Abstract
The bcr-abl oncogene plays a critical role in causing chronic myelogenous leukemia (CML). Effective laboratory animal models of CML are needed to study the molecular mechanisms by which thebcr-abl oncogene acts in the disease progression of CML. We used a murine stem cell retroviral vector (MSCV) to transduce thebcr-abl/p210 oncogene into mouse bone marrow cells and found that expression of Bcr-Abl/p210 induced a myeloproliferative disorder that resembled the chronic phase of human CML in 100% of bone marrow transplanted mice in about 3 weeks. This CML-like disease was readily transplanted to secondary recipient mice. Multiple clones of infected cells were expanded in the primary recipients, but the leukemia was primarily monoclonal in the secondary recipient mice. Mutation analysis demonstrated that the protein tyrosine kinase activity of Bcr-Abl/p210 was essential for its leukemogenic potential in vivo. Interestingly, we found that the leukemic cells expressed excess interleukin-3 (IL-3) and granulocyte-macrophage colony-stimulating factor (GM-CSF) in the diseased mice. These studies demonstrate that expression of Bcr-Abl can induce a CML-like leukemia in mice much more efficiently and reproducibly than in previously reported mouse CML models, probably due to efficient expression in the correct target cell(s). Our first use of this model for analysis of the molecular mechanisms involved in CML raises the possibility that excess expression of hematopoietic growth factors such as IL-3 and GM-CSF may contribute to the clinical phenotype of CML.
CHRONIC MYELOGENOUS leukemia (CML) is a clonal myeloproliferative disorder resulting from the neoplastic transformation of a hematopoietic stem cell.1-3 The disease usually has a biphasic or triphasic course, composed of chronic phase, accelerated phase, and blastic phase. The initial chronic phase is characterized by accumulation of large numbers of myeloid-lineage cells predominated by granulocytes in peripheral blood, bone marrow, and spleen. Progression of the disease after 3 to 5 years duration to terminal blast crisis stage is characterized by accelerated accumulation of immature myeloid or lymphoid cells. Greater than 90% of CML cases are associated with the presence of the Philadelphia chromosome (Ph+).4 The Philadelphia chromosome is a result of a reciprocal translocation between chromosomes 9 and 22 that fuses Bcr-encoded sequences to a truncated c-abl gene. The oncogene produces a fusion protein, Bcr-Abl, in which the protein tyrosine kinase activity of Abl is increased. Depending on the precise breakpoint within the bcr and c-abl genes, variousbcr-abl fusion genes can be generated and are associated with different types of leukemia.4 Bcr-Abl is apparently important in both initiation and maintenance of the neoplastic transformation. However, the progression of the disease to terminal blast crisis stage is thought to require additional mutations.
Bcr-Abl contains many functional domains, interacts with and/or phosphorylates a large number of proteins, and can potentially activate multiple signal transduction pathways.5-7 However, the roles and relative importance of the domains of Bcr-Abl, of its interacting proteins, and of Bcr-Abl-activated signaling pathways in developing CML remain to be elucidated. Furthermore, although Bcr-Abl has a pleiotropic effect on deregulating multiple cellular processes, CML cells remain dependent on growth factors.8-11Therefore, unbalanced growth factor production may also contribute to the massive expansion of myeloid cells seen in CML. Indeed, it has been shown that Bcr-Abl can induce production of granulocyte-macrophage colony-stimulating factor (GM-CSF) and/or interleukin-3 (IL-3) in myeloid cell lines12-14 and that excess expression of GM-CSF is often detected in CML patients.15-18 In mice, enforced expression of IL-3, GM-CSF, granulocyte colony-stimulating factor (G-CSF), or IL-6 can induce myeloproliferative disorders.19-23 Bcr-Abl has been shown to transform a variety of cell types in vitro.13 24-30 Although these in vitro transformation assays show the oncogenic potential of Bcr-Abl, they do not address the complex pathogenesis of CML. Therefore, an efficient, reproducible, and experimentally convenient in vivo CML model is needed to further elucidate the pathogenesis of CML, as well as the molecular mechanisms by which the bcr-abl oncogene acts in the pathogenesis of CML.
Previously, Bcr-Abl/p210, the major fusion protein form associated with CML, has been expressed in murine bone marrow cells by retroviral transduction. This resulted in a myeloproliferative disorder resembling CML.31-34 However, the efficiency of disease induction by this technique was low in terms of frequency, latency, and transplantability. This hindered the usefulness of retroviral transduction as a mean to study the biology of Bcr-Abl in CML. Other in vivo models used transgenic strains of mice expressing Bcr-Abl.35-38 Although these transgenic mice provide good models for Ph+ leukemias, most of them do not model CML. Recently, transgenic mice expressing Bcr-Abl/p210 driven by the promoter of the tec gene were developed.38 The founder mice developed acute lymphoblastic leukemia and the transgenic progeny developed a myeloproliferative disorder resembling CML. However, the latency of the disease was very long (∼1 year).
We report here a new in vivo model for CML. Bcr-Abl/p210 was expressed in bone marrow cells of mice by retrovirus transduction using a murine stem cell retroviral vector (MSCV), which can drive the expression of transduced genes in embryonic stem cells.39 This system efficiently, reproducibly, and with an experimentally convenient latency induced a myeloid leukemia that resembles the chronic phase of human CML. In analyzing this novel mouse model for CML, we found that the leukemic cells expressed excess hematopoietic growth factors IL-3 and GM-CSF. These studies demonstrate that expression of Bcr-Abl/p210 by this system produces an effective and experimentally useful in vivo model of CML in mice and raise the possibility that excess expression of IL-3 and GM-CSF may contribute to the clinical phenotype of CML.
MATERIALS AND METHODS
DNA constructs.
Retroviral transducing vector containing the green fluorescent protein (GFP) gene, MSCV-IRES-gfp, was constructed as follows: a modified humanized gfp gene40 was released from MSCVpuro-gfp (a generous gift from J. Jacob and B. Chen in Baltimore’s laboratory at MIT) with EcoRI and Nco I; this fragment was cloned into pCITE (Novagen, Madison, WI) between the Nco I andSal I sites with an EcoRI/Sal I adapter (the adapter was designed to include a Not I site and to destroy theEcoRI site after ligation); the IRES-gfp fragment was then released from the pCITE-gfp with EcoRI and Sal I and cloned into the MSCV vector. MSCV-bcr-abl/p210-IRES-gfp (Fig 1) was constructed by insertion of theEcoRI fragment containing coding sequences of the wild-typebcr-abl/p21031 into MSCV-IRES-gfp. MSCV-bcr-ablK1176R-IRES-gfp was constructed by insertion of theEcoRI fragment containing the coding sequences of the kinase-deficient K1176R mutant bcr-abl/p210 into MSCV-IRES-gfp.
The retrovirus construct used to transduce thebcr-abl/p210 and gfp genes. The construct MSCV-bcr-abl/p210-IRES-gfp was made as described in Material and Methods. LTR, long terminal repeat. Enzyme abbreviations: RI,EcoRI; N, Nco I; S, Sal I.
The retrovirus construct used to transduce thebcr-abl/p210 and gfp genes. The construct MSCV-bcr-abl/p210-IRES-gfp was made as described in Material and Methods. LTR, long terminal repeat. Enzyme abbreviations: RI,EcoRI; N, Nco I; S, Sal I.
Cell culture and retrovirus preparation.
NIH3T3 mouse fibroblasts were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% calf serum, 100 U/mL penicillin (GIBCO BRL, Grand Island, NY), and 100 μg/mL streptomycin (GIBCO BRL). Bosc23 cells41 were grown in DMEM containing 10% fetal calf serum (FCS), 100 U/mL penicillin, and 100 μg/mL streptomycin. Helper-free retroviruses were generated by transiently transfecting retroviral vectors into BOSC-23 cells as described.41
Bone marrow infection and transplantation.
Bone marrow cell infection and transplantation were performed as previously described,42 with modifications. Bone marrow cells from 5-fluorouracil (5-FU)–treated male BALB/c (Taconic farm) donor mice (6 to 9 weeks old) were infected at a concentration of 1 × 106 cells/mL for 24 hours in a cocktail consisting of DMEM, 15% FCS, 5% WEHI-conditioned medium, 30% viral supernatant, 3 μg/mL polybrene, 2 mmol/L L-glutamine (GIBCO BRL), 100 μg/mL streptomycin, 100 U/mL penicillin, 0.25 μg/mL amphotericin B (GIBCO BRL), 7 ng/mL IL-3 (Genzyme, Cambridge, MA), 12 ng/mL IL-6 (R&D System, Minneapolis, MN), and 56 ng/mL stem cell factor (SCF; R&D System). The infection was repeated once with freshly made retrovirus-containing cocktail as described above. The infected bone marrow cells were then washed once with phosphate-buffered saline (PBS; GIBCO BRL) and were injected into lethally irradiated (2 doses of 450 rads each dose administered 4 hours apart) syngeneic mice (female BALB/c; 6 to 8 weeks old) through the tail vein at 4 × 105 cells per mouse.
Pathological examination.
Tissues were fixed with 4% paraformaldehyde in PBS and processed for paraffin-embedded sectioning at 3 to 5 μm in thickness, followed by staining with hematoxylin and eosin (Fisher, Pittsburgh, PA). Tail-vein peripheral blood smears were stained with LeukoStat from Fisher according to the manufacturer’s instructions.
Flow cytometry.
Peripheral blood obtained from the orbital sinus and isolated bone marrow cells were treated with red blood cell (RBC) lysis solution ACK (0.15 mol/L NH4Cl, 1.0 mmol/L KHCO3, 0.1 mmol/L Na2EDTA, pH 7.3). Spleen, liver, and lung were dissected out from the diseased mice and rinsed with ice-cold PBS to remove excess blood. Dissociated cells from these tissues were also treated with ACK to lyse RBC. For flow cytometry analysis, all cells were blocked with antimouse CD16/CD32 (FcγIII/II receptor; Pharmingen, San Diego, CA); stained with phycoerythrin (PE)-conjugated antimouse CD45R/B220, CD90.2 (Thy-1.2), CD11b (Mac-1 α chain), Ly-6G (Gr-1), or TER-119; and then analyzed on FACScan (Becton Dickinson, Franklin Lakes, NJ) or sorted on FACSorter (Becton Dickinson).
Southern blot and genomic DNA polymerase chain reaction (PCR).
Peripheral blood obtained from the orbital sinus and dispersed cells from spleen or liver were treated with ACK. High molecular weight DNA from these cells was obtained by using the QIAamp Blood Kit (Qiagen, Santa Clara, CA). For Southern blots, 15 μg of this DNA was digested with EcoRI, separated on a 1% agarose gel, transferred to Hybond-N+ membrane (Amersham, Arlington Heights, IL), and hybridized with a probe of a 1.4-kbEcoRI-Not I fragment containing IRES-gfp sequences derived from the retroviral vector or a 1.2-kbSgrAI-Bgl II fragment from the 3′ end of the human c-abl cDNA. The washed membrane was exposed to x-ray film. For genomic DNA PCR, 0.2 μg of the DNA was used as a template in a 25 μL reaction, with primers 5′CTTGCAATAGGAACAAAACTC3′ and 5′CAGCCCATCAGTTCGCTGCAG3′ for the intron-3 of the mouse c-abl gene and primers 5′TTCCCCCCTTTTTCTGGAGAC3′ and 5′GGGGACGTGGTTTTCCTTTG3′ for the gfp gene. The PCR reaction was performed for 30 cycles at 94°C for 1 minute, 57°C for 2 minutes, and 72°C for 3 minutes, followed by 72°C for 5 minutes.
RNA isolation and reverse transcriptase-PCR (RT-PCR).
Total RNA was extracted from bone marrow or other tissues as indicated in the results section by using the ULTRASPEC RNA isolation system (Biotex, Houston, TX) according to the manufacturer’s instructions. Briefly, about 100 mg of each tissue was homogenized, or 1 × 107 cells were lysed directly in the reagent, then extracted with chloroform and precipitated with an equal volume of 2-propanol. Total RNA was dissolved in 2 mmol/L MgCl2, treated with RNase-free DNase (GIBCO BRL) for 10 minutes at room temperature, and then heated at 70°C for 10 minutes to inactivate the DNase. Two micrograms of RNA was used as template for RT reaction in 20 μL with Moloney murine leukemia virus (MMLV) reverse transcriptase (GIBCO BRL) using random hexamer oligonucleotides as primer. One to 2 μL of the RT reaction was used for PCR reaction with Taq DNA polymerase (Boehringer Mannheim, Indianapolis, IN). Primers 5′TCCAAGCTTCAATCAGTGGC3′ and 5′GTTCCACGGTTAGGAGAGAC3′ were used for examining IL-3 gene expression, 5′GCAGAATTTACTTTTCCTGGG3′ and 5′CATTCAAAGGGGATATCAGTC3′ for GM-CSF, 5′CTCATGCTTCTTAGGGCTAG3′ and 5′TAAGCCTCCGACTTGTGAAG3′ for IL-6, 5′GGGATGGATGTTTTGCCTAG3′ and 5′AAGGCTCCAAAAGCAAAGCC3′ for SCF, 5′CAAGTGAGGAAGATCCAGGC3′ and 5′CGGAAGTGGAGAGAATGATC3′ for G-CSF, and 5′CCATCACCATCATCCAGGAG3′ and 5′CCTGCTTCACCACCTTCTTG3′ for glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The PCR reaction was performed for 40 cycles at 94°C for 1 minute, 57°C (48°C in case of PCR IL-6 cDNA) for 2 minutes, and 72°C for 3 minutes, followed by 72°C for 5 minutes.
Immunoblotting.
NIH3T3 cells were collected and lysed in lysis buffer (50 mmol/L N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid [HEPES], pH 7.4, 150 mmol/L NaCl, 10% glycerol, 1% Triton X-100, 1 mmol/L EGTA, 1.5 mmol/L MgCl2, 10 mmol/L NaF, 1 mmol/L sodium orthovanadate, 1 mmol/L freshly made phenylmethylsulfonyl fluoride, 10 μg/mL aprotinin, and 10 μg/mL leupeptin). Lysates were adjusted to contain equal amounts of total protein using the Coomassie Protein Assay Reagent (Pierce, Rockford, IL) and were boiled for 5 minutes in an equal volume of 2× sodium dodecyl sulfate (SDS) sample buffer as described.43 Proteins were separated on 8% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and electrophoretically transferred to nitrocellulose filter (Schleicher & Schuell, Keene, NH). The filter was probed with anti-Abl monoclonal antibody Ab-3 (Oncogene Research Products, Cambridge, MA). Bound antibodies were visualized using horseradish peroxidase-conjugated antimouse IgG and ECL reagents as described by the manufacturer (Amersham).
Enzyme-linked immunospecific assay (ELISA) for IL-3 and GM-CSF.
Peripheral blood from the orbital sinus was collected into an eppendorf tube and incubated at room temperature for 4 hours and then 4°C overnight. The samples were spun at 16,000 rpm in a microcentrifuge at 4°C. The supernatant was transferred into a new tube and frozen at −70°C until use. The serum levels of IL-3 and GM-CSF were assayed using mouse IL-3 and GM-CSF ELISA kit from Endogen (Woburn, MA).
RESULTS
Bcr-Abl/p210 effectively induces a myeloid leukemia resembling human CML in mice.
CML is believed to be a neoplasm of hematopoietic stem cells. To target Bcr-Abl/p210 into hematopoietic stem cells, we used the retroviral vector, MSCV, to transduce the bcr-abl/p210 gene. MSCV was chosen because it is derived from the murine stem cell virus and can drive gene expression in embryonic carcinoma and embryonic stem cells.39 To facilitate identification of Bcr-Abl/p210–expressing cells and determination of virus titers, we cloned into MSCV the bcr-abl/p210 oncogene linked together with the gene encoding GFP by an internal ribosome entry site (IRES; Fig 1), which allows both Bcr-Abl and GFP to be translated from the same transcript. The 5-FU–treated primary bone marrow cells were infected with the retrovirus by incubating bone marrow cells in retrovirus-containing media and then transplanted to recipient mice (see Material and Methods). We found that expression of Bcr-Abl/p210 induced a myeloid leukemia that resembles the chronic phase of human CML in 100% of the bone marrow transplanted mice in about 3 weeks. The general features of the leukemia were similar to the Bcr-Abl–induced CML-like disease in mice described previously.31 Following are our observations of these mice.
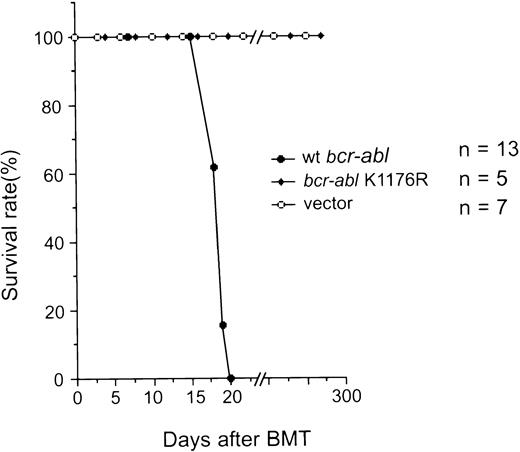
(1) Mice transplanted with bone marrow cells that were infected with the MSCV-bcr-abl/p210-IRES-gfp retrovirus (Bcr-Abl–BMT) became progressively moribund and died after about 3 weeks. We have examined a total of 47 Bcr-Abl–BMT mice in five experiments. A typical survival curve after bone marrow transplantation is shown in Fig 2. In contrast, none of the control mice transplanted with bone marrow cells that was infected with the MSCV-IRES-gfp retrovirus (vector-BMT) showed signs of the disease in the 10-month observation period. To examine the effect of different amounts of MSCV-bcr-abl/p210-IRES-gfp retrovirus-infected bone marrow cells on disease development, we diluted the infected bone marrow cells with various amounts of uninfected marrow cells. We found that even mice transplanted with the greatest dilution tested (10-fold dilution) still developed the disease with identical phenotypes, but had a longer latency (up to 2 weeks longer; data not shown). This indicates that the retroviruses used in our typical experiments were in excess for inducing the disease and that the disease latency can be affected by the numbers of transplanted cells.
Survival of recipient mice after transplantation of bone marrow cells that were infected with retroviruses containing various genes as indicated. The number of mice (n) used in the experiment for each retrovirus construct is indicated.
Survival of recipient mice after transplantation of bone marrow cells that were infected with retroviruses containing various genes as indicated. The number of mice (n) used in the experiment for each retrovirus construct is indicated.
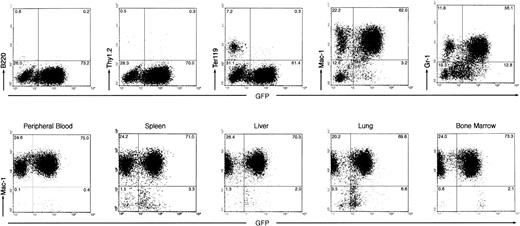
(2) The peripheral white blood cell (WBC) count of the diseased mice was drastically elevated, ranging from 200,000 to 600,000 cells/μL (as compared with the vector-BMT of ∼20,000 cells/μL). Examination of the peripheral blood smears showed that the majority of WBCs were granulocytes but contained some myeloblasts and basophils (Fig 3A). Differential counts of four diseased mice showed that cells in the peripheral blood were composed of 89% ± 2.3% granulocytes, 6.6% ± 3.5% lymphocytes, and 4.4% ± 3.6% myeloblasts. To further identify and quantify various types of WBCs, we analyzed the peripheral WBCs by flow cytometry for expression of lineage-specific antigens and GFP. As shown in Fig 4, the majority of the peripheral WBCs from the diseased mice were Mac-1 positive and, to a lesser extent, Gr-1 positive. A small fraction of Ter-119–positive cells (erythroid) were detected in the GFP-negative cell population. Few B220-positive or Thy-1–positive cells were detected. Interestingly, both GFP-positive and GFP-negative myeloid cells were massively expanded. These GFP-negative cells were shown not to harbor the provirus, as described in a later section.
Pathological analysis of the Bcr-Abl–BMT mice. (a) Peripheral blood smear from a primary Bcr-Abl–BMT mouse with the myeloproliferative disorder (original magnification × 600). The peripheral blood smear was stained with LeukoStat from Fisher. (B through D) Histological sections (original magnification × 400) of the spleen (b), liver (c), and lung (d) from a primary Bcr-Abl–BMT mice with the myeloproliferative disorder. Histological sections were stained with hematoxylin and eosin. Arrows in (c) point out mitotic hematopoietic cells.
Pathological analysis of the Bcr-Abl–BMT mice. (a) Peripheral blood smear from a primary Bcr-Abl–BMT mouse with the myeloproliferative disorder (original magnification × 600). The peripheral blood smear was stained with LeukoStat from Fisher. (B through D) Histological sections (original magnification × 400) of the spleen (b), liver (c), and lung (d) from a primary Bcr-Abl–BMT mice with the myeloproliferative disorder. Histological sections were stained with hematoxylin and eosin. Arrows in (c) point out mitotic hematopoietic cells.
Immunophenotyping of leukemic cells from Bcr-Abl–BMT mice by flow cytometry. Two-parameter dot-plots show expression of lineage-specific antigens versus GFP as indicated. (Top panel) The expression of B220, Thy1.2, Ter119, Mac-1, and Gr-1 versus GFP, as indicated, in the peripheral WBCs from a primary Bcr-Abl–BMT mouse with the myeloproliferative disorder. (Bottom panel) The expression of Mac-1 versus GFP in the peripheral WBCs, spleen, liver, lung, and bone marrow from a secondary Bcr-Abl–BMT mouse with the myeloproliferative disorder.
Immunophenotyping of leukemic cells from Bcr-Abl–BMT mice by flow cytometry. Two-parameter dot-plots show expression of lineage-specific antigens versus GFP as indicated. (Top panel) The expression of B220, Thy1.2, Ter119, Mac-1, and Gr-1 versus GFP, as indicated, in the peripheral WBCs from a primary Bcr-Abl–BMT mouse with the myeloproliferative disorder. (Bottom panel) The expression of Mac-1 versus GFP in the peripheral WBCs, spleen, liver, lung, and bone marrow from a secondary Bcr-Abl–BMT mouse with the myeloproliferative disorder.
(3) Examination of the abdomens of all the Bcr-Abl–BMT mice (dead or moribund mice) showed massive splenomegaly, weighing 0.6 to 1.1 g (as compared with the normal spleen of ∼0.1 g), and enlarged liver, weighing 1.9 to 2.7 g (as compared with the normal liver of ∼1 g). Lymphadenopathy was not observed. Pulmonary hemorrhages were seen in all the diseased Bcr-Abl–BMT mice when the thorax of these mice were examined. Thymic enlargement was not evident. Pathological examination of the Bcr-Abl–BMT mice showed massive infiltration of myeloid cells in the spleen, liver, and lung (Fig 3). Significant extramedullary hematopoiesis was observed in these tissues. The extensive accumulation of granulocytes in the spleen resulted in complete destruction of the normal splenic architecture (Fig 3B). Immature erythrocytes and megakaryocytes were also seen in the spleen. Infiltration of myeloid cells in liver was not as extensive as in the spleen, but paravasicular infiltration of granulocytes and foci of extramedullary erythropoiesis were observed throughout hepatic lobules (Fig 3C) and portal areas of the liver. In lung, excessive bleeding and infiltration of granulocytes in interstitial spaces was observed (Fig3D).
(4) To examine whether the myeloproliferative disease is transplantable, we transferred bone marrow cells (at 8 × 105 cells per recipient mouse) from two primary Bcr-Abl–BMT mice (BMT 5.2 and 5.3) with the CML-like myeloproliferative disorder into a set of lethally irradiated secondary recipient mice. Within 3 weeks, all nine mice transferred with the bone marrow cells from one of the primary Bcr-Abl–BMT mice, BMT5.3, developed a myeloproliferative disorder similar to the disease seen in the parental mouse. Flow cytometric analysis of dissociated cells of the spleen, liver, lung, and bone marrow from a secondary Bcr-Abl–BMT mouse demonstrated a massive expansion of myeloid cells in these tissues (Fig 4, bottom panel). The bone marrow cells from the other diseased primary Bcr-Abl–BMT mouse, BMT5.2, also generated transplantable myeloproliferative disease in three of nine secondary recipient mice within 5 weeks (the other 6 mice died of anemia with low WBC counts in peripheral blood, possibly due to the failure of transferring radioprotective hematopoietic stem cells; data not shown).
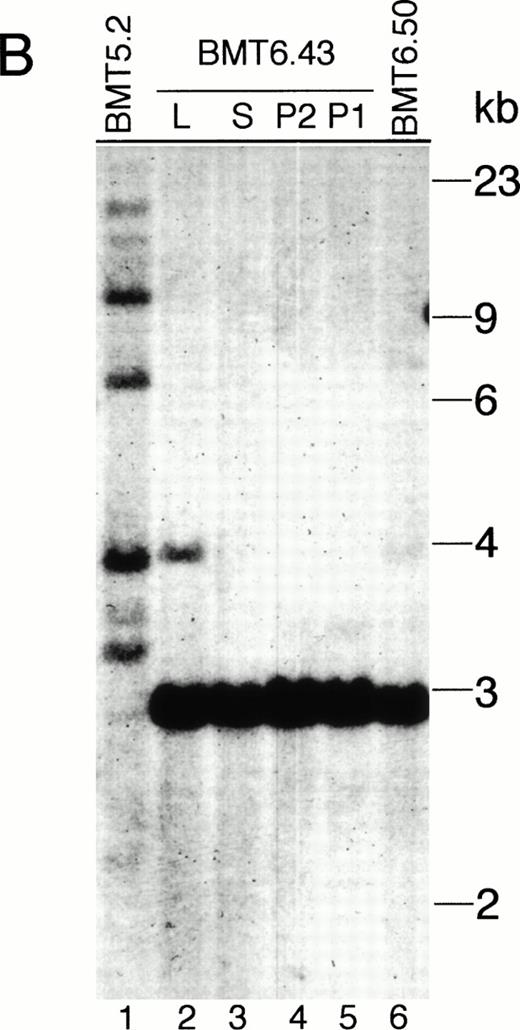
(5) The affected tissues of the Bcr-Abl–BMT mice containbcr-abl/gfp proviral DNA as analyzed by Southern blot using32P-labeled IRES-gfp sequences or a human c-ablcDNA fragment as a probe (Fig 5). No hybridization of both probes to normal murine genomic DNA was detected (data not shown). The endogenous murine c-abl was not detected, probably because the nucleotide sequence similarity between the murine and human c-abl gene was not sufficient for cross-hybridization under the high stringency conditions used in the experiment. Interestingly, all the primary Bcr-Abl–BMT mice analyzed showed multiple proviral integrates with different quantities (Fig 5A, B, and D), suggesting that multiple infected cells were expanded to various degrees. However, there were only one or two proviral integrates detected in the secondary recipient mice (Fig 5A and B). It is notable that the proviral DNA integration in the leukemic cell clone(s) found in the secondary recipient mice was not detected in the parental mouse. To further demonstrate that the expanded cells contained thebcr-abl/p210 gene and to confirm that the multiple bands hybridized with the IRES-gfp probe were not generated due to trivial reasons, such as partial digestion or DNA degradation, we stripped the IRES-gfp probe from the filters and reprobed them with32P-labeled abl cDNA. We found that a single 7.2-kb band (the EcoRI-digested bcr-abl/p210 DNA fragment) was detected for the affected tissues from all the primary and secondary Bcr-Abl–BMT mice (Fig 5C).
Analysis of MSCV-bcr-abl/p210-IRES-gfp proviral integration in Bcr-Abl–BMT mice. The genomic DNA isolated from peripheral WBCs or tissues of the Bcr-Abl–BMT mice was analyzed by Southern blot with 32P-labeled IRES-gfp sequences (A, B, and D) or a 1.2-kb SgrAI-Bgl II fragment from the 3′ end of the human c-abl cDNA (C) as a probe. (A) Peripheral WBCs of 5 primary Bcr-Abl–BMT (lanes 1 through 5) and 7 secondary recipient mice (lanes 6 through 12) transplanted with bone marrow cells of BMT5.3 (lane 5). (B) Peripheral WBCs of the primary Bcr-Abl–BMT mouse, BMT 5.2 (lane 1), and its secondary recipients, BMT6.43 (lanes 4 and 5) and BMT6.50 (lane 6), and the liver (lane 2) and spleen (lane 3) of BMT6.43. (C) The filter from (B) was stripped and reprobed with 32P-labeled abl cDNA. (D) Peripheral blood, spleen, and liver of the primary Bcr-Abl–BMT mice 4.27 and 4.28. P, peripheral WBCs; P1 and P2 specify the peripheral WBCs taken from BMT6.43 in two different days; L, liver; S, spleen.
Analysis of MSCV-bcr-abl/p210-IRES-gfp proviral integration in Bcr-Abl–BMT mice. The genomic DNA isolated from peripheral WBCs or tissues of the Bcr-Abl–BMT mice was analyzed by Southern blot with 32P-labeled IRES-gfp sequences (A, B, and D) or a 1.2-kb SgrAI-Bgl II fragment from the 3′ end of the human c-abl cDNA (C) as a probe. (A) Peripheral WBCs of 5 primary Bcr-Abl–BMT (lanes 1 through 5) and 7 secondary recipient mice (lanes 6 through 12) transplanted with bone marrow cells of BMT5.3 (lane 5). (B) Peripheral WBCs of the primary Bcr-Abl–BMT mouse, BMT 5.2 (lane 1), and its secondary recipients, BMT6.43 (lanes 4 and 5) and BMT6.50 (lane 6), and the liver (lane 2) and spleen (lane 3) of BMT6.43. (C) The filter from (B) was stripped and reprobed with 32P-labeled abl cDNA. (D) Peripheral blood, spleen, and liver of the primary Bcr-Abl–BMT mice 4.27 and 4.28. P, peripheral WBCs; P1 and P2 specify the peripheral WBCs taken from BMT6.43 in two different days; L, liver; S, spleen.
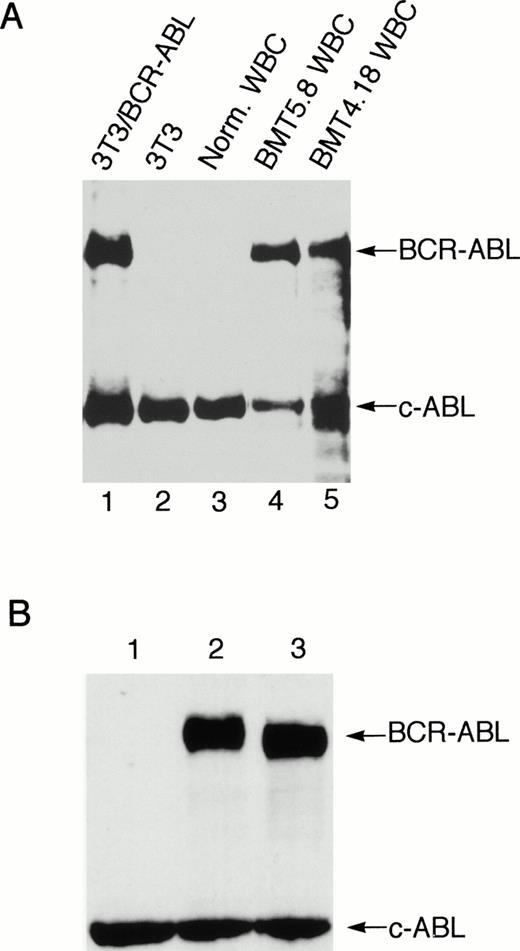
The provirus integration pattern of the genomic DNA extracted from the spleen, liver, and peripheral WBCs of the primary Bcr-Abl–BMT mice showed that the expanded myeloid cells in these tissues originated from the same infected clones (Fig 5D). Similarly, the most leukemic cells in the peripheral WBCs, spleen, and liver of the secondary recipient mouse BMT6.43 were also shown to originate from the same clone (Fig5B). However, a minor band appeared with the liver of BMT6.43 but not with its spleen and peripheral blood (Fig 5B). This second band was probably not generated by partial digestion, because there was only one band present when the abl probe was used (Fig 5C). This liver-specific band may be derived from expansion of a second leukemic progenitor cell. Expression of Bcr-Abl/p210 proteins in peripheral WBCs of the Bcr-Abl–BMT mice was detected by Western blot (Fig 6A) as well as indirect immunofluorescence (data not shown) using anti-Abl antibodies.
Expression of Bcr-Abl/p210 protein in the peripheral WBC from a Bcr-Abl–BMT mouse with the myeloproliferative disorder and expression of the wild-type and kinase-negative mutant Bcr-Abl/p210 proteins in NIH 3T3 cells. (A) Peripheral WBC from two Bcr-Abl–BMT mice with the myeloproliferative disorder (BMT4.18 and BMT5.8) were lysed directly in SDS sample buffer. The total cell lysates were subjected to Western blot analysis with anti-Abl monoclonal antibody Ab-3. The position of Bcr-abl/p210 and endogenous c-Abl is indicated. Total cell lysates from uninfected NIH3T3 and NIH3T3 infected with retrovirus containing the bcr-abl/p210 gene were used as negative and positive controls, respectively. (B) NIH 3T3 cells infected with equal amounts (1 mL) of MSCV-IRES-gfp (lane 1), MSCV-bcr-abl/p210-IRES-gfp (lane 2), and MSCV-K1176R-IRES-gfp (lane 3) retroviral supernatants of equal titer were subjected to Western blot analysis with anti-Abl monoclonal antibody Ab-3.
Expression of Bcr-Abl/p210 protein in the peripheral WBC from a Bcr-Abl–BMT mouse with the myeloproliferative disorder and expression of the wild-type and kinase-negative mutant Bcr-Abl/p210 proteins in NIH 3T3 cells. (A) Peripheral WBC from two Bcr-Abl–BMT mice with the myeloproliferative disorder (BMT4.18 and BMT5.8) were lysed directly in SDS sample buffer. The total cell lysates were subjected to Western blot analysis with anti-Abl monoclonal antibody Ab-3. The position of Bcr-abl/p210 and endogenous c-Abl is indicated. Total cell lysates from uninfected NIH3T3 and NIH3T3 infected with retrovirus containing the bcr-abl/p210 gene were used as negative and positive controls, respectively. (B) NIH 3T3 cells infected with equal amounts (1 mL) of MSCV-IRES-gfp (lane 1), MSCV-bcr-abl/p210-IRES-gfp (lane 2), and MSCV-K1176R-IRES-gfp (lane 3) retroviral supernatants of equal titer were subjected to Western blot analysis with anti-Abl monoclonal antibody Ab-3.
The protein tyrosine kinase (PTK) activity of Bcr-Abl is required for leukemogenesis.
It has been shown that the Abl PTK activity in Bcr-Abl is essential for transformation of cells in culture,44 suggesting that phosphorylation of proteins by Bcr-Abl is essential for activating oncogenic pathways. However, certain in vivo functions of PTKs, such as c-Src and c-Abl, have been shown to be kinase-independent.45,46 Furthermore, it was shown that Bcr-Abl was able to bind and activate the Src family protein tyrosine kinase Hck in a kinase-independent manner and that Hck could phosphorylate the kinase-negative mutant of Bcr-Abl and induce binding of Grb2 to Tyr177 of Bcr-Abl.47 To assess the role of the PTK activity of Bcr-Abl for inducing leukemia in vivo, we tested if Bcr-Abl/p210 with a point mutation that inactivates the PTK activity of Bcr-Abl (changing the lysine residue at position 1176 to arginine, referred to K1176R) can induce CML using the mouse model described above. The retroviral titer was quantified by measuring the expression of GFP after infection of NIH 3T3 cells by flow cytometry. The protein expression level of the K1176R mutant of Bcr-Abl/p210 was the same as the wild-type Bcr-Abl/p210 in NIH 3T3 cells infected with the same amounts of the viruses (Fig 6B). The wild-type Bcr-Abl/p210 migrated slower than the K1176R mutant, possibly due to autophosphorylation of the wild-type Bcr-Abl/p210. When mice were transplanted with bone marrow cells that were infected with the same amounts of either wild-type or K1176R mutantbcr-abl–containing viruses, the wild-type Bcr-Abl/p210 induced a lethal CML-like disease in approximately 3 weeks, whereas the K1176R mutant did not induce signs of disease over a 10-month observation period (Fig 2). Expression of the gfp transcripts was detected in the peripheral WBCs of the latter mice 10 months after transplantation by RT-PCR (data not shown), indicating that the hematopoietic stem cell(s) were successfully infected with the MSCV-bcr-ablK1176R-IRES-gfp retrovirus. These results demonstrated that the PTK activity is essential for Bcr-Abl/p210 to induce leukemia in vivo.
Both infected and noninfected myeloid cells were expanded in mice with Bcr-Abl/p210–induced myeloproliferative disorder.
As described above, both GFP-positive and GFP-negative myeloid cells in the Bcr-Abl–BMT mice with the CML-like myeloproliferative disorder were massively expanded. The percentage of expanded GFP-negative cells varied between 10% and 80% among the primary Bcr-Abl–BMT mice. In the secondary Bcr-Abl–BMT mice, there were generally more GFP-positive cells, ranging from 69% to 90% (data not shown). To distinguish whether the GFP-negative cells were expanded from noninfected bone marrow cells or were derived from some bcr-abl/gfpretrovirus-infected bone marrow cells that failed to express detectable GFP proteins, we sorted GFP-positive and GFP-negative cells by flow cytometry and tested for the presence of GFP-encoding sequences in these cells by PCR on their genomic DNA. As shown in Fig 7, while the total population of peripheral WBCs and the GFP-positive cell population from the primary Bcr-Abl–BMT mouse with the disease contains the gfp sequences, the GFP-negative cells from the same mouse did not harbor the gfp sequences in their genome. The same result was observed in three other primary Bcr-Abl–BMT mice examined (data not shown). These results indicate that both infected and noninfected myeloid cells can be expanded in mice with Bcr-Abl/p210–induced myeloproliferative disorder, but leukemic cells tend to outgrow bystander cells, as indicated by fewer GFP-negative cells in the secondary Bcr-Abl–BMT mice with the monoclonal myeloid leukemia.
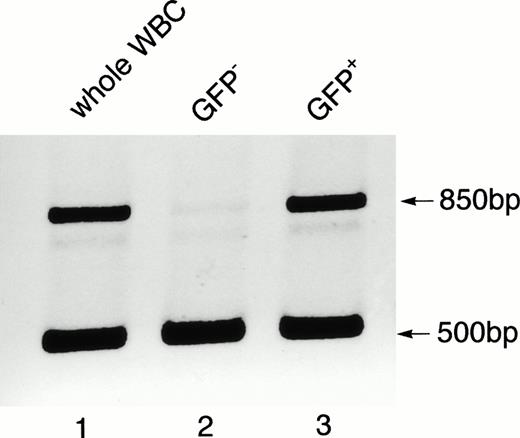
Detection of gfp in the genomic DNA of the peripheral WBCs from a Bcr-Abl–BMT mouse with the myeloproliferative disorder. Genomic DNA isolated from the unsorted WBCs (lane 1) and sorted GFP-negative (lane 2) and GFP-positive (lane 3) cells was subjected to PCR analysis using a mixture of primers that amplify the gfpgene (850 bp) and intron-3 of the mouse c-abl gene (500 bp). The amplified c-abl product was used as an internal positive control for the genomic DNA PCR.
Detection of gfp in the genomic DNA of the peripheral WBCs from a Bcr-Abl–BMT mouse with the myeloproliferative disorder. Genomic DNA isolated from the unsorted WBCs (lane 1) and sorted GFP-negative (lane 2) and GFP-positive (lane 3) cells was subjected to PCR analysis using a mixture of primers that amplify the gfpgene (850 bp) and intron-3 of the mouse c-abl gene (500 bp). The amplified c-abl product was used as an internal positive control for the genomic DNA PCR.
Bcr-Abl induces production of excess IL-3 and GM-CSF in mice.
The fact that the bcr-abl/p210-induced leukemia displayed a bystander effect suggested that excess growth factor(s) may be produced in Bcr-Abl–BMT mice. To test this possibility, we first examined the gene expression of growth factors known to promote survival, proliferation, and differentiation of myeloid cells, such as IL-3, GM-CSF, G-CSF, IL-6, and SCF, in the bone marrow of the Bcr-Abl–BMT mice and the vector-BMT mice by RT-PCR. Although no IL-3 gene expression was detected in the bone marrow cells of the vector-BMT mice, significant amounts of IL-3 transcripts were detected in the bone marrow cells of the Bcr-Abl–BMT mice (Fig8). Both primary (Fig 8) and secondary (data not shown) Bcr-Abl–BMT mice had IL-3 transcripts detected in their bone marrow. Several additional vector-BMT mice were examined, and none of them had IL-3 transcripts detected in their bone marrow (data not shown). For GM-CSF, low levels of GM-CSF transcripts were detected in the vector-BMT mice. However, GM-CSF gene expression was significantly increased in Bcr-Abl–BMT mice (Fig 8). In contrast, the same amounts of SCF (Fig 8) and G-CSF (data not shown) transcripts were detected in both Bcr-Abl–BMT mice and vector-BMT mice. IL-6 transcripts were not detected in all the transplanted mice examined, whereas they were detected in an IL-6–producing cell line, J558L/IL-6 (R. Gerstein, unpublished data), under the same RT-PCR conditions (data not shown).
Detection of gene expression of cytokines in bone marrow of transplanted mice by RT-PCR. Bone marrow cells from Bcr-Abl–BMT mice (BMT5.1 [lanes 1 and 2], BMT5.4 [lanes 3 and 4], and BMT5.6 [lanes 5 and 6]) and a vector-BMT mouse (BMT5.35 [lanes 7 and 8]) was collected, and RNA was extracted as described in Materials and Methods. The RT-PCR products generated with (RT+) or without (RT−) reverse transcriptase were subjected to agarose gel electrophoresis, stained with ethidium bromide, and photographed by Gel Doc 1000 (Bio-Rad, Hercules, CA) with inverse gray scale. The RT-PCR products of IL-3, GM-CSF, and SCF are indicated. GAPDH was used as a control for the quality and quantity of the RNA samples and the RT-PCR reactions.
Detection of gene expression of cytokines in bone marrow of transplanted mice by RT-PCR. Bone marrow cells from Bcr-Abl–BMT mice (BMT5.1 [lanes 1 and 2], BMT5.4 [lanes 3 and 4], and BMT5.6 [lanes 5 and 6]) and a vector-BMT mouse (BMT5.35 [lanes 7 and 8]) was collected, and RNA was extracted as described in Materials and Methods. The RT-PCR products generated with (RT+) or without (RT−) reverse transcriptase were subjected to agarose gel electrophoresis, stained with ethidium bromide, and photographed by Gel Doc 1000 (Bio-Rad, Hercules, CA) with inverse gray scale. The RT-PCR products of IL-3, GM-CSF, and SCF are indicated. GAPDH was used as a control for the quality and quantity of the RNA samples and the RT-PCR reactions.
It has been shown that, under physiological conditions, IL-3 is not produced by the bone marrow stroma and that there is no detectable IL-3 in the blood.48 Because significant amounts of IL-3 transcripts and increased amounts of GM-CSF transcripts were detected in the bone marrow of Bcr-Abl–BMT mice with the CML-like disease, we went on to examine the production of IL-3 and GM-CSF proteins in these mice. We examined the serum level of IL-3 and GM-CSF in the Bcr-Abl–BMT mice with ELISA. As expected, the vector-BMT mice examined had a below-detection level of IL-3 and a very low level of GM-CSF in their sera, but the Bcr-Abl–BMT mice contained a significant amount of IL-3 and GM-CSF in their sera (Tables 1 and2).
IL-3 in Sera of Bone Marrow Transplanted Mice
| Mice . | No. of Animals . | IL-3* (pg/mL) . |
|---|---|---|
| Vector-BMT | 1 | ND |
| Primary Bcr-Abl–BMT | 2 | 6.29 ± 4.45 |
| Secondary Bcr-Abl–BMT | 6 | 8.15 ± 2.27 |
| Mice . | No. of Animals . | IL-3* (pg/mL) . |
|---|---|---|
| Vector-BMT | 1 | ND |
| Primary Bcr-Abl–BMT | 2 | 6.29 ± 4.45 |
| Secondary Bcr-Abl–BMT | 6 | 8.15 ± 2.27 |
Abbreviation: ND, not detectable.
Results are shown as the mean ± SD.
To determine whether IL-3 and GM-CSF are produced by leukemic cells in response to Bcr-Abl/p210 signaling or by noninfected cells in response to leukemia and/or tissue destruction, we sorted GFP-positive and GFP-negative bone marrow cells from Bcr-Abl–BMT mice by flow cytometry and tested for the presence of IL-3 and GM-CSF transcripts in these cells by RT-PCR. As shown in Fig 9, while the GFP-positive cells from Bcr-Abl–BMT mice with the CML-like disease contained IL-3 and GM-CSF transcripts, the GFP-negative cells from the same mice had undetectable IL-3 and very low levels of GM-CSF transcripts. This result demonstrated that the MSCV-bcr-abl/p210-IRES-gfp retrovirus-infected cells express excess IL-3 and GM-CSF.
Detection of gene expression of IL-3 and GM-CSF in GFP-positive and GFP-negative bone marrow cells from Bcr-Abl–BMT mice. Bone marrow cells from Bcr-Abl–BMT mice (BMT8.40 [lanes 1 through 4] and BMT8.41 [lanes 5 through 8]) were isolated and treated with ACK to lyse RBC. GFP-positive and GFP-negative bone marrow cells were sorted by flow cytometry and their RNA was extracted as described in Materials and Methods. The RT-PCR was performed and analyzed as in Fig8. The RT-PCR products of IL-3, GM-CSF, GFP, and GAPDH are indicated.
Detection of gene expression of IL-3 and GM-CSF in GFP-positive and GFP-negative bone marrow cells from Bcr-Abl–BMT mice. Bone marrow cells from Bcr-Abl–BMT mice (BMT8.40 [lanes 1 through 4] and BMT8.41 [lanes 5 through 8]) were isolated and treated with ACK to lyse RBC. GFP-positive and GFP-negative bone marrow cells were sorted by flow cytometry and their RNA was extracted as described in Materials and Methods. The RT-PCR was performed and analyzed as in Fig8. The RT-PCR products of IL-3, GM-CSF, GFP, and GAPDH are indicated.
DISCUSSION
In this study we describe an improved murine model for CML. Expression of Bcr-Abl/p210 can induce a transplantable myeloproliferative disorder resembling CML efficiently, reproducibly, and with an experimentally convenient latency. This efficient murine model for CML will be useful for delineating the molecular mechanisms by which the bcr-abloncogene acts in the pathogenesis of CML. In particular, the model system will help to assess the roles and relative importance of the domains of Bcr-Abl, of its interacting proteins, of Bcr-Abl–activated signaling pathways, and of host factors such as cytokines in developing CML. This model will also be useful for identifying the target cell(s) of Bcr-Abl that give rise to the clinical phenotypes of CML by analyzing the oncogenic potential of Bcr-Abl in various hematopoietic cell types and the subsequent pathology that arises from each cell type. Our first use of this model for analysis of the molecular mechanisms involved in CML demonstrated that the protein tyrosine kinase activity of Bcr-Abl/p210 was essential for its leukemogenic potential in vivo and that Bcr-Abl/p210 induced production of excess IL-3 and GM-CSF in the diseased mice.
Our new model for CML, based on previously described retroviral transduction methods, is much more effective and efficient than earlier methods. This is probably due to the use of a different retroviral vector. Because CML is believed to be a neoplasm of hematopoietic stem cells, it is likely that CML-specific disease can only be induced when Bcr-Abl expression is targeted into hematopoietic stem cells or pluripotent progenitor cells. The MSCV vector we now use for this procedure can drive expression of transduced genes in embryonic stem cells.39 It is clear that the retroviruses were targeted into the hematopoietic stem cells, because we can detect by RT-PCR the expression of gfp in the peripheral WBCs of mice transplanted with bone marrow cells infected with either MSCV-IRES-gfp or MSCV-bcr-ablK1176R-IRES-gfp 10 months after transplantation (data not shown). It is possible that the ability of MSCV to target gene expression in stem cells makes the induction of CML-like disease more effective. Systematic comparison of the disease phenotypes that result from targeting Bcr-Abl into different hematopoietic cell types will help to address this question.
Human CML is a clonal myeloproliferative disorder. However, it is not known how that clonal disease develops. It is striking that, in our model system, there is an apparent change of clonality from the polyclonal disease in the primary recipient mice to primarily monoclonal disease in the secondary recipients. Interestingly, the leukemic cell clone(s) found in the secondary recipients was not even expanded sufficiently in the parental Bcr-Abl–BMT mice to be detected by Southern blot analysis. These results suggest that Bcr-Abl/p210 can promote proliferation and/or survival of many cell types, but only certain Bcr-Abl/p210–expressing cells are capable of continuous expansion and there is a delay of the expansion of such cells. One possible explanation is that malignant transformation requires secondary mutations. Therefore, only one or two clones became leukemic and thus capable of continued expansion in the secondary recipients. For human CML, it is generally believed that Bcr-Abl is necessary but not sufficient to cause CML. A mathematical model based on epidemiological data predicts that three mutations in a stem cell are necessary for the disease and progression to blast crisis is caused by only one more stem cell mutation or mutations restricted to committed cells.49 However, by this notion it is hard to explain why the expanded clone in the secondary recipients was not derived from one of the already massively expanded clones seen in the primary recipient (more cells should have more chances to acquire secondary mutations). A second possibility is that multiple expanded clones were malignantly transformed and capable of continued expansion, but only one or two clones outgrew all other clones in the secondary recipients due to an unknown selective process. A third possibility is that Bcr-Abl/p210 does not immortalize cells when it causes disease, but rather it promotes proliferation, differentiation, and/or survival of many types of hematopoietic cells, including rare hematopoietic stem cells and committed precursor cells, without changing the self-renewal potential of the original cell type. The myeloid precursor cells may expand even earlier and faster than stem cells, but lack the ability to repopulate secondary recipients. As a result, only the Bcr-Abl/p210–expressing hematopoietic stem cell(s) or primitive progenitor cell(s) can continue to expand in secondary recipients. Such a change of clonality may occur within one animal if it can live long enough. Under this scenario, the reason that the disease became monoclonal is at least partly due to limited targeting of Bcr-Abl/p210 into the hematopoietic stem cells. The phenomenon we observed here that expansion of the leukemic progenitor cell clone(s) is delayed in the primary recipient mice as compared with the largely expanded clones that are not transplantable to the second recipients is consistent with the clinical phenotype of CML, in which the amplification of CML stem cells is constrained but the differentiation of the CML stem cells and the expansion of maturational compartments is enhanced.50-52 The development of clonal myeloproliferative disorder may be a complex process. The possibilities discussed above are not mutually exclusive. For example, assuming more than one stem cell is initially infected, an additional process must occur for all the secondary recipient mice to develop the same monoclonal disease. In any case, the leukemic clone(s) that repopulates in the secondary recipient animals in our system must bear additional unique properties. Identifying the target cell(s) that can be malignantly transformed by Bcr-Abl/p210 and give rise to a transplantable CML-like disease should provide important understanding about the pathogenesis of CML.
The cardinal features of the myeloproliferative disorder in our model resemble the chronic phase of human CML. However, differences still exist between our model and human CML. The development of the myeloproliferative disorder in the mouse model is very fast and the leukemic mice die without progression to blast phase. These phenotypes could be due to the overexpression of Bcr-Abl/p210 from a retroviral promoter, rather than expression of Bcr-Abl from the endogenous c-Bcr promoter as in human CML. It has been shown in cultured cells that Bcr-Abl induced different biological effects in a dose-dependent manner.53 The lack of blast transformation in the mouse model may be due to insufficient time for acquiring secondary genetic abnormalities. Another difference is that pulmonary hemorrhages, which may be a main cause of death of the leukemic mice, were consistently observed in the CML mouse model. The cause of the pulmonary hemorrhage in the leukemic mice is not known. In humans, pulmonary hemorrhage occurs, although rarely, in patients with acute leukemias, with acute promyelocytic leukemia treated with all-trans retinoic acid, and with allogeneic bone marrow transplantation.54-58 It was suggested that pulmonary hemorrhage in humans may be caused by blast or basophilic degranulation.56 In addition, development of pulmonary leukostasis, which can lead to extensive pulmonary hemorrhages in experimental myelocytic leukemia in the Brown-Norway rat, has been reported.59 Similarly, the pulmonary hemorrhage observed in the CML mouse model may be caused by rapid development of leukostasis or blast or basophilic degranulation.
An expansion of macrophages in the liver was frequently observed in association with the myeloproliferative disease in the previous CML model.31 However, such macrophage tumors were not observed in our system. Pear et al59a found that the macrophage expansion was more frequently associated with the cocultivation method used to infect bone marrow cells and that the macrophage expansion disappeared in secondary recipients, suggesting that the macrophage expansion is not a characteristic of murine CML. We infect bone marrow cells with retroviruses simply by incubating the cells in retrovirus-containing media, which may have eliminated the problem of the concomitant macrophage tumors not seen in human CML.
The excess expression of IL-3 and GM-CSF in Bcr-Abl–BMT mice raises the possibility that abnormal expression of hematopoietic growth factors may contribute to the genesis and/or clinical phenotypes of CML. IL-3 and GM-CSF are multilineage acting hematopoietic growth factors.48,60-62 IL-3 is a cytokine produced in response to certain pathological conditions and is not required for normal hematopoiesis.61 It was shown that IL-3 plays an important role in mast and basophil development and immunity under parasitic infection.63 GM-CSF is not essential for basal hematopoiesis either, except for the proper function of alveolar macrophages. GM-CSF–deficient mice show no major perturbation of hematopoiesis but develop pulmonary alveolar proteinosis.64,65 Although IL-3 and GM-CSF are not essential for basal hematopoiesis, they are potent cytokines that can support proliferation, survival, and differentiation of a broad spectrum of hematopoietic lineages. Abnormal expression of IL-3 and GM-CSF may cause an imbalance of hematopoiesis. It has been shown that enforced expression of IL-3 or GM-CSF can induce myeloproliferative disorders in mice.19,22 In juvenile myelomonocytic leukemia (JMML), the autocrine production of GM-CSF seems to play a pivotal role in leukemogenesis.66 67 The production of IL-3 and GM-CSF induced by Bcr-Abl/p210 may also play an important role in development of CML.
The production of excess GM-CSF in the murine CML model is in agreement with results obtained in human CML. Excess production of GM-CSF is often detected in CML patients.15-18 However, CML patients have not been demonstrated to have increased serum levels of IL-3. It is notable that the half-life of both IL-3 mRNA in cells and protein in sera is relatively short.48 68 So, it is possible that human CML cells produce low levels of IL-3 that is below detection. Such IL-3 production may have been exaggerated in the murine CML model. Clearly, the murine CML is a more aggressive disease, in which many disease phenotypes of CML are exaggerated. This may be caused partially by overexpression of Bcr-Abl/p210 in cells. In addition, although IL-3 was not detected in sera in human CML, it may still play an important role in disease development if local production of IL-3 occurs in bone marrow, the site of most hematopoietic target cells. In any case, the role of the abnormal production of hematopoietic growth factors in the genesis of CML can be examined experimentally now with the development of this murine CML model.
IL-3 and GM-CSF are not normally expressed in resting cells.48,60 Upon activation, T lymphocytes, natural killer cells, and mast cells are normal resources of IL-3, whereas these cells and macrophages, megakaryocytes, as well as fibroblast and endothelial cells can produce GM-CSF.60 Certain leukemic cells also produce GM-CSF or IL-3. Juvenile CML cells and acute myeloid leukemia (AML) blasts produce GM-CSF and ALL blasts with t(5:14) produce IL-3.60 Our results demonstrated that the MSCV-bcr-abl/p210-IRES-gfp retrovirus-infected cells express excess IL-3 and GM-CSF. Because the expanded cells infected with the MSCV-bcr-abl/p210-IRES-gfp retrovirus are predominantly, if not all, myeloid cells, it is very likely that Bcr-Abl/p210 induces the gene expression of IL-3 and GM-CSF in myeloid cells. This is in agreement with the in vitro results that Bcr-Abl can induce production of GM-CSF and/or IL-3 in myeloid cell lines.12-14 In vivo, it has previously been shown that Bcr-Abl induced GM-CSF gene expression in reticulum cell sarcomas of macrophage origin, which induced a CML-like syndrome due to a bystander effect.32
The IL-3 and GM-CSF genes are located near each other. A DNase hypersensitive site, which contains a NF-AT binding site, is located between the IL-3 and GM-CSF genes. Several transcription regulatory elements are common between IL-3 and GM-CSF promoters. These include Ets, CBF/AML-1, CK-1, and CK-2 binding sites and Octamer-1.60 It is possible that Bcr-Abl–activated signaling pathway(s) activates certain transcription factor(s) that leads to activation of gene expression of both IL-3 and GM-CSF. Further study of these signaling processes and evaluating the functional role of excess hematopoietic growth factor(s) in development of CML will help to elucidate the molecular mechanism by which Bcr-Abl induces CML. Our finding of abnormal production of IL-3 and GM-CSF induced by Bcr-Abl/p210 raises the possibility that abnormal cytokine production may contribute to the clinical phenotypes of human CML; thus, correcting such abnormal cytokine production may have therapeutic values.
ACKNOWLEDGMENT
The authors thank Dr W. Pear for technical assistance and for sharing results before publication, Drs M. Scott and H. Dong for pathological consultations, Dr R. Gerstein for kindly providing the IL-6–expressing cell line, Dr J. Jacob and Dr B. Chen in Dr Baltimore’s laboratory for kindly providing the modified GFP construct, Dr J. Chen and G. Paradis for helping on flow cytometry, P. Skourides for analyzing the expression of Bcr-Abl/p210 in leukemic cells by indirect immunofluorescence, and A. Gross for critically reading this manuscript. The flow cytometric analyses were performed in the flow cytometry core facility at the Massachusetts Institute of Technology center for cancer research.
Supported by National Cancer Institute Grant No. CA68008 (to R.R.). R.R. is a recipient of American Cancer Society Junior Faculty Research Award.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Ruibao Ren, MD, PhD, Rosenstiel Center, Brandeis University, Waltham, MA 02254; e-mail:ren@hydra.rose.brandeis.edu.






![Fig. 8. Detection of gene expression of cytokines in bone marrow of transplanted mice by RT-PCR. Bone marrow cells from Bcr-Abl–BMT mice (BMT5.1 [lanes 1 and 2], BMT5.4 [lanes 3 and 4], and BMT5.6 [lanes 5 and 6]) and a vector-BMT mouse (BMT5.35 [lanes 7 and 8]) was collected, and RNA was extracted as described in Materials and Methods. The RT-PCR products generated with (RT+) or without (RT−) reverse transcriptase were subjected to agarose gel electrophoresis, stained with ethidium bromide, and photographed by Gel Doc 1000 (Bio-Rad, Hercules, CA) with inverse gray scale. The RT-PCR products of IL-3, GM-CSF, and SCF are indicated. GAPDH was used as a control for the quality and quantity of the RNA samples and the RT-PCR reactions.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/10/10.1182_blood.v92.10.3829/4/m_blod42214008w.jpeg?Expires=1764955076&Signature=hHxIA1VNJKFD-obpjuzuyMlO3nIHl4~3IxzHYqgOEHAjz2-jjaPmAmbfCsLSSv04GxYZ9KTxAXiqqhZXHZwCOtDKbYpR2X~ABM9TmyjagK1a4FHb~dW68wV8mYfqO~uczKNlO4Lb5t~ncJP-FiujkBsJX4-PUW8ALFJ73dwgF7facV0weJIx7XBm5n4y72VhvzK4s90xRM3OpW4vCD5zboagH91T3bKfYEKK7Ray8vYg41wMBvyJWfp8nGXZQlJHZ0EcsqQ8rC7-~wZOEHBPHeekKLRcOm1kf9OCy0ueRQ725SDZf9N005IDOYKnskTGj0v~vpg3cPyWYRlrkChCrA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 9. Detection of gene expression of IL-3 and GM-CSF in GFP-positive and GFP-negative bone marrow cells from Bcr-Abl–BMT mice. Bone marrow cells from Bcr-Abl–BMT mice (BMT8.40 [lanes 1 through 4] and BMT8.41 [lanes 5 through 8]) were isolated and treated with ACK to lyse RBC. GFP-positive and GFP-negative bone marrow cells were sorted by flow cytometry and their RNA was extracted as described in Materials and Methods. The RT-PCR was performed and analyzed as in Fig8. The RT-PCR products of IL-3, GM-CSF, GFP, and GAPDH are indicated.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/10/10.1182_blood.v92.10.3829/4/m_blod42214009w.jpeg?Expires=1764955076&Signature=p3yMKqIkXFRkixQ3Z88gTrWpRqSX5andrVJR~WMAG3R4hfRtRDhV6Qh5vQDWcWSSSF15jlukplWRRrcqwPIDgfEGn0Ux~Jlu-YsnU62GEkYwz1prKW9P-5h~2yhc6bVD29PObldGGOJqXZnxvZbvp8vXH-7amG2eeU25CS9DRty~aBMmlYJIZ3-sg8mRmEJQrsAUEZdD0V9Qit8GdqndWWNrlZEfVVYQexeVkONgYGOgPFRcT5mJlLLOdSR1XMaWWwpsAeaW-WakUqvStoCF3SqJbBSOpa6TjKvdGfCotGkTH22rHHMNYl~tW03~D8u4s9NBSR6ETxmxDw51mJwDCA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal