Abstract
Recent observations indicate that chromosome aberrations are important prognostic factors in patients with multiple myeloma (MM) treated with high-dose chemotherapy. Nevertheless, the inherent problems of conventional cytogenetics have hampered the systematic evaluation of this parameter in series of patients treated with conventional chemotherapy. Fluorescence in situ hybridization (FISH) analysis is an attractive alternative for evaluation of numerical chromosomal changes. In the present study, we analyze the relationship between aneuploidies of 15 different chromosomes assessed by FISH and prognosis in a series of 63 patients with MM treated with conventional chemotherapy. After a median follow-up of 61 months (range, 6 to 109), 49% of patients are still alive with a median survival of 33 months. The overall incidence of numerical chromosome abnormalities was 70%. This incidence significantly increased when seven or more chromosomes were analyzed (53 patients), reaching 81%. Trisomies of chromosomes 6, 9, and 17 were associated with prolonged survival (P = .033, P = .035, and P = .026, respectively); by contrast, overall survival (OS) was lower in cases with monosomy 13 (as assessed by deletion of Rb gene,P = .0012). From the clinical point of view, loss of Rb gene was associated with a poor performance status; low hemoglobin levels; high creatinine, C-reactive protein, and lactic dehydrogenase serum levels; high percentage of bone marrow plasma cells (BMPC); extensive bone lytic lesions; and advanced clinical stage. Other chromosome abnormalities such as trisomy of chromosome 9 and 17 were associated with good prognostic features including high hemoglobin levels, early clinical stage, β2microglobulin less than 6 μg/mL, and low percentage of BMPC. A multivariate analysis for OS showed that S-phase PC greater than 3% (P = .010) and β2microglobulin serum levels greater than 6 μg/mL (P = .024), together with monosomy of chromosome 13 (P = .031) and nontrisomy of chromosome 6 (P = .048) was the best combination of independent parameters for predicting survival in patients with MM. According to these results, chromosomal analysis is of great use in patients with MM at diagnosis to have a correct prognostic evaluation for clinical decision making.
THE SURVIVAL OF patients with multiple myeloma (MM) ranges from a few months to more than 10 years (median, 2.5 to 3 years). For the last 30 years, investigators have attempted to identify clinical and laboratory features affecting survival of patients with MM. At present, β2microglobulin and C-reactive protein serum levels, together with the proliferative activity of plasma cells (labeling index or proportion of S-phase plasma cells [PC]) are considered to be the best prognostic indicators for predicting survival.1-6 Recently, a poor prognostic value has been shown in patients with monosomy and/or deletions of chromosome 13 treated with high-dose chemotherapy.7,8 Nevertheless, the availability of cytogenetic information in MM has long been hampered by the low mitotic activity of the myelomatous PC as well as by their poor growth in cell culture, which make it difficult to obtain metaphases from neoplastic cells for conventional chromosome analysis. Cytogenetic studies have shown a 20% to 50% incidence of abnormal karyotypes in patients with MM.9-12 This incidence increases up to 60% when bone marrow (BM) cells are incubated with interleukin-3 (IL-3) and IL-6.13 However, it is important to note that the majority of these studies have failed to obtain metaphases in many patients; therefore, it is not possible to say whether or not they have an abnormal karyotype.9-12 Procedures that can be applied to both metaphase cells and interphase nuclei, such as the flow cytometric measurement of cell DNA content and fluorescence in situ hybridization (FISH), are very well suited to the analysis of cytogenetic abnormalities in tumor cells. Studies using FISH and conventional cytogenetics simultaneously have shown that FISH can detect chromosome abnormalities which are hidden to metaphase cytogenetic analysis.14 Thus, FISH studies have shown an incidence of aneuploidy in 67% to 90% of patients14,15—higher than that obtained by conventional cytogenetics (20% to 50%). Despite these limitations, cytogenetic abnormalities, and particularly deletions of chromosome 13 and abnormalities involving the 11q chromosome, have been proven to be associated with poor survival9,10 in patients undergoing autologous transplantation.7 8 However, until now, the prognostic influence of chromosome aberrations detected by FISH in patients treated with conventional chemotherapy has not been explored.
In the present study, we analyze the relationship between aneuploidy of 15 different chromosomes assessed by FISH and prognosis in a series of 63 untreated patients with MM. Deletion of chromosome 13 was found to be an independent prognostic factor, associated with a significantly poor outcome, whereas trisomies of chromosomes 6, 9, 11, and 17 were related to a better prognosis.
MATERIALS AND METHODS
Patients.
Sixty-three previously untreated patients with MM diagnosed according to the criteria of the Chronic-Leukemia-Myeloma Task Force were included in the study.2 The mean age of the series was 66 ± 8 years (range, 46 to 82). According to Durie & Salmon's clinical staging system, the patients were distributed as follows: stage I, 5.5%; stage II, 32.7%; stage III, 61.8%.2 The monoclonal component was IgG in 50% of cases, IgA in 28%, IgD in 3%, and Bence Jones in the remaining 19% of patients. In one case no monoclonal serum protein was found. The serum monoclonal light chain was Κ in 58% and λ in 42%. Forty patients were men and 23 were women. Three patients presented plasmocytomas at diagnosis. All patients were treated according to the protocols of the PETHEMA (Spanish Cooperative Group for the Treatment of Malignant Hemopathies) group.16
Immunophenotypic identification of PC.
The percentage of BMPC was assessed by two different observers on May-Grünwald-Giemsa stained smears, the mean value being 49% ± 25%. The immunophenotypic identification of PC was based on their strong reactivity for the CD38 (Leu17; Becton Dickinson, San José, CA) and CD138 (Imico, Madrid, Spain) monoclonal antibodies (MoAbs) whose specificity has been described elsewhere.17-19 The immunophenotypic characterization of the PC was performed by direct immunofluorescence using the following MoAbs: Leu19 (CD56), Leu16 (CD20), antiHLA-DR (HLA DR), LeuM7 (CD13), LeuM1 (CD15; Becton Dickinson), FMC56 (CD9; FMC, Adelaide, Australia), and C-kit (CD117; Imico), together with the previously mentioned CD38 and CD138 reagents. For data acquisition and analysis, a FACScan flow cytometer (Becton Dickinson), equipped with an argon ion laser tuned at 488 nm and 15 mW was used. Results were stored and analyzed for at least 10,000 cells per test, using the LYSYS-II and PAINT-A-GATE PRO software programs (Becton Dickinson), respectively.17 18
FISH studies.
FISH analysis of numerical chromosome abnormalities was performed on erythrocyte-lysed whole BM samples obtained before cytotoxic treatment, according to previously described methods.15-18 Samples were hybridized as previously described with either a biotinylated, fluoresceinated, or digoxigenin-labeled alpha-satellite DNA probes20-22 specific for either the centromeric or the pericentromeric regions of the following chromosomes: 1 (pUC1.77; Boehringer Mannheim), 3 (pAE0.68; Boehringer Mannheim), 6 (D6Z1; Oncor, Gaithersburg, MD), 7 (pZ7.6B; Boehringer Mannheim), 8 (pZ8.4; Boehringer Mannheim), 9 (D9Z1; Oncor), 10 (CEP10; Vysis, Framingham, MA), 11 (CEP11; Vysis), 12 (D12Z3; Oncor), 15 (pMC15; Boehringer Mannheim), 17 (pZ17-1.6A; Boehringer Mannheim), 18 (pZXba; Boehringer Mannheim), X (pDMX1; Boehringer Mannheim), and Y (pHY2.1; Boehringer Mannheim). In addition, a locus-specific DNA probe for the Rb gene sequence in chromosome 13 was used (LSI13; Vysis). To probe the efficacy of the hybridization for chromosome 13, cohybridization was simultaneously performed using a chromosome 10 probe; only cells with clear signals for the control probe (chromosome 10) were scored. The number of hybridization spots were evaluated using a DMRB fluorescence microscope (Leitz, Wetzlar, Germany) equipped with a 100× oil-objective, which was used for counting hybridization spots per cell in at least 200 cells per sample. In all slides analyzed, the number of unhybridized cells in the areas assessed was lower than 1% and only those spots with a similar size, a strong intensity, and a round shape were counted. The mean percentage of trisomic/monosomic cells in control samples ranged between 0% to 2% for trisomies and 0% to 5% for monosomies.15
A patient was considered to carry a numerical chromosomal abnormality for a certain chromosome when the percentage of cells displaying an abnormal number of spots was at percentages higher than the mean value plus two SDs of the percentages obtained for that specific chromosome in normal controls.
The expected amount of total DNA cell content according to the FISH results was assessed by the chromosome index calculated as previously reported.15
Flow cytometry DNA measurements.
DNA measurements by flow cytometry were performed as previously described.15,18 22 Briefly, BM cells were incubated for 15 minutes with 10 μL of the GR7A4 (CD38) and BB4 (CD138) MoAbs, washed once (5 minutes, 1,900 rpm) in phosphate-buffered saline, and incubated for another 15 minutes with an MoAb anti-mouse Igs (F[ab′]2 fragments; Dakopatts, Copenhagen, Denmark). Afterwards, 2 mL of ammonium chloride were added and cells were incubated in the dark for 10 minutes. After lysing the erythrocytes, cells were washed once in 1 mL of sodium citrate buffer and resuspended in 200 μL of the same buffer. Then, 1.5 mL of solution containing RNAse and Nonident P40 were added and the cells were incubated for 10 minutes. Finally, 1.5 mL of solution containing propidium iodide (PI) was added and another incubation period was performed for at least 15 minutes in the dark.
In all cases, measurements were performed within 1 hour on a FACScan flow cytometer (Becton Dickinson) using the Lysis II software program (Becton Dickinson), for at least 10,000 cells per sample. The electronics of the instrument were adjusted so that the modal channel for the G0/G1 diploid nuclei was 200 (fluorescence scaled from channel 0 to 1023). Fluorescence compensation between fluorescein isothiocyanate (FITC) and PI was established using a mixture of PI-stained chicken erythrocyte nuclei and FITC-labeled beads (CALIBRITE beads; Becton Dickinson). The percentage of CD38/CD138 strong positive PC was calculated after gating out cell doublets on a FL2A/FL2W dot plot using the PAINT-A-GATE PRO software (Becton Dickinson).
The criteria for flow cytometry DNA aneuploidy was defined by the presence of two distinct peaks of G0/G1-phase cells in the DNA/propidium iodide histogram. The positivity for the CD38/CD138 MoAbs was used to identify which of the G0/G1 peaks corresponded to myelomatous PC (CD38/CD138 strong positive events) and the normal residual hemopoietic cells (CD38/CD138 negative or dim/intermediate positive events). The analysis of the cell cycle was performed on CD38/CD138 strong positive gated events using the MOD FIT software (Verity Software Home, Topsham, ME).
Statistical methods.
To estimate the significance of the differences observed between means, the Student's t-test was used (t-test, SPSS). The Chi-square test (SPSS, chi-square) was used for dichotomic variables. Overall survival (OS) curves were plotted according to the method of Kaplan and Meier, and compared using the Mantel-Cox, Peto-Prentice, and Breslow tests.
The different clinical and biological characteristics were considered individually for their relationship with OS by univariate tests (t-test, Chi-square, correlation, and nonparametric tests, SPSS). Subsequently, a multivariate analysis—stepwise regression—(regression, SPSS)23 24 was performed to examine the simultaneous effect of the different variables. Variables considered for possible inclusion in the regression analysis were those displaying a significant association with survival in the univariate analysis (P < .05) or for which prior studies had suggested a possible prognostic value. The stepwise regression method was discontinued when the P value for entering an additional factor was greater than .05. The model was tested both by including the variables in a continuous fashion (continuous model) and by grouping them into categories (dichotomous model).
RESULTS
After a median follow-up of 61 months (range, 6 to 109), 49% of patients remain alive, with a median survival of 33 months (range, 12 days to 109 months).
The overall incidence of aneuploidies assessed by FISH analysis in patients with MM included in the present study was 70%. This incidence increased to 81% when only those cases in which seven or more chromosomes were analyzed (53 patients). Overall, trisomies were more frequent than monosomies (84% v 16%). Table1 shows the distribution of numerical chromosomal abnormalities according to each of the 15 chromosomes analyzed in the present study. Chromosome 9 (55.8%), chromosome 1 (44.9%), and chromosome 15 (43.7%) were the most frequently altered. Of the remaining chromosomes analyzed, most displayed trisomies either as the only chromosomal abnormality detected (chromosomes 3, 6, 7, 10, 12, and 17) or the most frequent one (chromosomes 8, 11, and 18). In contrast, monosomies were the only numerical abnormality detected for chromosome 13 (33.3%). There was a frequent association between chromosome abnormalities. Thus, trisomy 6 was significantly associated with gains of chromosomes 7 (P = .0001), 9 (P = .001), 10 (P = .0002), and 17 (P = .005). In addition, abnormalities of chromosomes 1, 7, and 11 were associated with those of chromosomes 3 (P = .0005), 6 (P = .0002), and 17 (P = .0006), respectively.
Incidence of Numerical Chromosome Changes in Patients With MM According to the Different Chromosomes Analyzed
| Chromosome Abnormality . | Chromosome . | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 . | 3 . | 6 . | 7 . | 8 . | 9 . | 10 . | 11 . | 12 . | 13 . | 15 . | 17 . | 18 . | Xm . | Xf . | |
| Trisomy | 19/49 | 10/30 | 15/55 | 14/47 | 3/39 | 29/52 | 3/38 | 13/40 | 1/39 | 13/32 | 10/46 | 9/35 | 1/39 | ||
| 38.8 | 33.3 | 27.3 | 29.8 | 7.7 | 55.8 | 7.9 | 32.5 | 2.6 | 40.6 | 21.7 | 25.7 | 4.5 | |||
| Monosomy | 3/49 | 2/39 | 1/40 | 16/48 | 1/32 | 2/35 | 6/19 | ||||||||
| 6.1 | 5.1 | 2.5 | 33.3 | 3.1 | 5.7 | 31.5 | |||||||||
| Total | 22/49 | 10/30 | 15/55 | 14/47 | 5/39 | 29/52 | 3/38 | 14/40 | 1/39 | 16/48 | 14/32 | 10/46 | 11/35 | 1/39 | 6/19 |
| 44.9 | 33.3 | 27.3 | 29.8 | 12.8 | 55.8 | 7.9 | 35 | 2.6 | 33.3 | 43.7 | 21.7 | 31.4 | 4.5 | 31.5 | |
| Chromosome Abnormality . | Chromosome . | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 . | 3 . | 6 . | 7 . | 8 . | 9 . | 10 . | 11 . | 12 . | 13 . | 15 . | 17 . | 18 . | Xm . | Xf . | |
| Trisomy | 19/49 | 10/30 | 15/55 | 14/47 | 3/39 | 29/52 | 3/38 | 13/40 | 1/39 | 13/32 | 10/46 | 9/35 | 1/39 | ||
| 38.8 | 33.3 | 27.3 | 29.8 | 7.7 | 55.8 | 7.9 | 32.5 | 2.6 | 40.6 | 21.7 | 25.7 | 4.5 | |||
| Monosomy | 3/49 | 2/39 | 1/40 | 16/48 | 1/32 | 2/35 | 6/19 | ||||||||
| 6.1 | 5.1 | 2.5 | 33.3 | 3.1 | 5.7 | 31.5 | |||||||||
| Total | 22/49 | 10/30 | 15/55 | 14/47 | 5/39 | 29/52 | 3/38 | 14/40 | 1/39 | 16/48 | 14/32 | 10/46 | 11/35 | 1/39 | 6/19 |
| 44.9 | 33.3 | 27.3 | 29.8 | 12.8 | 55.8 | 7.9 | 35 | 2.6 | 33.3 | 43.7 | 21.7 | 31.4 | 4.5 | 31.5 | |
Results expressed as number of abnormalities found/number of cases studied (percentage below). Chromosome Y was normal in all men studied (n = 36).
Abbreviations: Xm, chromosome X in men; Xf, chromosome X in women.
Concerning the clinical implications of numerical chromosome abnormalities, it was found that monosomy of chromosome 13 was significantly associated with a poor performance status; low hemoglobin levels; high creatinine, C-reactive protein, and lactic dehydrogenase (LDH) serum levels; high percentage of BMPC; extensive bone lytic lesions; and advanced stage. This relationship with poor prognostic features was confirmed in the survival analysis (see below). Other chromosome abnormalities such as trisomies of chromosome 9 and 17 were associated with good prognostic features including high hemoglobin levels, early clinical stage, β2microglobulin less than 6 μg/mL, and low percentage of BMPC. Other associations between numerical chromosome abnormalities and clinical characteristics are listed in Table2.
MM Relationship Between the Presence of Numerical Chromosome Abnormalities and Clinico-Biological Characteristics of the Disease
| Chr . | Age . | PS . | Hb . | Ca . | Creat . | Bone . | Stage . | β2M . | Crp . | LDH . | PC . | BJ . | Resp . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | — | — | — | — | — | — | — | — | ↑ | — | — | — | Better |
| 3 | — | — | — | — | ↓ | ↓ | — | — | — | — | — | — | — |
| 6 | — | — | — | ↓ | — | — | — | — | ↓ | — | — | — | — |
| 7 | — | — | — | — | — | — | Early | — | — | — | — | — | — |
| 8 | — | — | — | — | — | — | — | — | — | — | — | Absent | — |
| 9 | — | — | — | ↓ | — | — | — | ↓ | — | — | — | — | — |
| 10 | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 11 | — | — | — | — | ↓ | — | — | — | — | — | — | — | — |
| 12 | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 13 | — | ↑ | ↓ | — | ↑ | ↑ | Advanced | — | ↑ | ↑ | ↑ | — | — |
| 15 | — | — | — | — | — | — | — | — | — | — | — | Present | — |
| 17 | — | — | ↑ | — | — | ↓ | Early | ↓ | — | — | ↓ | — | — |
| 18 | — | — | — | — | ↓ | — | — | — | — | — | ↓ | — | — |
| X | — | — | — | — | — | — | — | — | — | — | — | — | — |
| Chr . | Age . | PS . | Hb . | Ca . | Creat . | Bone . | Stage . | β2M . | Crp . | LDH . | PC . | BJ . | Resp . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | — | — | — | — | — | — | — | — | ↑ | — | — | — | Better |
| 3 | — | — | — | — | ↓ | ↓ | — | — | — | — | — | — | — |
| 6 | — | — | — | ↓ | — | — | — | — | ↓ | — | — | — | — |
| 7 | — | — | — | — | — | — | Early | — | — | — | — | — | — |
| 8 | — | — | — | — | — | — | — | — | — | — | — | Absent | — |
| 9 | — | — | — | ↓ | — | — | — | ↓ | — | — | — | — | — |
| 10 | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 11 | — | — | — | — | ↓ | — | — | — | — | — | — | — | — |
| 12 | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 13 | — | ↑ | ↓ | — | ↑ | ↑ | Advanced | — | ↑ | ↑ | ↑ | — | — |
| 15 | — | — | — | — | — | — | — | — | — | — | — | Present | — |
| 17 | — | — | ↑ | — | — | ↓ | Early | ↓ | — | — | ↓ | — | — |
| 18 | — | — | — | — | ↓ | — | — | — | — | — | ↓ | — | — |
| X | — | — | — | — | — | — | — | — | — | — | — | — | — |
Abbreviations: Chr, chromosome; PS, performance status according to the Eastern Cooperative Oncology Group scale; Hb, hemoglobin levels; Ca, calcium levels; Creat, creatinine levels; Bone, lytic bone lesions; Stage, Durie & Salmon clinical stage; β2M, β2 microglobulin serum levels; Crp, C-reactive protein levels; PC, percentage of BM plasma cells (morphology); BJ, Bence Jones protein; Resp, response to first line of treatment; —, no relationship; ↑, higher in cases with the chromosome abnormality; ↓, lower in cases with the chromosome abnormality (with statistical significance [P < .05]).
As far as the response to treatment was concerned, no major differences were observed regarding the presence of specific abnormalities with the exception of a lower rate of responses in patients with abnormalities of chromosome 1 (47% in trisomic cases v 78% in cases without trisomy 1; P = .04).
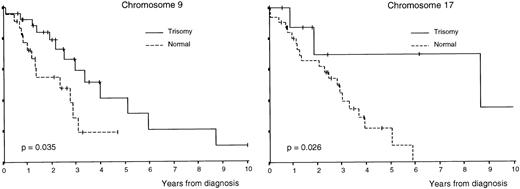
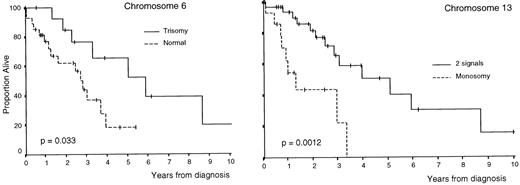
OS was superior in patients with trisomies of chromosomes 6, 9, and 17 compared with that of cases without trisomy (P = .033,P = .035, and P = .026, respectively; Table3 and Figs 1 and2); by contrast, OS was lower in cases with monosomy of chromosome 13 (P = .0012; Table 3 and Fig 2). In addition, all clinical and biological disease characteristics were considered individually for their relationship with OS by univariate tests. The variables for which a statistically significant relationship with OS was found are summarized in Table 4.
Univariate Analysis of the Prognostic Value for the OS of the Different Chromosome Abnormalities
| Chromosome . | N . | Events . | Survival (mo) . | PUnivariate . | |
|---|---|---|---|---|---|
| 1 | Trisomy | 19 | 11 | 29 | .49 |
| Nontrisomy | 30 | 15 | 34 | ||
| 3 | Trisomy | 10 | 5 | 35 | .84 |
| Nontrisomy | 20 | 11 | 39 | ||
| 6 | Trisomy | 15 | 8 | 70 | .033* |
| Nontrisomy | 40 | 16 | 33 | ||
| 7 | Trisomy | 14 | 6 | 46 | .11 |
| Nontrisomy | 33 | 20 | 32 | ||
| 8 | Trisomy | 3 | 3 | 39 | .86 |
| Nontrisomy | 36 | 20 | 33 | ||
| 9 | Trisomy | 29 | 13 | 46 | .035* |
| Nontrisomy | 23 | 13 | 26 | ||
| 10 | Trisomy | 3 | 0 | — | .15 |
| Nontrisomy | 35 | 24 | 33 | ||
| 11 | Trisomy | 13 | 6 | 70 | .060 |
| Nontrisomy | 27 | 14 | 33 | ||
| 12 | Trisomy | 1 | 1 | 22 | .21 |
| Nontrisomy | 38 | 22 | 34 | ||
| 13 | 1 signal | 16 | 9 | 14 | .0012* |
| Nontrisomy | 32 | 13 | 60 | ||
| 15 | Trisomy | 13 | 7 | 34 | .99 |
| Nontrisomy | 19 | 8 | 60 | ||
| 17 | Trisomy | 10 | 3 | 103 | .026* |
| Nontrisomy | 36 | 22 | 33 | ||
| 18 | Trisomy | 9 | 6 | 60 | .50 |
| Nontrisomy | 26 | 14 | 35 | ||
| X | Monosomy† | 6 | 4 | 29 | .75 |
| Nontrisomy | 13 | 9 | 35 | ||
| Chromosome . | N . | Events . | Survival (mo) . | PUnivariate . | |
|---|---|---|---|---|---|
| 1 | Trisomy | 19 | 11 | 29 | .49 |
| Nontrisomy | 30 | 15 | 34 | ||
| 3 | Trisomy | 10 | 5 | 35 | .84 |
| Nontrisomy | 20 | 11 | 39 | ||
| 6 | Trisomy | 15 | 8 | 70 | .033* |
| Nontrisomy | 40 | 16 | 33 | ||
| 7 | Trisomy | 14 | 6 | 46 | .11 |
| Nontrisomy | 33 | 20 | 32 | ||
| 8 | Trisomy | 3 | 3 | 39 | .86 |
| Nontrisomy | 36 | 20 | 33 | ||
| 9 | Trisomy | 29 | 13 | 46 | .035* |
| Nontrisomy | 23 | 13 | 26 | ||
| 10 | Trisomy | 3 | 0 | — | .15 |
| Nontrisomy | 35 | 24 | 33 | ||
| 11 | Trisomy | 13 | 6 | 70 | .060 |
| Nontrisomy | 27 | 14 | 33 | ||
| 12 | Trisomy | 1 | 1 | 22 | .21 |
| Nontrisomy | 38 | 22 | 34 | ||
| 13 | 1 signal | 16 | 9 | 14 | .0012* |
| Nontrisomy | 32 | 13 | 60 | ||
| 15 | Trisomy | 13 | 7 | 34 | .99 |
| Nontrisomy | 19 | 8 | 60 | ||
| 17 | Trisomy | 10 | 3 | 103 | .026* |
| Nontrisomy | 36 | 22 | 33 | ||
| 18 | Trisomy | 9 | 6 | 60 | .50 |
| Nontrisomy | 26 | 14 | 35 | ||
| X | Monosomy† | 6 | 4 | 29 | .75 |
| Nontrisomy | 13 | 9 | 35 | ||
*P < .05.
Presence of monosomy X in women.
Survival curves of patients with MM according to the number of copies of chromosomes 9 and 17 per nuclei.
Survival curves of patients with MM according to the number of copies of chromosomes 9 and 17 per nuclei.
Survival curves of patients with MM according to the number of copies of chromosomes 6 and 13 (deletion of Rb) per nuclei.
Survival curves of patients with MM according to the number of copies of chromosomes 6 and 13 (deletion of Rb) per nuclei.
Univariate and Multivariate Analysis of the Prognostic Value of the Different Clinico-Biological Characteristics in MM With Respect to OS
| Variable . | Percentage of Cases . | Median OS (mo) . | P Value Univariate Analysis . | P Value Multivariate Analysis . |
|---|---|---|---|---|
| S-phase PC | ||||
| <3% | 68 | 39 | <.0001 | .010 |
| ≥3% | 32 | 15 | ||
| β2microglobulin | ||||
| <6 mg/L | 69 | 44 | .0030 | .024 |
| ≥6 mg/L | 31 | 15 | ||
| Chromosome 13 | ||||
| Monosomy | 33 | 14 | .0012 | .031 |
| Normal | 67 | 60 | ||
| Chromosome 6 | ||||
| Trisomy | 27 | 70 | .0330 | .048 |
| Nontrisomy | 73 | 33 | ||
| C–reactive Protein | ||||
| <6 mg/dL | 77 | 32 | .0026 | NS |
| ≥6 mg/dL | 23 | 11 | ||
| ECOG | ||||
| <2 | 48 | 39 | .0040 | NS |
| ≥2 | 52 | 16 | ||
| Calcium | ||||
| <11.5 mg/dL | 87 | 36 | <.0001 | NS |
| ≥11.5 mg/dL | 13 | 9 | ||
| Creatinine | ||||
| <2 mg/dL | 78 | 36 | <.0001 | NS |
| ≥2 mg/dL | 22 | 9 | ||
| Albumin | ||||
| ≤3.5 mg/dL | 57 | 39 | .0198 | NS |
| >3.5 mg/dL | 43 | 22 | ||
| Clinical Stage | ||||
| I & II | 38 | 60 | .0010 | NS |
| III | 62 | 19 | ||
| LDH | ||||
| <320 U/mL | 65 | 34 | .0295 | NS |
| ≥320 U/mL | 35 | 16 | ||
| DNA index | ||||
| >1 | 62 | 39 | .0088 | NS |
| ≤1 | 38 | 15 | ||
| Hemoglobin | ||||
| ≤9.5 mg/dL | 54 | 60 | .0123 | NS |
| >9.5 mg/dL | 46 | 19 | ||
| Chromosome 9 | ||||
| Trisomy | 56 | 46 | .0350 | NS |
| Nontrisomy | 44 | 26 | ||
| Chromosome 17 | ||||
| Trisomy | 22 | 103 | .0260 | NS |
| Nontrisomy | 78 | 33 |
| Variable . | Percentage of Cases . | Median OS (mo) . | P Value Univariate Analysis . | P Value Multivariate Analysis . |
|---|---|---|---|---|
| S-phase PC | ||||
| <3% | 68 | 39 | <.0001 | .010 |
| ≥3% | 32 | 15 | ||
| β2microglobulin | ||||
| <6 mg/L | 69 | 44 | .0030 | .024 |
| ≥6 mg/L | 31 | 15 | ||
| Chromosome 13 | ||||
| Monosomy | 33 | 14 | .0012 | .031 |
| Normal | 67 | 60 | ||
| Chromosome 6 | ||||
| Trisomy | 27 | 70 | .0330 | .048 |
| Nontrisomy | 73 | 33 | ||
| C–reactive Protein | ||||
| <6 mg/dL | 77 | 32 | .0026 | NS |
| ≥6 mg/dL | 23 | 11 | ||
| ECOG | ||||
| <2 | 48 | 39 | .0040 | NS |
| ≥2 | 52 | 16 | ||
| Calcium | ||||
| <11.5 mg/dL | 87 | 36 | <.0001 | NS |
| ≥11.5 mg/dL | 13 | 9 | ||
| Creatinine | ||||
| <2 mg/dL | 78 | 36 | <.0001 | NS |
| ≥2 mg/dL | 22 | 9 | ||
| Albumin | ||||
| ≤3.5 mg/dL | 57 | 39 | .0198 | NS |
| >3.5 mg/dL | 43 | 22 | ||
| Clinical Stage | ||||
| I & II | 38 | 60 | .0010 | NS |
| III | 62 | 19 | ||
| LDH | ||||
| <320 U/mL | 65 | 34 | .0295 | NS |
| ≥320 U/mL | 35 | 16 | ||
| DNA index | ||||
| >1 | 62 | 39 | .0088 | NS |
| ≤1 | 38 | 15 | ||
| Hemoglobin | ||||
| ≤9.5 mg/dL | 54 | 60 | .0123 | NS |
| >9.5 mg/dL | 46 | 19 | ||
| Chromosome 9 | ||||
| Trisomy | 56 | 46 | .0350 | NS |
| Nontrisomy | 44 | 26 | ||
| Chromosome 17 | ||||
| Trisomy | 22 | 103 | .0260 | NS |
| Nontrisomy | 78 | 33 |
Abbreviations: ECOG, Eastern Cooperative Oncology Group scale; NS, not significant (P > .05).
A multivariate analysis for OS was performed using all variables with significant prognostic influence on univariate analysis (Table 4). This analysis showed that S-phase PC greater than 3% (P = .010) and β2microglobulin serum levels greater than 6 μg/mL (P = .024), together with monosomy of chromosome 13 (P = .031) and nontrisomy of chromosome 6 (P = .048) was the best combination of independent parameters for predicting survival in patients with MM. Although trisomy of chromosome 17 was associated with a better outcome, its impact on OS in the multivariate analysis was on the limit of the statistical significance (P = .06).
DISCUSSION
In the present study we have explored the clinical impact of aneuploidies for 15 different human chromosomes assessed by FISH in a series of 63 consecutive patients with MM. The presence of monosomy 13 was associated with poor prognostic features and a shorter survival, whereas the existence of trisomy for chromosomes 6, 9, and 17 was associated with a better outcome.
The favorable prognostic impact observed in patients with trisomy of chromosomes 6, 9, and 17 would be in accordance with the longer median survival observed in patients with MM displaying DNA hyperploidy (DNA index > 1) by conventional flow cytometry5,25,26 and would also support the high incidence of hyperploidy observed in patients with monoclonal gammopathy of undetermined significance.27 Although the favorable prognosis of hyperploidy has often been reported, the biological mechanisms responsible for this positive influence are yet to be clarified. Nevertheless, it could be speculated that the DNA damage occurring in these patients is partially offset by an increase in the number of copies of tumor suppressor genes located in those specific chromosomes. This could be the case of the p16 and p15 tumor suppressor genes, placed at 9p21,28,29 which have recently been considered to have an important role in MM because of the high frequency of hypermethylation of their CpG islands in this disease.30 The products of both genes are capable of inhibiting the interaction between CDK4/6 and D1 cyclins, and as a consequence, cells are arrested at the G1-phase of the cell cycle. If these genes are hypermethylated in MM, their function would be hampered, thus allowing clonal plasma cells to enter S-phase. However, the presence of trisomy of chromosome 9 would provide the presence of a normal extra copy of the p16 and p15 genes that would increase the probability of a nonhypermethylated gene copy continuing to work normally despite the hypermethylation of other copies; this would help to explain the better survival observed in MM cases with trisomy 9. A similar hypothesis could be used to explain the favorable outcome observed in patients with MM with trisomy 17, because the p53 tumor suppressor gene is located in this chromosome at 17p13.31-34 In a similar way, recent findings in endometrium carcinoma or melanoma suggest the presence of tumor suppressor genes in chromosome 6.35-37 The loss of heterozygosity at 6q25 has been found in several solid tumors such as ovarian, breast, kidney, or mesothelial cancers.35-37 This would suggest the presence of tumor suppressor genes at this level.
Tricot et al8 reported that monosomy 13 is associated with poor outcome in patients treated with high-dose chemotherapy. In the present study conducted in patients treated with conventional chemotherapy it was also confirmed that monosomy 13, as assessed by deletion of the Rb gene, has an adverse prognostic influence. Moreover, this chromosomal deletion retained the independent value on multivariate analysis. The Rb tumor suppressor gene located at chromosome 13 plays a central role in the control of the cell cycle, because its phosphorylation by cyclin-dependent kinases relieves the G1 block and promotes the transition of cells into S-phase.38-43
According to these results, chromosomal analysis is of great use in patients with MM at diagnosis to have a correct prognostic evaluation for clinical decision making. Because FISH analysis is a reliable technique that may afford accurate information on numerical changes, overcoming the problems of conventional cytogenetics, FISH evaluation of chromosomes 6, 9, 13, and 17 should be recommended in all patients with MM at diagnosis.
In summary, this report shows the high prognostic value of the FISH analysis to evaluate the presence of aneuploidies in patients with MM. In addition, our findings show a potential important role of the p16/CDK4/Rb axis in the pathogenesis of the MM and suggest the existence of new tumor suppressor genes, placed at chromosome 6, that could be involved in the disease.
Supported in part by a grant from the Spanish Fondo de Investigaciones Sanitárias (95/1475) and Dirección General de Investigación Cientifica y Tecnológica (PB93-0614). J.A. was supported by a grant from the Spanish Fondo de Investigaciones Sanitárias de la Seguridad Social (97/3537).
Address reprint requests to J.F. San Miguel, MD, PhD, Servicio de Hematologı́a, Hospital Universitario de Salamanca, Paseo de San Vicente, 58-182, 37007, Salamanca, Spain.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal