Abstract
We investigated a new method using fluorescence in situ hybridization and DNA probes that span the common breakpoints of t(9;22)(q34;q11.2) and that detect double BCR/ABL fusion (D-FISH) in bone marrow cells with this translocation, one on the abnormal chromosome 9 and one on the Philadelphia chromosome (Ph chromosome). D-FISH patterns were abnormal in 30 of 30 specimens with classic, simple, complex, and masked Ph chromosomes. Based on 200 nuclei from each of 30 normal specimens, the mean percentage of false-positive cells was 0.25 ± 0.39. Thirty-seven specimens from 10 patients were studied before treatment and two or more times at 4-month intervals after treatment with interferon-α2b (IFN-α2b) with or without ara-C. Based on 200 nuclei, the results of D-FISH in these specimens correlated closely with quantitative cytogenetics and accurately quantified disease within a few percent. We studied 6,000 nuclei for each of six specimens, three normal and three from patients with chronic myeloid leukemia (CML) in cytogenetic remission. The normal cutoff for 6,000 nuclei was 0.079% and patients in cytogenetic remission had residual disease ranging from 7 (0.117%) to 53 (0.883%) Ph-positive nuclei. We conclude that D-FISH can detect the Ph chromosome and its variant translocations and accurately quantify disease in CML at diagnosis and at all times after treatment, including cytogenetic remission.
MULTIPLE GENETIC TESTING methods are used in clinical practice to assess response to therapy in chronic myeloid leukemia (CML), but no one technique accurately detects and quantifies disease at diagnosis and at all times during treatment.1-6A new method, using fluorescence in situ hybridization (FISH), now appears to meet this need in clinical practice. This new method uses FISH and commercially available differently colored BCR and ABL probes that span the common breakpoints of t(9;22)(q34;q11.2) and show double BCR/ABL fusion (D-FISH) in cells with this translocation, one on the abnormal chromosome 9 and one on the Ph chromosome. With D-FISH, the number of false-positive and false-negative cells approaches zero.
Conventional FISH methods with differently colored BCR and ABL DNA probes detect a single BCR/ABL fusion signal on the Ph chromosome (S-FISH).7 S-FISH is highly accurate for analysis of metaphases but is imprecise for the study of interphase cells because BCR and ABL signals coincidentally overlap in about 4% of normal nuclei (false-positive cells).8 Moreover, BCR/ABL fusion signals can be incorrectly scored in many Ph-positive nuclei (false-negative cells) because scoring fusion signals with S-FISH is subjective.8 These technical artifacts limit the potential of S-FISH to detect and quantify minimal residual disease accurately and to measure small fluctuations in the percentage of Ph-positive cells in response to therapy.
Dewald et al7 established that the normal range of S-FISH for interphase bone marrow nuclei is 10% or less, and the abnormal reference range for most untreated patients with CML is 69% to 92%. The statistical method used to determine the normal cutoff was intentionally calculated in a conservative fashion to avoid making false-positive diagnoses of untreated CML. In our experience, this cutoff has worked well in clinical practice to diagnose CML but is high for assessing residual disease in some patients with CML who are receiving therapy. Cox Froncillo et al9 used a more liberal normal cutoff to monitor patients with CML who have been treated, suggesting that detection of 7% or greater of cells with BCR/ABL fusion indicates residual disease.9 Alternative FISH strategies that use either three differently colored DNA probes for BCR, ABL, and ASS (a locus on the centromeric side of ABL on chromosome 9) or differently colored probes that span either the breakpoints in BCR or ABL have been reported to be useful to detect Ph-positive cells in less than 1% of nuclei.10-12 The efficacy of these techniques to detect variant Ph chromosomes and the clinical value of these methods have not been tested.
In a three-part study, we tested D-FISH (1) on various kinds of Ph anomalies and determined its normal range, (2) to monitor patients on treatment for CML, and (3) to detect and quantify low levels of residual disease. The kinds of Ph chromosomes tested included classic t(9;22)(q34;q11.2) anomalies, as well as simple, complex, and masked Ph chromosomes. We also studied specimens with t(9;22)(q34;q11.2) and a breakpoint in the minor BCR region. To test the efficacy of D-FISH for monitoring patients receiving therapy, we studied specimens from 10 patients with CML before and after treatment with interferon-α2b (IFN-α2b) with or without cytosine arabinoside (ara-C). We found the results of D-FISH to correlate well with quantitative cytogenetic methods and were able to determine the percentage of abnormal nuclei accurately within a few percentage points. In testing the limits of resolution to detect Ph-positive nuclei with D-FISH in three normal specimens and in two patients in cytogenetic remission, we detected residual disease as low as 0.117%. We believe that D-FISH is a significant technological advancement in monitoring therapy for CML. D-FISH detects all variant translocations of the Ph chromosome and accurately quantifies disease in patients with CML at diagnosis, and at all times during treatment, even in patients in cytogenetic remission.
MATERIALS AND METHODS
This study was performed using differently colored, directly labeled BCR and ABL probes developed by Oncor (Gaithersburg, MD) to produce two fusion signals in cells with a t(9;22)(q34;q11.2). The BCR and ABL probes were derived from DNA sequences that span the common breakpoints on chromosomes 9 and 22 in this translocation. The ABL probe set included several DNA sequences that hybridized to 9q34 and spanned the 200-kb breakpoint region of ABL. The BCR probe included several DNA sequences that hybridized to 22q11.2 and spanned the common breakpoints in both the major and minor BCR.
In part I of the investigation, we used D-FISH to study bone marrow specimens from 30 patients with CML or acute lymphocytic leukemia (ALL) and who collectively had classic, simple, complex, and masked Ph chromosomes. We studied most of these same Ph-positive specimens in 1993, using S-FISH.7 Specimens from 35 normal subjects were also studied, 30 used to establish the normal cutoff and 5 to test this cutoff.
In part II of the investigation, we studied 37 bone marrow specimens from 10 patients enrolled in the CML National Study Group who were randomly receiving treatment with IFN-α2b with or without ara-C. As a quality control procedure, we included a set of serial dilutions created by mixing cells from a normal individual and an untreated patient with CML.
In part III of the investigation, we attempted to establish the limits of resolution for D-FISH to detect minimal residual disease. To do this, we scored 6,000 nuclei from three normal specimens from part I and three specimens in part II from patients in cytogenetic remission. We calculated the normal range from the normal specimens and tested the cutoffs on three specimens from patients with CML.
Bone marrow samples were processed by conventional cytogenetic procedures and stored at −70°C in methanol/glacial acetic acid (3:1). Before slide preparation, cells were washed with two changes of fresh methanol/glacial acetic acid (3:1), dropped on microscope slides, and allowed to air dry. To aid in slide preparation, a CDS-5 cytogenetic drying chamber (Thermotron, Holland, MI) was used to prepare slides at 50% relative humidity and 25°C.13Slides were treated with 2× standard saline citrate (SSC) (300 mmol/L sodium chloride, 30 mmol/L sodium citrate) for 1 hour at 37°C; dehydrated with cold 70%, 85%, and 100% ethanol for 2 minutes each; and air dried. Chromosomal DNA was denatured in 70% formamide/2× SSC for 2 minutes at 70°C. Slides were dehydrated with a cold ethanol series (70%, 85%, and 100%) for 2 minutes each and air dried. To denature the probe DNA, 10 μL of probe mixture was aliquoted into a micropipette tube and placed in a water bath at 70°C for 5 minutes. After the probe was added to each slide, a 22 × 22-mm coverslip was placed over each hybridization site and sealed with rubber cement. Slides were hybridized for 18 to 20 hours at 37°C in a humidified chamber. Coverslips were removed and slides were washed for 2 minutes in a 0.5× SSC solution at 70°C, and transferred to 1× phosphate-buffered detergent (PBD) for 2 minutes. Chromosomes were counterstained with a mixture of 10 μl 4′-6′-diamidine-2-phenylindole dihydrochloride (DAPI) and Vectashield antifade in a ratio of 1:100.
Cells were viewed with a fluorescent microscope equipped with a dual-bandpass filter for fluoroisothiocyanate (FITC) and Texas Red (Chromatechnology, Brattleboro, VT) or a triple-bandpass filter for DAPI, FITC, and Texas Red (Chromatechnology). The BCR probe had a red signal and the ABL probe a green signal, and the background chromatin was blue.
For the purposes of this discussion, we refer to red BCR signals as R, green ABL signals as G, and BCR/ABL fusion signals as F. For scoring purposes, fusion signals were defined as merging or touching R and G signals. The scoring process was limited to normal nuclei with 2R2G, and abnormal nuclei with 1R1G2F or 2R2G1F (one Ph chromosome), and 1R1G3F or 2R2G2F (two Ph chromosomes) (Fig 1A, B, D, and E). During the investigation, we noted the number of nuclei that did not meet the scoring criteria. Representative cells were captured using a computer-based imaging system (Quips XL Genetics Workstation; Vysis, Downers Grove, IL).
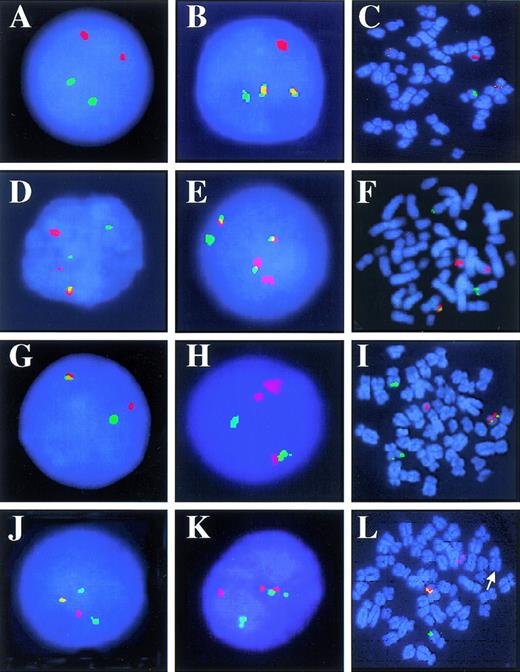
Representative D-FISH patterns in interphase nuclei and metaphase cells. (A) Normal, 2R2G. (B) One copy of t(9;22)(q34;q11.2), 1R1G2F. (C) t(9;22)(q34;q11.2), 1R1G2F. (D) One copy of t(9;22)(q34;q11.2), 2R2G1F caused by a split BCR and ABL signal on abnormal chromosome 9. (E) One copy of t(9;22)(q34;q11.2) and an additional Ph chromosome, 1R1G3F. (F) t(9;2210)(q34;q11.2;q22), 2R2G1F. (G) Atypical pattern 1R1G1F, t(9;22)(q34;q11.2). (H) Atypical pattern 2R1G1F, t(9;22)(q34;q11.2). (I) 1R2G1F, metaphase with a masked Ph chromosome caused by insertion translocation of ABL into BCR. (J) Atypical pattern 1R2G1F, t(9;22)(q34;q11.2). (K) Nucleus with a false-positive pattern due to signal separation and coincidental overlap of BCR and ABL signals. (L) 1R1G1F, metaphase with no BCR or ABL on the abnormal chromosome 9 (arrow).
Representative D-FISH patterns in interphase nuclei and metaphase cells. (A) Normal, 2R2G. (B) One copy of t(9;22)(q34;q11.2), 1R1G2F. (C) t(9;22)(q34;q11.2), 1R1G2F. (D) One copy of t(9;22)(q34;q11.2), 2R2G1F caused by a split BCR and ABL signal on abnormal chromosome 9. (E) One copy of t(9;22)(q34;q11.2) and an additional Ph chromosome, 1R1G3F. (F) t(9;2210)(q34;q11.2;q22), 2R2G1F. (G) Atypical pattern 1R1G1F, t(9;22)(q34;q11.2). (H) Atypical pattern 2R1G1F, t(9;22)(q34;q11.2). (I) 1R2G1F, metaphase with a masked Ph chromosome caused by insertion translocation of ABL into BCR. (J) Atypical pattern 1R2G1F, t(9;22)(q34;q11.2). (K) Nucleus with a false-positive pattern due to signal separation and coincidental overlap of BCR and ABL signals. (L) 1R1G1F, metaphase with no BCR or ABL on the abnormal chromosome 9 (arrow).
In parts I and II of the investigation, specimens were studied in random order and in a blind fashion by two microscopists (W.A.W. and A.L.J.). Each microscopist scored 100 consecutive qualifying interphase nuclei from different areas of the same slide. We considered the intermicroscopist agreement was good enough to average their results on each specimen in subsequent analyses of the data. In part III of the investigation, one microscopist (W.A.W.) studied specimens in random order in a blind fashion: 6,000 consecutive qualifying interphase nuclei were scored for each specimen.
In part I of the investigation, 15 specimens underwent Southern blot analysis of the MBCR region.7 In part II, except for the serial dilutions, each specimen had quantitative cytogenetic studies (Q-cytogenetics) and S-FISH analyses with the Oncor probe for the minor BCR region. Q-cytogenetic studies were done by analysis of up to 25 consecutive metaphases for a Ph chromosome.
RESULTS
Preliminary Studies With Various Kinds of Ph Chromosomes
Preliminary studies were done on metaphases and nuclei from bone marrow of four patients with several types of Ph chromosomes: a t(9;22)(q34;q11.2) with a breakpoint in the major BCR region, a t(9;22)(q34;q11.2) with a breakpoint in the minor BCR region, a t(5;9;22)(q31;q34;q11.2), and a masked Ph chromosome.
In metaphases with a t(9;22)(q34;q11.2) and either a major or minor BCR, we observed a BCR/ABL fusion signal at 22q11.2 on the Ph chromosome, an ABL/BCR fusion signal at 9q34 on the abnormal chromosome 9, a BCR signal at 22q11.2 on the normal chromosome 22, and an ABL signal at 9q34 on the normal chromosome 9 (Fig 1C). In interphase, the D-FISH patterns were mostly 1R1G2F or 2R2G1F. Nuclei with 2R2G1F resulted from separation of the BCR and ABL probes on the abnormal chromosome 9. We noted a predominance of nuclei with 1R1G2F in t(9;22)(q34;q11.2) with a minor BCR breakpoint. In t(9;22)(q34;q11.2) with a major BCR breakpoint, nuclei with 2R2G1F were prevalent.
In metaphases with a t(5;9;22)(q31;q34;q11.2), we observed a BCR/ABL fusion signal at 22q11.2 on the Ph chromosome, a small ABL signal on the abnormal chromosome 9, a small BCR signal on the abnormal chromosome 5, a BCR signal at 22q11.2 on the normal chromosome 22, and an ABL signal at 9q34 on the normal chromosome 9 (Fig 1F). In interphase, the predominant D-FISH pattern was 2R2G1F.
In metaphases with a masked Ph chromosome, we observed a BCR/ABL fusion signal at 22q11.2 on the abnormal chromosome 22, a small ABL signal on the abnormal chromosome 9, a BCR signal at 22q11.2 on the normal chromosome 22, and an ABL signal at 9q34 on the normal chromosome 9 (Fig 1I). In interphase, the predominant D-FISH pattern was 1R2G1F. This signal pattern is consistent with a masked Ph chromosome derived by insertion of a portion of ABL into BCR and represents an atypical D-FISH pattern.
To determine the initial scoring criteria, we recorded all the signal patterns in 100 consecutive nuclei for each of the preliminary specimens and recognized 34 different signal patterns. These patterns included the five we used in the scoring criteria and two that we expected to encounter for masked Ph chromosomes. The remaining 27 patterns represented various types of technical problems, such as signal overlaps, signal separation, and lack of probe hybridization.
Typical D-FISH Patterns
In part I of the investigation, we tested the scoring criteria for 65 bone marrow specimens, including 35 that were normal and 30 with various kinds of Ph chromosomes (Table 1).We found the nuclei in 57 of 65 specimens met the scoring criteria (Fig1A, B, D, and E), but eight had nuclei with atypical but abnormal D-FISH patterns.
D-FISH for Patients With Various Kinds of Ph Chromosomes and Normal Subjects Compared With Conventional Cytogenetics, S-FISH on Interphase and Metaphase, and Southern Blot Analysis
| Category . | No. . | Karyotype-150 . | Cyto Meta (%) . | S-FISH Nuclei (%) . | S-FISH Meta (%) . | Sth blt Mbcr . | D-FISH Nuclei (%) . | Atypical D-FISH Pattern . |
|---|---|---|---|---|---|---|---|---|
| t(9;22) only | 1 | 46,XY,t(9;22)[20] | 100 | 85.5 | 100.0 | + | 95.0 | |
| 2 | 46,XX,t(9;22)[20] | 100 | 74.0 | + | 97.5 | |||
| 3 | 46,XY,t(9;22)[20] | 100 | 70.5 | 95.0 | 98.5 | |||
| 4 | 46,XY,t(9;22)[25] | 100 | 76.5 | 97.5 | ||||
| 5 | 46,XY,t(9;22)[20] | 100 | 69.0 | 97.5 | 98.5 | |||
| t(9;22) + other | 6 | 46,XY,t(9;22)[18]/47,idem,+8[2] | 100 | 69.5 | 100.0 | + | 97.5 | |
| 7 | 47,XY,t(9;22),+Ph[20] | 100 | 80.0 | 100.0 | 95.5 | 1R1G1F | ||
| 8 | 46,XY,t(9;22)[28]/47,idem,+Ph[2] | 100 | 85.0 | 100.0 | 92.5 | |||
| 9 | 46,XY,t(9;22)[14]/47,idem,+8[5]/46,XY[1] | 95 | 76.5 | 95.0 | 76.5 | 1R1G1F | ||
| 10 | 46,XY,t(9;22)[19]/47,idem,+Ph[1] | 100 | 84.0 | 100.0 | 96.5 | |||
| Complex Ph | 11 | 46,XY,t(9;22;10)(q34;q11.2;q22)[19]/46,XY[1] | 95 | 81.5 | 100.0 | 99.5 | ||
| 12 | 46,XY,t(9;22;19)(q34;q11.2;q13.3)[20] | 100 | 91.5 | 100.0 | 94.0 | 1R2G1F | ||
| 13 | 46,XX,del(2)(q23q31),t(5;9;22)(q31;q34;q11.2)[25] | 100 | 65.0 | 97.0 | ||||
| 14 | 46,XY,t(3;17;9;22)(q26.2;q21;q34;q11.2)[20] | 100 | 69.0 | 100.0 | 98.5 | |||
| 15 | 46,XX,t(9;22;12)(q34;q11.2;q15)[20] | 100 | 84.0 | 100.0 | 96.5 | |||
| Simple Ph | 16 | 46,XY,der(22)t(19;22)(q13.3;q11.2), + other anomalies[20] | 100 | 68.0 | 95.0 | + | 91.5 | |
| 17 | 46,XY,t(3;22)(q29;q11.2)[20] | 100 | 83.5 | 100.0 | 95.5 | |||
| 18 | 46,XX,t(7;22)(p22;q11.2)[25] | 100 | 84.5 | 100.0 | 95.5 | |||
| 19 | 46,XX,t(1;22)(p26;q11.2)[2]/46,XX[18] | 10 | 8.5 | 12.5 | + | 12.0 | ||
| 20 | 46,XX,t(17;22)(q25;q11.2)[20] | 100 | 76.0 | 100.0 | 90.0 | |||
| Masked Ph | 21 | 46,XY[20] | 0 | 74.0 | 100.0 | + | 94.5 | 1R1G1F |
| 22 | 46,XX[20] | 0 | 75.5 | 97.5 | + | 93.0 | 2R1G1F | |
| 23 | 46,XY[20] | 0 | 54.0 | 58.8 | + | 10.5 | 1R2G1F-151 | |
| 24 | 46,XY[20] | 0 | 77.5 | 92.5 | + | 78.0 | 1R1G1F | |
| 25 | 46,XY[20] | 0 | 83.0 | 97.5 | + | 74.0 | 1R2G1F | |
| Ph+ ALL | 26 | 46,XY,t(9;22)[20] | 100 | 74.5 | 99.5 | |||
| 27 | 46,XX,t(9;22)[2]/46,XX[18] | 10 | 6.5 | 0-152 | − | 3.5 | ||
| 28 | 46,XY,t(9;22)[5]/46,XY[15] | 25 | 50.5 | 62.5 | 60.5 | |||
| 29 | 47,XX,t(9;22), + mar[7]/46,XX[3] | 70 | 12.5 | 0-152 | − | 95.0 | ||
| 30 | 49,XY,+4,i(8q)x2,t(9;22), + Ph[12]/46,XY[8] | 60 | 77.5-153 | − | 69.5 | |||
| Normal | 31 | 46,XX[20] | 0 | 0.0 | ||||
| 32 | 46,XX[20] | 0 | 0.0 | |||||
| 33 | 46,XX[20] | 0 | 0.0 | |||||
| 34 | 46,XX[20] | 0 | 0.0 | |||||
| 35 | 46,XX[20] | 0 | 0.0 |
| Category . | No. . | Karyotype-150 . | Cyto Meta (%) . | S-FISH Nuclei (%) . | S-FISH Meta (%) . | Sth blt Mbcr . | D-FISH Nuclei (%) . | Atypical D-FISH Pattern . |
|---|---|---|---|---|---|---|---|---|
| t(9;22) only | 1 | 46,XY,t(9;22)[20] | 100 | 85.5 | 100.0 | + | 95.0 | |
| 2 | 46,XX,t(9;22)[20] | 100 | 74.0 | + | 97.5 | |||
| 3 | 46,XY,t(9;22)[20] | 100 | 70.5 | 95.0 | 98.5 | |||
| 4 | 46,XY,t(9;22)[25] | 100 | 76.5 | 97.5 | ||||
| 5 | 46,XY,t(9;22)[20] | 100 | 69.0 | 97.5 | 98.5 | |||
| t(9;22) + other | 6 | 46,XY,t(9;22)[18]/47,idem,+8[2] | 100 | 69.5 | 100.0 | + | 97.5 | |
| 7 | 47,XY,t(9;22),+Ph[20] | 100 | 80.0 | 100.0 | 95.5 | 1R1G1F | ||
| 8 | 46,XY,t(9;22)[28]/47,idem,+Ph[2] | 100 | 85.0 | 100.0 | 92.5 | |||
| 9 | 46,XY,t(9;22)[14]/47,idem,+8[5]/46,XY[1] | 95 | 76.5 | 95.0 | 76.5 | 1R1G1F | ||
| 10 | 46,XY,t(9;22)[19]/47,idem,+Ph[1] | 100 | 84.0 | 100.0 | 96.5 | |||
| Complex Ph | 11 | 46,XY,t(9;22;10)(q34;q11.2;q22)[19]/46,XY[1] | 95 | 81.5 | 100.0 | 99.5 | ||
| 12 | 46,XY,t(9;22;19)(q34;q11.2;q13.3)[20] | 100 | 91.5 | 100.0 | 94.0 | 1R2G1F | ||
| 13 | 46,XX,del(2)(q23q31),t(5;9;22)(q31;q34;q11.2)[25] | 100 | 65.0 | 97.0 | ||||
| 14 | 46,XY,t(3;17;9;22)(q26.2;q21;q34;q11.2)[20] | 100 | 69.0 | 100.0 | 98.5 | |||
| 15 | 46,XX,t(9;22;12)(q34;q11.2;q15)[20] | 100 | 84.0 | 100.0 | 96.5 | |||
| Simple Ph | 16 | 46,XY,der(22)t(19;22)(q13.3;q11.2), + other anomalies[20] | 100 | 68.0 | 95.0 | + | 91.5 | |
| 17 | 46,XY,t(3;22)(q29;q11.2)[20] | 100 | 83.5 | 100.0 | 95.5 | |||
| 18 | 46,XX,t(7;22)(p22;q11.2)[25] | 100 | 84.5 | 100.0 | 95.5 | |||
| 19 | 46,XX,t(1;22)(p26;q11.2)[2]/46,XX[18] | 10 | 8.5 | 12.5 | + | 12.0 | ||
| 20 | 46,XX,t(17;22)(q25;q11.2)[20] | 100 | 76.0 | 100.0 | 90.0 | |||
| Masked Ph | 21 | 46,XY[20] | 0 | 74.0 | 100.0 | + | 94.5 | 1R1G1F |
| 22 | 46,XX[20] | 0 | 75.5 | 97.5 | + | 93.0 | 2R1G1F | |
| 23 | 46,XY[20] | 0 | 54.0 | 58.8 | + | 10.5 | 1R2G1F-151 | |
| 24 | 46,XY[20] | 0 | 77.5 | 92.5 | + | 78.0 | 1R1G1F | |
| 25 | 46,XY[20] | 0 | 83.0 | 97.5 | + | 74.0 | 1R2G1F | |
| Ph+ ALL | 26 | 46,XY,t(9;22)[20] | 100 | 74.5 | 99.5 | |||
| 27 | 46,XX,t(9;22)[2]/46,XX[18] | 10 | 6.5 | 0-152 | − | 3.5 | ||
| 28 | 46,XY,t(9;22)[5]/46,XY[15] | 25 | 50.5 | 62.5 | 60.5 | |||
| 29 | 47,XX,t(9;22), + mar[7]/46,XX[3] | 70 | 12.5 | 0-152 | − | 95.0 | ||
| 30 | 49,XY,+4,i(8q)x2,t(9;22), + Ph[12]/46,XY[8] | 60 | 77.5-153 | − | 69.5 | |||
| Normal | 31 | 46,XX[20] | 0 | 0.0 | ||||
| 32 | 46,XX[20] | 0 | 0.0 | |||||
| 33 | 46,XX[20] | 0 | 0.0 | |||||
| 34 | 46,XX[20] | 0 | 0.0 | |||||
| 35 | 46,XX[20] | 0 | 0.0 |
Abbreviations: Cyto, cytogenetic; Meta, metaphases; Sth blt Mbcr, Southern blot test for breakpoint in the major region of BCR; Ph, Ph chromosome; Ph+ ALL, patients with acute lymphocytic leukemia and metaphases with a t(9;22)(q34;q11.2).
Breakpoints for 9;22 translocations are q34 and q11.2 unless otherwise specified.
The discrepancy between S-FISH and D-FISH may be produced by different ABL probes.
Patient had a t(9;22) with a breakpoint in the BCR minor region that does not produce BCR/ABL fusion with a probe for the BCR major region.
Patient had a t(9;22)(q34;q11.2) with additional Ph chromosome and a minor BCR breakpoint. The high number of aberrant nuclei is attributable to cells with extra Ph chromosomes.
Normal specimens.
We studied 35 normal specimens, and each one met the scoring criteria. Among the 7,000 nuclei from these specimens, 6,985 (99.79%) were 2R2G. Each of the 15 nuclei with abnormal D-FISH patterns involved split BCR and ABL signals, suggesting that these cells were in G2 of the cell cycle (Fig 1K, false-positive).
t(9;22) only.
We studied five specimens with only a t(9;22) (Table 1). Each of these specimens met the scoring criteria. Among the collective 1,000 nuclei from these specimens, the D-FISH patterns were 2R2G (2.6%), 1R1G2F (72.8%), and 2R2G1F (24.6%). Thus, 97.4% of nuclei from these five untreated patients were Ph positive and 2.6% were normal.
t(9;22) plus other anomalies.
We studied five specimens with a t(9;22) and either an extra Ph chromosome or trisomy 8 (Table 1). Three of these specimens (nos. 6, 8, and 10) met the scoring criteria. Among the 600 nuclei from these specimens, the D-FISH patterns were 2R2G (4.5%), 1R1G2F (72.3%), 2R2G1F (20.5%), and either 1R1G3F or 2R2G2F (2.7%). Thus, 95.5% of nuclei from these three specimens were Ph positive and 4.5% were normal.
Complex Ph chromosomes.
We studied five specimens with complex Ph chromosomes (Table 1). Four of these specimens (nos. 11, 13, 14, and 15) met the scoring criteria. Among the 800 nuclei from these specimens, the D-FISH patterns were 2R2G (2.1%), 1R1G2F (8.9%), and 2R2G1F (89.0%). Thus, 97.9% of nuclei from these four specimens were Ph positive, and 2.1% were normal.
Simple Ph chromosomes.
We studied five specimens with simple Ph chromosomes (Table 1). Each of these specimens met the scoring criteria. Specimen 19 was mosaic as 2 of 20 metaphases had a Ph chromosome, and 12.0% of nuclei were Ph positive by D-FISH analysis. Among the 800 nuclei from the nonmosaic specimens, the D-FISH patterns were 2R2G (6.9%), 1R1G2F (11.1%), and 2R2G1F (82.0%). Thus, 93.1% of nuclei were Ph positive, and 6.9% of nuclei were normal in these four nonmosaic specimens. The D-FISH patterns in these specimens were similar to four of the specimens with a complex translocation.
Ph chromosomes in ALL.
We studied five specimens from patients with Ph-positive ALL (Table 1). Each of these specimens met the scoring criteria, regardless of whether the BCR breakpoint was in the major (nos. 26 and 28) or minor (nos. 27, 29, and 30) region. Four of these specimens (nos. 27 through 30) were mosaic for normal and Ph-positive metaphases by cytogenetics. Specimen 26 was nonmosaic, and the D-FISH patterns were 2R2G (0.5%), 1R1G2F (70.0%), and 2R2G1F (29.5%). Thus, 99.5% of nuclei from this specimen were Ph positive and 0.5% were normal.
Atypical D-FISH Patterns
In part I of the investigation, eight specimens had nuclei with an abnormal D-FISH pattern (Table 1), but the signal pattern was different from those used in the scoring criteria. Three of the specimens (nos. 9, 7, and 12) with an atypical D-FISH pattern were in the category of t(9;22) plus other anomalies and one was classified with the complex Ph chromosomes. The five other specimens with atypical D-FISH patterns all had masked Ph chromosomes (nos. 21 through 25).
t(9;22) plus other anomalies.
In specimen 9, 76.5% of nuclei were 1R1G1F (Fig 1G). Metaphases from this specimen had BCR/ABL fusion on the Ph chromosome, but no ABL or BCR signal on the abnormal chromosome 9 (Fig 1L). In specimen 7, D-FISH patterns were 2R2G (5%), 1R1G1F (27%), 1R1G2F (55%), and 2R2G1F (14%). In each Ph-positive metaphase, no ABL or BCR signal was observed on the abnormal chromosome 9. Metaphases from this specimen demonstrated an atypical D-FISH pattern. Some metaphases had one Ph chromosome, but most had two. Thus, nuclei with 1R1G2F or 2R2G1F signals most likely represent cells with two Ph chromosomes.
Complex Ph chromosomes.
Specimen 12 had a t(9;22;19)(q34;q11.2;q13.3); D-FISH patterns were 2R2G (6.0%) and 1R2G1F (94.0%). Metaphases from this specimen had BCR/ABL fusion on the Ph chromosome and an ABL signal on the abnormal chromosome 9. No BCR or ABL signal was apparent on the abnormal chromosome 19. We studied this specimen with FISH using probes for D22S75 (22q11.2) and D22S39 (22q13.3). We observed a D22S75 signal on the Ph chromosome and a D22S39 signal on the abnormal chromosome 19.
Masked Ph chromosomes.
We studied five specimens with a masked Ph chromosome and found each to have an atypical D-FISH pattern. Specimens 21 and 24 had 94.5% and 78.0% nuclei with 1R1G1F signals, respectively (Fig 1G). Metaphases from each of these specimens had BCR/ABL on the Ph chromosome, but no BCR or ABL signal on the abnormal chromosome 9.
Specimens 23 and 25 had 10.5% and 74.0% nuclei with 1R2G1F signals, respectively (Fig 1J). Metaphases from each of these specimens had a BCR/ABL fusion on the Ph chromosome and a small residual ABL signal on the abnormal chromosome 9 (Fig 1I). This finding is consistent with an insertion of a portion of ABL into the BCR locus.
Specimen 22 had 93.0% nuclei with 2R1G1F signals (Fig 1H). Metaphases from this specimen had an ABL/BCR fusion on the abnormal chromosome 9 and a small residual BCR signal on the abnormal chromosome 22. This is consistent with an insertion of a portion of BCR into the ABL locus.
Normal Range
Based on 200 nuclei from each of 30 normal specimens, the mean percentage and standard deviation of nuclei with scorable D-FISH patterns other than 2R2G patterns was 0.25 ± 0.39, within a range of 0 to 1.5. We calculated the upper bound of a one-sided 95% confidence interval for observing 3 of 200 (1.5%) aberrant cells using the binomial distribution. This implied a cutoff of greater than 7 (3.83%) nuclei with scorable D-FISH patterns other than 2R2G in 200 nuclei to classify any specimen as abnormal.
We classified each of the 35 other specimens in part I of the investigation as normal or abnormal based on the 3.83% normal cutoff. Each of the five normal specimens used to test the cutoff was classified correctly. Among the 30 specimens with a Ph chromosome, 29 were classified as abnormal. The results of specimen 27 were within normal limits based on finding 3.5% abnormal nuclei. This specimen had 2 of 20 (10%) metaphases with a Ph chromosome by conventional cytogenetics.
Abnormal Reference Range
We used the observed values from the D-FISH findings in the 16 patients (nos. 1-6, 8, 10, 13-18, 20, 26) with only a t(9;22) or a variant Ph chromosome in all metaphases, to establish an abnormal reference range. Among these specimens, the percentage of abnormal nuclei ranged from 90.0 to 99.5, with a mean of 96.1.
We tested the precision of D-FISH to identify abnormal nuclei in cells collected by apheresis from a patient with CML in blast crisis and a 9;22 translocation. We processed and analyzed samples from this specimen on 10 occasions and found the percentage of abnormal nuclei ranged from 97 to 100, with a mean of about 98.2.
Unscorable Nuclei
Among the 35 normal specimens from part I of the investigation, the mean number of unscorable cells encountered to acquire 200 scorable nuclei was 152 ± 21.9, within a range of 70 to 240. This number was comparable for each of the microscopists. Among the 30 abnormal specimens from part I, the mean number of unscorable cells encountered to acquire 200 scorable nuclei was 287 ± 77.7, within a range of 98 to 698.
Monitoring Response to Therapy for CML
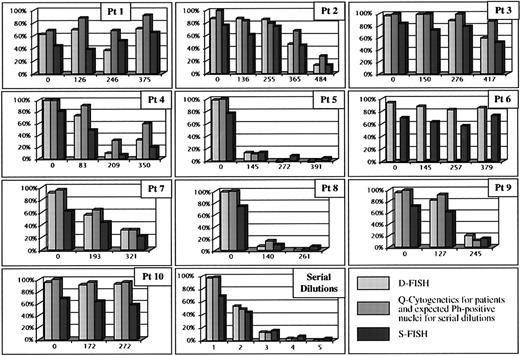
In part II of the investigation, we used Q-cytogenetics, S-FISH, and D-FISH to study 37 bone marrow specimens from 10 patients (Fig2). For each patient, we studied one specimen collected before treatment and two or more specimens at approximately 4-month intervals after treatment with IFN-α2b with or without ara-C, according to protocol. These 10 patients included seven with a classic t(9;22)(q34;q11.2) and a typical D-FISH pattern.
The y-axis shows the percentage of Ph-positive cells in each graph. The x-axis for patients 1 through 10 is the number of days treated with IFN-α2b with or without ara-C. The x-axis for serial dilutions represents different mixtures of normal and Ph-positive cells: the expected percentage of Ph-positive nuclei is 98.2, 49.1, 12.3, 3.1, and 0.0 for specimens 1 through 5, respectively. Patients 2 and 7 had atypical D-FISH patterns. Patient 6 had a masked Ph chromosome.
The y-axis shows the percentage of Ph-positive cells in each graph. The x-axis for patients 1 through 10 is the number of days treated with IFN-α2b with or without ara-C. The x-axis for serial dilutions represents different mixtures of normal and Ph-positive cells: the expected percentage of Ph-positive nuclei is 98.2, 49.1, 12.3, 3.1, and 0.0 for specimens 1 through 5, respectively. Patients 2 and 7 had atypical D-FISH patterns. Patient 6 had a masked Ph chromosome.
Three patients in part II of the investigation had an atypical D-FISH pattern. Patient 2 had nuclei with 1R2G1F resulting from BCR/ABL fusion on the Ph chromosome and a small residual ABL signal, but no BCR signal, on abnormal chromosome 9 (Fig 1J). Patient 7 had nuclei with 1R1G1F resulting from BCR/ABL fusion on the Ph chromosome, but no BCR or ABL signal on the abnormal chromosome 9 (Fig 1G). Patient 6 had a masked Ph chromosome, and the D-FISH pattern was consistent with an insertion of a portion of ABL into the BCR locus (Fig 1J).
This series included samples of five different artificial mosaics created by mixing cells from normal specimens with cells from a specimen with a classic t(9;22)(q34;q11.2). The percentage of abnormal nuclei in each of these samples as determined by D-FISH was closer to the expected proportion of Ph-positive nuclei than S-FISH (Fig 2, serial dilutions).
Based on cytogenetic studies during the course of therapy, six patients (nos. 2, 4, 5, 7, 8, and 9) attained less than 33% Ph-positive cells and 3 (nos. 1, 3, and 10) showed no notable change in percentage of Ph-positive metaphases (Fig 2). In assessing the results in seven specimens with typical D-FISH patterns, we found the results of D-FISH to be a better predictor of the findings by Q-cytogenetics than S-FISH based on a mixed model regression analysis, adjusting for within subject correlations.
Patients 2 and 7 had atypical D-FISH patterns and eventually attained more than 67% normal metaphases in response to therapy. In each of these specimens, the percentage of abnormal nuclei by D-FISH was somewhat comparable to S-FISH but still correlated strongly with changing proportions of abnormal nuclei after therapy. Patient 6 had a masked Ph chromosome that was not detected by conventional cytogenetics but that was detected by both D-FISH and S-FISH methods. The percentage of abnormal nuclei for this patient never fell below 80% based on D-FISH. The results of patients 2, 6, and 7 show that D-FISH can be used to monitor response to therapy, even when the Ph anomaly is associated with an atypical signal pattern.
Resolution Limits to Detect Residual Disease
In part III of the investigation, we tested the hypothesis that analysis of more than 200 nuclei with D-FISH would significantly increase the ability to detect low levels of Ph-positive nuclei. To test this possibility, 6,000 nuclei from each of six specimens were studied in a blind fashion. Three were from normal specimens from part I of the investigation and three specimens were from two patients (nos. 5 and 8) with CML in cytogenetic remission from part II.
We observed one aberrant cell among 6,000 nuclei in only one of the three specimens from normal individuals (Table2). On the basis of this finding, we calculated the upper bound of a one-sided 95% confidence interval for observing 1 of 6,000 (0.0167%) aberrant cells using the binomial distribution. This implied a normal cutoff of greater than .079% (>4) aberrant cells in 6,000 nuclei to classify any specimen as abnormal.
Limits of D-FISH Resolution for Detection of Ph-Positive Cells in 6,000 Nuclei
| Specimen . | Diagnosis . | Treatment (days) . | Abnormal Nuclei* . | Abnormal Nuclei* (%) . | Analysis Time (h:min) . |
|---|---|---|---|---|---|
| Part I, no. 40 | Normal | 0 | 1 | 0.017 | 2:38 |
| Part I, no. 44 | Normal | 0 | 0 | 0.000 | 2:29 |
| Part I, no. 46 | Normal | 0 | 0 | 0.000 | 2:37 |
| Part II, pt 5, no. 3 | Cytogenetic remission | 272 | 21 | 0.350 | 3:03 |
| Part II, pt 5, no. 4 | Cytogenetic remission | 391 | 7 | 0.117 | 3:25 |
| Part II, pt 8, no. 3 | Cytogenetic remission | 261 | 53 | 0.883 | 2:41 |
| Specimen . | Diagnosis . | Treatment (days) . | Abnormal Nuclei* . | Abnormal Nuclei* (%) . | Analysis Time (h:min) . |
|---|---|---|---|---|---|
| Part I, no. 40 | Normal | 0 | 1 | 0.017 | 2:38 |
| Part I, no. 44 | Normal | 0 | 0 | 0.000 | 2:29 |
| Part I, no. 46 | Normal | 0 | 0 | 0.000 | 2:37 |
| Part II, pt 5, no. 3 | Cytogenetic remission | 272 | 21 | 0.350 | 3:03 |
| Part II, pt 5, no. 4 | Cytogenetic remission | 391 | 7 | 0.117 | 3:25 |
| Part II, pt 8, no. 3 | Cytogenetic remission | 261 | 53 | 0.883 | 2:41 |
*Normal cutoff is >4 abnormal nuclei or more precisely ≥0.079%.
We compared this cutoff with the results for each of the three specimens from patients with CML in cytogenetic remission (Table 2). One specimen, from patient 8, was within normal D-FISH limits for 200 nuclei after 261 days of treatment. In 6,000 cells, we found 53 (0.883%) abnormal nuclei. Patient 5 had two specimens within normal D-FISH limits for 200 nuclei after 272 days and 391 days of treatment. In 6,000 nuclei, we found 21 (0.350%) abnormal nuclei in the first of these two specimens and 7 (0.117%) in the other. Thus, in each specimen these results exceeded the normal cutoff and were consistent with residual disease.
DISCUSSION
True D-FISH
A true D-FISH method would detect a BCR/ABL fusion on the Ph chromosome and an ABL/BCR fusion on the abnormal chromosome 9 in cells with a t(9;22)(q34;q11.2). Thus, normal cells would have four signals, 2R2G, and nuclei with a t(9;22)(q34;q11.2) would have six signals, 1R1G2F, with fusion signals counting as two hybridization sites.
With the current D-FISH strategy, the ABL/BCR fusion on chromosome 9 was observed as separate ABL and BCR signals in as many as 24% of nuclei with a 9;22 translocation. We scored this pattern as 2R2G1F because we defined a fusion signal as touching or merging BCR and ABL signals. Nevertheless, we classified nuclei with 2R2G1F as Ph positive because they had six BCR and ABL signals; the same as the true D-FISH pattern of 1R1G2F.
With D-FISH, incorrectly scoring a normal nucleus (2R2G) as abnormal (1R1G2F or 2R2G1F) would be rare. Such false-positive observations would require multiple events, including separation of BCR and ABL signals and random overlap of at least one BCR with an ABL signal. Our results are consistent with this conclusion, as we observed only one false-positive cell in 18,000 nuclei in part III of this investigation and 15 of 7,000 nuclei in part I.
The accuracy of scoring with D-FISH improved with experience. In part I of the investigation, we found 15 false-positive nuclei among 7,000 cells from 35 normal individuals. In part III, we found only 1 false-positive nucleus in 18,000 cells from three normal specimens. Part I was performed first, when our technologists had the least experience with scoring D-FISH signals. Part III was performed last and benefited from the learning experience of scoring cells in parts I and II.
Incorrectly scoring abnormal nuclei (1R1G2F or 2R2G1F) as normal (2R2G) would also be rare. Observation of a false-negative nuclei would require multiple events, including the separation of all BCR/ABL fusion signals in a nucleus and random overlap of two BCR signals with each other and two ABL signals with each other. Scoring true-positive cells is reproducible with D-FISH, as the percentage of Ph-positive nuclei that we found in the same specimen processed on 10 separate occasions ranged from 97 to 100, with a mean of 98.2.
D-FISH is a good method for quantifying disease even in untreated patients with CML. The frequency of normal cells in five untreated patients from part I of the investigation with only a 9;22 translocation ranged from 95.0 to 98.5, with a mean of about 97.4. We believe that the few nuclei with a normal pattern represent true normal cells, rather than false-negative observations, because of the objectivity of scoring D-FISH, and because not all cells in each hematopoietic compartment are usually involved in the neoplastic process in CML.
We intentionally used strict scoring criteria in this investigation to reduce the chances of false-positive and false-negative signals. This resulted in rejecting approximately 45% of nuclei in normal specimens and approximately 60% of nuclei in abnormal specimens in the scoring process. The higher incidence of unscorable nuclei in abnormal specimens may be attributed to the greater number of hybridization sites with potential for technical problems. We do not believe this finding significantly biased the data toward a normal or abnormal outcome.
D-FISH Patterns in Various Kinds of Ph Chromosomes
In 57 of 65 specimens in part I of the investigation, the D-FISH patterns met the scoring criteria. Except for masked Ph chromosomes, typical D-FISH patterns were found in specimens from each of the different kinds of Ph chromosomes (Table 1). With D-FISH, complex Ph chromosomes can be distinguished from classic 9;22 translocations by virtue of a higher proportion of abnormal nuclei with 2R2G1F. In complex translocations, a BCR/ABL fusion occurs on the Ph chromosome, and no reciprocal ABL/BCR fusion occurs because the telomeric portion of BCR is translocated to another chromosome. We saw similar proportions of nuclei with 2R2G1F in specimens with simple and complex Ph chromosomes. This is consistent with the findings and conclusions of others who suggest that simple Ph-chromosomes are actually subtle, complex Ph chromosomes.7 14
Thus, excluding the five specimens with a masked Ph chromosome, in part I of the investigation, 22 of 25 specimens with a Ph chromosome met the scoring criteria. In part II, we studied 10 patients selected because we had three or more specimens to test the efficacy of D-FISH to monitor therapy. Excluding the single patient with a masked Ph chromosome, seven of nine patients met the scoring criteria. These findings may suggest that approximately 78% to 88% of patients with CML in clinical practice would have a D-FISH pattern that would meet these scoring criteria.
Atypical D-FISH Patterns
In this study, eight specimens from part I and three patients from part II had abnormal but atypical D-FISH patterns. The scoring criteria were not designed for masked Ph chromosomes. Nevertheless, we expected to observe nuclei with either 1R2G1F or 2R1G1F in these specimens, because masked Ph chromosomes result from the submicroscopic insertion translocation of either a portion of ABL into BCR or vice versa.7 The masked Ph chromosome in specimens 23 and 25 of part I, and patient 6 in part II, had a portion of ABL translocated into the BCR locus.
Specimen 22 from part I, had a D-FISH pattern that was consistent with an insertion of a portion of BCR into the ABL locus.
Two other specimens from part I of the investigation (nos. 21 and 24) had a masked Ph chromosome with no BCR or ABL signal on a chromosome 9, but they did have a BCR/ABL fusion on a chromosome 22. This could result by an insertion translocation of ABL into BCR, but the two breakpoints to excise ABL would have had to occur outside the hybridization sites. Moreover, specimens 7 and 9 from Part I, and patient 7 from part II, had typical 9;22 translocations, but lacked a BCR and ABL signal on the abnormal chromosome 9. This could result from a break on chromosome 9 on the centromeric side of the ABL hybridization site and a break on chromosome 22 on the telomeric side of BCR.
The aforementioned theories for the origin of atypical D-FISH patterns in masked and 9;22 translocations are problematic, as each of these mechanisms should produce a BCR/ABL fusion signal on the Ph chromosome approximately twice the usual size. Subjectively, the BCR/ABL fusion signal in these specimens did not appear larger than usual, favoring an alternative theory. It is possible that the breakpoints are within the usual BCR and ABL locus, but the ABL/BCR hybridization site for chromosome 9 may have been lost by a microdeletion process during the formation of some masked Ph chromosomes and 9;22 translocations.
Patient 2 in part II of the investigation had a typical 9;22 translocation, but the D-FISH pattern was 1R2G1F. Metaphases had no BCR signal on chromosome 9, a small residual ABL signal was present on the abnormal chromosome 9. This could result from either a microdeletion of a portion of the BCR hybridization site or a break on the Ph chromosome that was on the telomeric side of the BCR hybridization site.
Specimen 12 from part I of the investigation had a t(9;22;19) anomaly and an atypical D-FISH pattern, 1R2G1F. Metaphases had a BCR/ABL fusion on the Ph chromosome, a small residual ABL on the abnormal chromosome 9, but no BCR signal on the abnormal chromosome 19. This could result from a break in the usual ABL locus and a break on the telomeric side of BCR or a microdeletion process that resulted in loss of a portion of the BCR locus.
Although the scoring criteria did not include these atypical patterns, these specimens were clearly abnormal and required special scoring criteria. For investigators who will use D-FISH, it will be important to account for these atypical patterns on a patient-to-patient basis. For example, it appears from patients 2, 6, and 7 in part II that the atypical patterns persist in cells before treatment and throughout the treatment period. We did not attempt to calculate normal or abnormal reference ranges for specimens with atypical patterns, because they varied and the sample size was small. We expect the percentage of false-positive and false-negative nuclei to be higher in these atypical cases than for patients who meet the D-FISH scoring criteria.
Monitoring Response to Treatment
Based on analysis of 200 nuclei from each of the 30 normal specimens in part I of the investigation, we calculated an upper limit of the normal range at 3.83% abnormal nuclei. This is a conservative estimate of the normal cutoff, because it is based on using the maximum number of false-positive cells in any one patient in a series (eg, 1.5%) and then calculating the 95th percentile for all normals using the binomial distribution. We used a similar statistical method to calculate a normal cutoff for S-FISH at 10%,7 a figure that has served us and others well in clinical practice to arrive at an accurate diagnosis of CML.
The percentage of Ph-positive metaphases as determined by conventional cytogenetic methods is considered the current “gold standard” for measuring response to therapy in CML. Most cytogenetic laboratories analyze 20 metaphases selected primarily on the basis of metaphase morphology. This approach is adequate to detect cells with a Ph chromosome but is susceptible to sample error and bias toward metaphases with good morphology. These factors can interfere with the reliability of conventional cytogenetics to quantify residual disease in response to therapy. Consequently, we have modified the conventional method to select consecutive metaphases in which the 9q34 and 22q11.2 regions of chromosomes 9 and 22 are visible, placing less emphasis on quality of metaphase morphology. We refer to this method as quantitative cytogenetics or Q-cytogenetics.15 In comparing the results of Q-cytogenetics–based analysis of 25 metaphases with the results of analysis of 200 consecutive metaphases using S-FISH, we have found that the results are usually within approximately 5%. Thus, we believe Q-cytogenetics is an efficacious and cost-effective method to quantify proliferating disease in CML.
Each of the specimens from patients in part II of the investigation was studied using Q-cytogenetics, S-FISH, and D-FISH. For the seven patients who had “true D-FISH” patterns, the results of D-FISH were a significantly better predictor of the results of Q-cytogenetics than S-FISH. Based on analysis of only 200 nuclei, D-FISH is comparable to cytogenetics before and after treatment. However, D-FISH would be far superior to S-FISH to monitor patients with CML on therapy (Fig 2).
We did not attempt to correlate the results of D-FISH with any clinical parameter or survival. Because the results correlate well with cytogenetics, D-FISH should prove to have comparable prognostic value, but this will not be known until D-FISH is used to study a large series of patients over a long period of time.
Limits of Resolution to Detect Residual Disease
Based on analysis of 200 nuclei with D-FISH, in part I of the investigation we found that it is possible to detect residual disease corresponding to 3.83% or greater abnormal cells. In part III, we found that, with D-FISH, the limits of detecting residual disease could be reduced to 0.079% by analysis of 6,000 nuclei. For patients in part II of the investigation, only three specimens fell within normal limits for D-FISH, based on analysis of 200 nuclei. Each of these specimens had only normal metaphases by Q-cytogenetics. However, based on analysis of 6,000 nuclei, using D-FISH, each specimen had residual disease ranging from 7 (0.117%) nuclei in one specimen to 53 (0.883%) nuclei in another.
The effort needed to score 6,000 nuclei ranged from approximately 2.5 to 3.25 hours, compared with 5 to 10 minutes to score 200 nuclei. Perhaps a cost-effective strategy to use D-FISH for monitoring patients with CML receiving therapy would be to study 200 nuclei in bone marrow specimens collected before treatment to establish a baseline for the percentage of Ph-positive cells and to determine whether the D-FISH pattern is typical or atypical. If the patient has a typical D-FISH pattern for a Ph chromosome, D-FISH can be used after treatment until the patient achieves a level of less than 3.83% abnormal nuclei. The normal cutoff for 200 nuclei approximates cytogenetic remission, ie, 25 of 25 normal metaphases by Q-cytogenetics. For specimens within normal limits for 200 nuclei, it may be useful to study up to 6,000 nuclei to detect residual disease as low as 0.079% Ph-positive cells. If the patient has an atypical D-FISH pattern, a different normal cutoff would have to be established.
In conclusion, D-FISH is a significant technological advancement in monitoring therapy for CML. D-FISH detects all variant translocations of the Ph chromosome and accurately quantifies disease in CML at diagnosis and at all times during treatment, even for patients in cytogenetic remission.
Supported in part by a grant from Oncor Inc, Gaithersburg, MD. The Chronic Myeloid Leukemia National Study Group provided specimens from some patients and is supported in part by Integrated Therapeutics Group Inc. Research support was also provided by the United Leukemia Fund, Inc., and the Cancer Research and Treatment Fund, Inc.
Address reprint requests to Gordon W. Dewald, PhD, Cytogenetics Laboratory, Mayo Clinic, 200 First St SW, Rochester, MN 55905.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal