dic(9;20) and the BM stromal cell requirements of this cell line is currently unknown. Additional cell lines containing a dic(9;20) would need to be established and studied to address this question. Given that the breakpoints involved in the formation of the dic(9;20) occur within heterochromatin regions, it is highly unlikely that a chimeric gene is created at the point of fusion. More likely, the significant ramification of this rearrangement is the resulting monosomy for 9p and 20q. Of particular relevance may be the loss of the entireINK4a locus mapped to 9p21.
The INK4a locus on chromosome 9p21 includes the cell cycle inhibitor genes p15INK4b,p16INK4a, andp19ARF.27-31 As shown in Fig 10, BLIN-2 has both alleles of p16INK4a andp19ARF deleted. The PCR results do not allow us to determine whether one or both alleles of p15INK4bare intact. However, all of the chromosomal DNA telomeric to 9p11 has been deleted on one copy of chromosome 9, making it likely that only one copy of p15INK4b is present in BLIN-2 cells. Chromosomal 9p21 rearrangements or deletions in ALL can result in the loss of both alleles of p16INK4a, but preserve one or both alleles of p15INK4b.49-51Although less is known about the disposition ofp19ARF in ALL, Gardie et al52 recently reported an evaluation of this gene in 87 cases of T-lineage ALL. These investigators found that p19ARF was deleted or disrupted in 75 cases of T-lineage ALL harboring recombination events in the INK4a locus; yet, in 4 of the 75 cases,p16INK4a was not deleted or altered. On the basis of these data, they proposed that p19ARF may be targeted by the genetic events that occur at the INK4 locus in T-lineage ALL. To our knowledge, a comparable analysis ofp16INK4a and p19ARF in B-cell precursor ALL has not been reported.
Two recent reports have demonstrated thatp16INK4a-deficient leukemic cell lines reconstituted with wild-type p16INK4a undergo growth arrest.53,54 BLIN-2 cell contact with BM stromal cells induces Rb phosphorylation in BLIN-2 (Figs 11 and 12), whereas BLIN-1 (also p16INK4a-deficient) cell contact with BM stromal cells does not induce Rb phosphorylation in BLIN-1 (Fig 11). These data are consistent with a model wherein continuous growth of BLIN-2 on BM stromal cells can at least be partially explained by the lack of CDK4/CDK6 inhibition by p16INK4a, resulting in continuous Rb phosphorylation and G1 to S-phase entry. BLIN-2 provides the opportunity to compare the individual functions of p16INK4a and p19ARF proteins by transduction with retroviral vectors containing wild-typep16INK4a or p19ARF, studies that are currently in progress in this laboratory. The role ofp19ARF as a tumor suppressor has been shown by targeted disruption of p19ARF exon 1β,55 and very recent evidence supports a role for p19ARF protein in regulating the p53 pathway by binding to MDM2.56 57 Thus, BLIN-2 could provide a model system for further elucidation of the potentially complementary functions of p16INK4a and p19ARF in regulating the survival and growth of a contact-dependent human leukemia.
ACKNOWLEDGMENT
The authors express their gratitude to Lisa Jarvis and Daitaro Kurosaka for technical advice and Dan Billadeau and Brian Van Ness for assistance with DNA sequencing. They also thank Diane Arthur for assistance with karyotyping and Betsy Hirsch for cytogenetic interpretation and comments on the manuscript. Karen Nelson provided excellent word processing support.
Supported in part by National Institutes of Health Grants No. R01 CA31685 and R01 CA76055.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Tucker W. LeBien, PhD, Box 806 FUMC, University of Minnesota Cancer Center, 420 Delaware St, SE, Minneapolis, MN 55455; e-mail: lebie001@tc.umn.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal