Abstract
The role of Epstein-Barr virus (EBV) in the pathogenesis of severe, chronic active EBV infection and its complications is unclear. We investigated two Japanese patients diagnosed with severe, chronic active EBV infection who subsequently developed EBV–positive T-cell lymphoma. The patients displayed abnormally high antibody titers to EBV antigens, and had evidence of peripheral blood CD4+T-cell infection with EBV, 19 months and 3 months, respectively, before the diagnosis of EBV–positive T-cell lymphoma. The lymphomas were infected with monoclonal EBV and expressed the EBV latency genes EBNA-1, LMP-1, and LMP-2. Genetic studies showed that the virus detected in the T-cell lymphoma was indistinguishable, with respect to type and previously defined LMP-1 and EBNA-1 gene variations, from virus detected in the peripheral blood T cells 19 months earlier. These studies support an important pathogenetic role of T-cell infection with EBV in chronic active EBV infection and in the EBV–positive T-cell lymphoma that followed.
EPSTEIN-BARR VIRUS (EBV) is a ubiquitous herpes virus in the human population. Most primary EBV infections are inapparent, but occasionally, EBV may cause acute infectious mononucleosis (IM), and in association with severe immunodeficiency, B-cell lymphoproliferative disorders.1 Infection with EBV has also been linked with a variety of malignancies, including Burkitt's lymphoma, nasopharyngeal carcinoma,2 Hodgkin's disease,3,4 smooth muscle tumors,5,6 and gastric carcinoma.7,8 Recently, the virus has been associated with lymphoproliferative disorders of T and NK cells, including cases of fulminant infectious mononucleosis,9chronic active EBV infections,10,11 nasal or nasal-type lymphomas,12,13 and nodal and extranodal lymphomas of various histologies.14-17
Studies in vitro have generally failed to show normal T-cell immortalization by exposure to EBV. In one study, transfection of EBV DNA into normal T cells was reported to induce T-cell immortalization.18 In other studies, exposure to EBV caused transient infection of normal peripheral blood T cells and thymocytes.19 In vivo T-cell infection with EBV has only rarely been documented in otherwise normal individuals. Recently, EBV was found to infect circulating T cells and NK cells in some patients with chronic active EBV infection,10,11,20-22 and spontaneously outgrowing EBV–infected T-cell lines were derived from three patients diagnosed with this illness.23
The difficulties with which normal T cells are infected with EBV in vitro and the rare occurrence of benign T-cell infections with EBV in vivo raise unresolved questions on the pathogenesis of EBV–positive lymphoproliferative diseases of T-cell lineage. Most importantly, it is unclear what role, if any, the virus might play when it infects the unusual T-cell target. We report here on two patients diagnosed with severe, chronic active EBV infection who subsequently developed EBV–positive T-cell lymphoma.
MATERIALS AND METHODS
Study subjects.
Case 1 is a 2-year-old girl who visited a local hospital, complaining of fever and cough, in May 1994. She was treated for pneumonia. Laboratory tests showed elevated liver function tests. In October 1994, liver function tests had deteriorated and hepatosplenomegaly was diagnosed. In November 1994, physical examination showed cervical lymphadenopathy and hepatosplenomegaly, and laboratory tests detected high titer antibodies to EBV antigens. The blood count included a white cell count of 3700/μL with 48% neutrophils, 2% monocytes, and 50% lymphocytes; a red cell count of 391 × 104/μL, hemoglobin of 9.9 g/dL, hematocrit of 30.2%, and a platelet count of 36.8 x 104/μL. Serum alkaline phosphatase (ALP), aspartate aminotransferase (AST), alanine aminotransferase (ALT), and lactate dehydrogenase (LDH) were all abnormally elevated (1897, 314, 204, and 1044 IU/L, respectively). Total protein was also elevated (7.8 g/dl), with hypergammaglobulinemia. All other chemistries were within normal limits. In May 1996, 2 years after the onset of symptoms, the lymphadenopathy deteriorated and a lymph node biopsy was diagnostic for T-cell lymphoma. Combination chemotherapy with vincristine and prednisone resulted in clinical remission.
Case 2 was an 11-year-old boy who was hospitalized in August 1993 because of fever, hepatosplenomegaly, and left parotid swelling that were first noted 2 to 3 months earlier. Physical examination showed hepatosplenomegaly and lymphadenopathy. The blood count included a white cell count of 1500/μL with 57% neutrophils, 2% monocytes, and 41% lymphocytes; a red cell count of 439 × 104/μL, hemoglobin of 11.9 g/dL, and a platelet count of 13.5 × 104/μL. Serum AST, ALT, and LDH were elevated (119, 141, and 2160 IU/L, respectively), and IgG levels were also abnormally elevated (3218 mg/dL). In October 1993, a lymph node biopsy showed disruption of the normal lymph node structure by lymphocytes with plasma cell infiltration, and a diagnosis of lymphoma was made. Fever and hepatosplenomegaly transiently improved on α-interferon treatment, but this was stopped after 2 weeks because of evidence of hepatic toxicity and emotional changes. Splenectomy and treatment with cyclosporin A, etoposide, and prednisone did not control the disease, and the patient died with multiple organ failure and systemic fungal infection, 2 years and 4 months after the onset of symptoms.
Virological studies.
Serum antibodies to viral capsid antigen (VCA), early antigen (EA), and nuclear antigen (EBNA) of EBV were determined by conventional immunofluorescence methods. Serum antibodies to hepatitis A, B, and C were detected by passive hemagglutination tests. Antibodies to cytomegalovirus were determined by complement fixation or immunofluorescence.
In situ hybridization for EBV RNA.
The presence of EBV in cell populations was assessed by in situ hybridization for EBV–encoded small nuclear RNAs (EBER).22 24 CD4+ T, CD8+ T, B, and NK cells were separated from the patient's peripheral blood mononuclear cells by incubating with monoclonal antibodies to CD4, CD8, CD20, and CD16, followed by isolation of each subpopulation by electronic cell sorting, using an Epics Elite flow cytometer (Coulter Immunology, Hialeah, PA) or a FACStar Plus (Becton Dickinson, San Jose, CA). Each sorted population was more than 97% pure. Subpopulations of CD4+ T, CD8+ T, B, NK cells, and control cells were centrifuged on 3-aminopropyltriethoxy-silane–coated glass slides and fixed in 4% formaldehyde in 0.1 mol/L phosphate buffer. After rinsing and rehydration, hybridization was performed with an ALP-conjugated sense and antisense oligonucleotide probe to EBER-1, as described.22, 24
Southern blot analysis for EBV clonality.
DNA was prepared from frozen patient tissues and from B95-8, Raji, and Louckes cell lines with the QIAamp Tissue Kit (Qiagen, Inc, Chatsworth, CA) according to the manufacturer's protocol. Ten μg of genomic DNA was digested with BamHI separated on a 0.7% agarose gel and transferred to a nitrocellulose membrane. The membrane was hybridized with a 32P-labeled B95-8 DNA fragment (167,129-169, 566) that contains the LMP-1 open reading frame and detects the right terminal repeats.25 The membrane was washed with 0.2% SSC and 1% SDS at 68 oC and visualized by autoradiography.
Polymerase chain reaction (PCR) for EBV DNA.
The oligonucleotide sequences for amplification of the EBV U2 region encoding EBNA-2 were 5′-TTTCACCAATACATGAACC-3′ and 5′-TGGCAAAGTGCTGAAAGCAA-3′. Amplification was performed as described previously.22,26 Expected lengths of the amplified products derived from type 1 and type 2 EBV were 378 bp and 483 bp, respectively. Sequences within the C-terminal region of LMP-1 gene were amplified, as described,27 by using the following primers: 5′-GCGACTCTGCTGGAAATGAT-3′ and 5′-GACATGGTAATGCCTAGAAG-3′. The expected size of the amplified products from wild-type EBV is 260 bp; the expected size from the 30 bp LMP-1 deletion mutant is 230 bp. EBNA-1 typing by PCR was performed as described.28 Two EBNA-1 fragments were amplified including a 284 bp fragment (nucleotides 50-334) and a 330 bp fragment (nucleotides 1341-1671). The primer pairs used for amplifications were: ACAGGACCTGGGAAATGGCCTA and CCTCCCTGCTCCTGCCCCTC (284 bp fragment), and CCCGCAGATGACCCAGGAGA and GGGTCCAGGGGCCATTCCAA (330 bp fragment). The amplified fragments were directly sequenced from PCR amplified products by using the Sequenase (US Biochem, Cleveland, OH) protocol, as described.28
RNA preparation and reverse transcriptase-mediated PCR (RT-PCR).
Total RNA was extracted from cell pellets or tissue, by using guanidine isothiocyanate-phenol (Trizol, Life Technologies). The first-strand cDNA was synthesized from 5 μg of total RNA by using the Superscript preamplification system (Life Technologies). For the detection of EBV latent gene expression (EBNA-1,-2, and LMP-1, -2A, -2B), nested PCR was performed essentially as described elsewhere.26 29Expression of the other EBV genes (EBER1, BZLF1, vIL-10) and hIL-10 was also assessed by RT-PCR. The sequences of the primer pair were 5′-AAAACATGCGGACCACCAGC and 5′-AGGACCTACGCTGCCCTAGAA (EBER1); 5′-TTCCACAGCCTGCACCAGTG and 5′-GGCAGCAGCCACCTCACGGT (BZLF1); 5′-ATGGAGCGAAGGTTAGTGGTCACT and 5′-AATTGGATCATTTCTGACAGCGCC (vIL-10); and 5′-CTTCGAGATCTCCGAGATGCCTTC and 5′-ATTCTTCACCTGCTCCACGGCCTT (hIL-10).
RESULTS
EBV serologies from the two patients were consistent with chronic active EBV infection 19 months (case 1) and 3 months (case 2) before the diagnosis of T-cell lymphoma. Positive IgG, but not IgM, anti-VCA antibodies and positive anti-EBNA antibodies indicated past infection with EBV (Table 1). Abnormally elevated IgG antibodies to VCA and EA and detection of IgA antibodies to VCA and EA suggested ongoing viral replication. In both patients antibody titers to EBV antigens did not change substantially during the following 2 years. Antibodies to hepatitis virus and cytomegalovirus were all negative (data not shown).
Antibodies to EBV Antigens from Patients with Chronic Active EBV Infection
| . | IgG . | VCA− . | EA− . | EBNA . | ||
|---|---|---|---|---|---|---|
| IgM . | IgA . | IgG . | IgA . | |||
| Case 1 | 20480 | <10 | 320 | 5120 | 40 | 160 |
| Case 2 | 5120 | <10 | 80 | 5120 | 160 | 160 |
| . | IgG . | VCA− . | EA− . | EBNA . | ||
|---|---|---|---|---|---|---|
| IgM . | IgA . | IgG . | IgA . | |||
| Case 1 | 20480 | <10 | 320 | 5120 | 40 | 160 |
| Case 2 | 5120 | <10 | 80 | 5120 | 160 | 160 |
Antibody titers to EBV viral capsid antigen (VCA), early antigen (EA), and nuclear antigen (EBNA) were determined by immunofluorescence in patient serum at presentation.
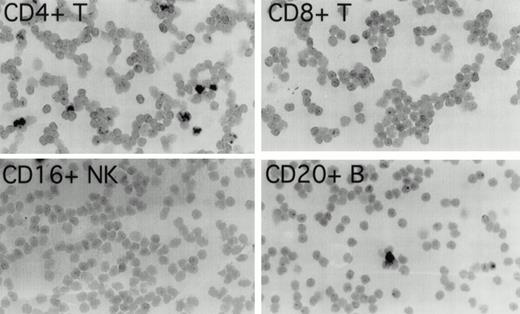
To determine which cell populations were infected with EBV in these patients, highly purified (>97% pure) populations of CD4+ T cells, CD8+ T cells, CD16+NK cells, and CD20+ B cells, obtained by cell sorting of peripheral blood mononuclear cells, were evaluated for the presence of EBV by in situ hybridization for EBER1. In patient 1 (Fig 1), 3.4% of CD4+ T cells were EBER1 positive 19 months before the diagnosis of T-cell lymphoma was made. At the same time point, occasional EBER1-positive cells were also detected in the purified CD20+ B-cell population, but no EBER1-positive cells were detected in CD8+ and CD16+ cell populations (Fig 1). In patient 2 (not shown), 3.9% of peripheral blood CD4+ T cells were found to be EBER1 positive, but no EBER1-positive cells were detected in the CD20+ B cells, CD8+ T cells or CD16+ NK cells 3 months before the diagnosis of T-cell lymphoma. As expected, more than 95% of Daudi cells used as a positive control were EBER1 positive, whereas all control purified peripheral blood cell populations from two EBV–seropositive normal individuals were EBER1 negative (data not shown).
Detection of EBER1 expression by in situ hybridization. Purified populations of CD4+ T cells, CD8+T cells, CD16+ NK cells, and CD20+ B cells obtained from patient 1, 19 months before the development of lymphoma, were cytocentrifuged and hybridized with an alkaline phosphatase-conjugated EBER1 antisense oligonucleotide probe.
Detection of EBER1 expression by in situ hybridization. Purified populations of CD4+ T cells, CD8+T cells, CD16+ NK cells, and CD20+ B cells obtained from patient 1, 19 months before the development of lymphoma, were cytocentrifuged and hybridized with an alkaline phosphatase-conjugated EBER1 antisense oligonucleotide probe.
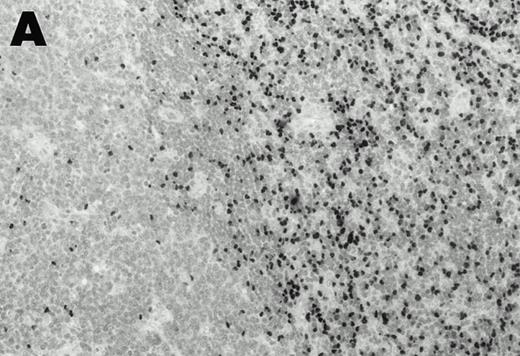
Lymphomas were diagnosed in the lymph node from patient 1 and in the spleen from patient 2 at 19 months and 3 months, respectively, after peripheral blood CD4+ T-cell infection with EBV was documented. Both lymphomas were EBV positive by EBER1 in situ hybridization (Fig 2), and were of T-cell lineage as determined by immunohistochemistry with an anti-CD45RO and anti-CD4 MoAb, and Southern analysis that detected rearranged T-cell receptor γ- and β-chain genes with germline immunoglobulin JH genes (not shown).
Detection of EBER1 expression by in situ hybridization of lymphoma tissues. (A) Lymph node from case 1 (100X magnification); (B) spleen from case 2 (200X magnification).
Detection of EBER1 expression by in situ hybridization of lymphoma tissues. (A) Lymph node from case 1 (100X magnification); (B) spleen from case 2 (200X magnification).
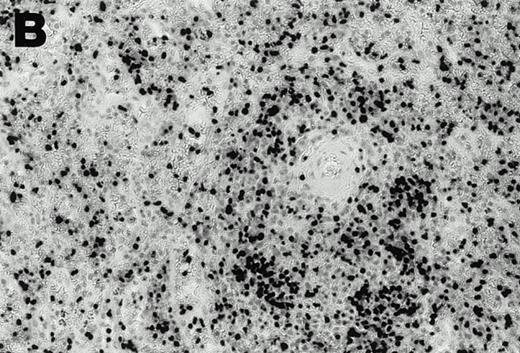
Both tumors contained monoclonal EBV determined by Southern analysis of EBV terminal repeats (Fig 3). Two tumor samples from patient 1, derived from distinct lymph nodes, displayed indistinguishable clonality. The monoclonal EBV in the tumor sample from patient 2 was distinct from that found in patient 1. As expected, EBV in the B95-8 cell line was present at a high copy number in both episomal and linear forms, whereas EBV was monoclonal in the Raji cell line; no EBV was detected in the EBV-negative Louckes cell line (Fig3).
EBV clonality in lymphoma tissues assessed by Southern analysis. Genomic DNA was digested with BamHI, separated on an agarose gel, and transferred to a nitrocellulose membrane. The membrane was hybridized with 32P-labeled B95-8 fragment containing the LMP open reading frame, washed, and visualized by autoradiography. Tumors 1 and 2 were derived from different lymph nodes of case 1; one tumor specimen was derived from case 2. B95-8 and Raji cell lines were used as EBV–positive controls; Louckes cell line was used as an EBV–negative control.
EBV clonality in lymphoma tissues assessed by Southern analysis. Genomic DNA was digested with BamHI, separated on an agarose gel, and transferred to a nitrocellulose membrane. The membrane was hybridized with 32P-labeled B95-8 fragment containing the LMP open reading frame, washed, and visualized by autoradiography. Tumors 1 and 2 were derived from different lymph nodes of case 1; one tumor specimen was derived from case 2. B95-8 and Raji cell lines were used as EBV–positive controls; Louckes cell line was used as an EBV–negative control.
RT-PCR analysis for EBV–gene expression in the lymphoma tissue specimens showed a type II form of EBV latency. EBER-1, EBNA-1, LMP-1, and LMP-2A transcripts could be amplified readily from both lymphoma specimens, whereas no EBNA-2 or LMP-2B transcripts were derived from these tissues (Fig 4). The mRNAs for the EBV replication genes BZLF1 and BCRF1 (vIL-10) were amplified from both lymphomas, consistent with the occurrence of viral replication in these tissues. The mRNA for the cellular genes hIL-10 and GAPDH were amplified from all samples tested. As expected, the EBV–producing marmoset cell line B95-8 expressed all EBV genes, and the EBV–negative lymphoma cell line BJAB expressed the cellular hIL-10 and GAPDH genes, but no EBV genes.
EBV gene expression in lymphoma tissues assessed by RT-PCR analysis. Total RNA extracted from lymphoma tissues of case 1 (lymph node) and case 2 (spleen), and from control EBV–negative BJAB and EBV–positive B95-8 cell lines, was reverse transcribed and subjected to PCR amplification with specific primers. The amplified products containing α-[32P]dCTP were electrophoresed through 6% acrylamide Tris-borated EDTA gels, followed by autoradiography. For GAPDH control, the amplified products were electrophoresed through a 1.5% agarose gel prestained by ethidium bromide.
EBV gene expression in lymphoma tissues assessed by RT-PCR analysis. Total RNA extracted from lymphoma tissues of case 1 (lymph node) and case 2 (spleen), and from control EBV–negative BJAB and EBV–positive B95-8 cell lines, was reverse transcribed and subjected to PCR amplification with specific primers. The amplified products containing α-[32P]dCTP were electrophoresed through 6% acrylamide Tris-borated EDTA gels, followed by autoradiography. For GAPDH control, the amplified products were electrophoresed through a 1.5% agarose gel prestained by ethidium bromide.
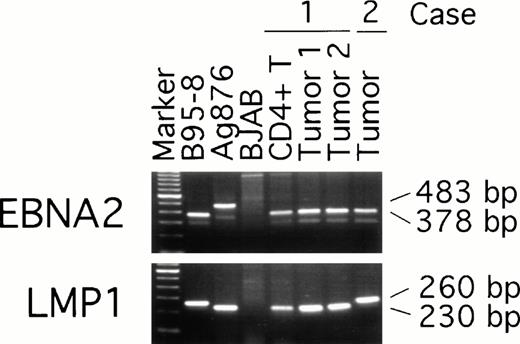
To examine possible links between peripheral blood T-cell infection with EBV and the subsequent development of EBV–positive T-cell lymphoma, we compared EBNA-2, LMP, and EBNA-1 virus variants in the purified CD4+ peripheral blood T cells from patient 1 (obtained 19 months before the diagnosis of T-cell lymphoma was made) with the virus detected in two T-cell lymphoma tissue specimens from the same patient (obtained at the time the diagnosis of lymphoma was made). Two types of EBV, type 1 and 2, have been defined by differences in the U2 region encoding EBNA-2 resulting in distinct serologic reactivities.30 PCR amplification with EBNA-2 specific primers indicated that all samples from patient 1, including the peripheral blood CD4+ T cells and the two lymphoma tissue specimens, were infected with type-1 EBV (Fig 5). Also infected with type-1 EBV was the lymphoma tissue from patient 2 (Fig 5). As expected, type-1 EBV was detected in the control B95-8 cells (378 bp amplification product), whereas type-2 EBV was detected in Ag876 cells (483 bp amplification product).
EBV typing of virus-infected patient peripheral blood and lymphoma tissues. DNA, extracted from CD4+ peripheral blood T cells and lymphoma specimens (from distinct lymph nodes) of patient 1, from a lymphoma specimen (spleen) of patient 2, and from control cell lines, was subjected to PCR amplification with specific probes. The EBNA-2 primers were designed to distinguish type-1 and type-2 EBV; the LMP primers were designed to distinguish full length from deleted LMP-1 gene.
EBV typing of virus-infected patient peripheral blood and lymphoma tissues. DNA, extracted from CD4+ peripheral blood T cells and lymphoma specimens (from distinct lymph nodes) of patient 1, from a lymphoma specimen (spleen) of patient 2, and from control cell lines, was subjected to PCR amplification with specific probes. The EBNA-2 primers were designed to distinguish type-1 and type-2 EBV; the LMP primers were designed to distinguish full length from deleted LMP-1 gene.
The presence of a 30 bp deletion in the LMP1 gene has defined a variant EBV isolate,31 detectable in the Ag876 cell line (230 bp amplification product), that can be distinguished from the full-length product (260 bp amplification product) of the prototype B95-8 cell line (Fig 5). All samples from patient 1, including peripheral blood CD4-positive cells and lymphoma tissue from the two lymph nodes, yielded the 230 bp PCR product (Fig 5) indicative of the presence of a deleted LMP1 gene. In contrast, the lymphoma tissue from patient 2 yielded a 260 bp PCR product indicative of the presence of a full-length LMP1 gene (Fig 5).
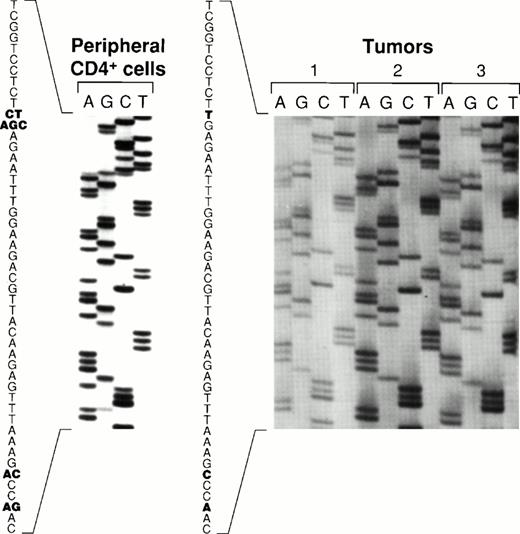
Sequence variations of the carboxy terminal region of EBNA-128 have defined five EBV subtypes distinguished on the basis of several amino acid substitutions, including substitutions at position 487 (alanine, detected in the prototype B95-8 virus, and substitutions with threonine, valine, proline, or leucine). Using PCR to amplify EBNA-1 specific fragments, followed by DNA sequencing, all five previously identified EBNA-1 subtypes were recovered from the peripheral blood CD4+ T cells from patient 1 (Fig 6). In contrast, only the EBNA-1 subtype identified by valine at position 487 was detected in the lymphoma specimens from patient 1 (Fig 6). The same EBNA-1 subtype was also detected in the lymphoma specimen from patient 2 (Fig 6). Thus, patient 1 harbored EBV in the peripheral blood CD4+ T cells that was indistinguishable from the virus detected 19 months later in the T-cell lymphoma, with respect to previously defined EBNA-1, EBNA-2, and LMP-1 variants.
Comparative sequence analysis of EBNA-1 from peripheral blood CD4+ T cells and lymphoma specimens. An EBNA-1 carboxy terminal fragment comprising nucleotides 1341-1671 (330 bp fragment) was PCR-amplified with DNA extracted from peripheral blood CD4+ T cells from patient 1, from two lymphoma specimens from patient 1 (distinct lymph nodes tumors 1, and 2), and from a lymphoma specimen from patient 2 (spleen, tumor 3). Shown are the results of direct sequencing of EBNA-1 codons 475 to 490. Nucleotide substitutions compared to the prototype EBNA-1(B95-8) are highlighted in bold. Multiple substitutions at the same position are indicative of multiple EBNA-1 variants.
Comparative sequence analysis of EBNA-1 from peripheral blood CD4+ T cells and lymphoma specimens. An EBNA-1 carboxy terminal fragment comprising nucleotides 1341-1671 (330 bp fragment) was PCR-amplified with DNA extracted from peripheral blood CD4+ T cells from patient 1, from two lymphoma specimens from patient 1 (distinct lymph nodes tumors 1, and 2), and from a lymphoma specimen from patient 2 (spleen, tumor 3). Shown are the results of direct sequencing of EBNA-1 codons 475 to 490. Nucleotide substitutions compared to the prototype EBNA-1(B95-8) are highlighted in bold. Multiple substitutions at the same position are indicative of multiple EBNA-1 variants.
DISCUSSION
In the present studies, we show that approximately 4% of the peripheral blood CD4+ T cells from two Japanese patients with serologic evidence of severe, chronic, active EBV infection were infected with EBV at 19 months and 3 months, respectively, before the development of EBV–positive T-cell lymphoma. Both T-cell lymphomas contained monoclonal EBV and expressed the transforming EBV gene LMP-1,32 suggesting that the virus might play a role in the pathogenesis of these tumors. Genetic studies showed that EBV infecting both the peripheral blood CD4+ T cells and the T-cell lymphoma diagnosed 19 months later was of type 1 and displayed a 30 bp deletion of LMP-1. In addition, the virus recovered from the T-cell lymphoma tissue was characterized by a substitution at position 487 in the EBNA-1 gene. The same virus variant was also detected in the peripheral blood T cells, albeit in conjunction with four other EBNA-1 subtypes. Thus, we describe the previously unrecognized occurrence of peripheral blood T-cell infection with EBV preceding the development of malignant EBV–-positive T-cell lymphoma. Genetic evidence in one of the patients linked the virus from the peripheral blood CD4+ T cells with the monoclonal virus later detected in the T-cell lymphoma in which the pattern of viral gene expression suggested a continued role for EBV as a transforming agent. Thus, peripheral blood T-cell infection with EBV may be important to the pathogenesis of certain EBV–positive T-cell lymphomas.
Recently, T-cell infection with EBV was reported with increasing frequency in the context of certain leukemias and lymphomas, particularly in the Orient.9-17 Some of these cases have presented as fatal lymphoproliferative disorders associated with primary EBV infection that rapidly progressed toward multiple organ failure, sepsis, and death.9-11 Other cases have presented as extranodal lymphomas localized to the upper respiratory tract exhibiting characteristic histological features of tissue necrosis, vascular damage, and infiltration with inflammatory cells, and have been variously identified as nasal or nasal-type T/NK cell lymphomas, lymphomatoid granulomatosis, lethal midline granuloma, or angiocentric lymphomas.12,13 Other cases included nodal or extranodal T-cell lymphomas with various histologies and phenotypes.14-17 Recently, four EBV–infected T-cell lines were derived from culture of peripheral blood from three patients with severe, chronic active EBV infection.23 In addition, circulating T cells from two patients with severe, chronic active EBV infection were reported to be infected with EBV, raising the possibility that T-cell infection with this virus might be important to the pathogenesis of this illness.23 However, the occurrence of peripheral blood T-cell infection with EBV preceding the development of EBV–positive T-cell lymphoma was not previously described.
In the patients described here, the relationship between the EBV–infected circulating CD4+ T cells and the malignant CD4+ T cells in the lymphomas that subsequently developed could not be established. It is possible that the circulating EBV–infected CD4+ cells were premalignant and contributed to lymphomagenesis. Alternatively, they could have been normal lymphocytes serving as a reservoir for virus later detected in the lymphoma. T-cell receptor clonality analysis could not be performed on the rare circulating CD4+ T cells that were infected with EBV, and thus, cell clonality relationships could not be established.
Normal T-cells are not easily infected with EBV in vitro or in vivo, and generally do not express the EBV receptor, CD21. Some studies have documented expression of CD21 in a proportion of circulating CD4 and CD8+ T cells, and on immature thymic T cells.19,33,34 Some reports mention transient infection of T cell with EBV, but no EBV–immortalized T-cell lines have been generated in vitro by exposure to the virus.35,36 Recently, an HTLV-1–infected T-cell line, MT-2, was developed that expresses functional EBV receptors, and can be persistently infected with EBV.37 Benign T-cell infections with EBV in vivo have rarely been documented. One report describes a case of transient polyclonal benign proliferation of EBV-infected T cells in a Japanese individual presenting with an infectious mononucleosis-like syndrome.38 Another documented the presence of EBV–infected T cells in the lymph nodes of individuals with acute infectious mononucleosis.39 Thus, T-cell infection with EBV was reported rarely, and mostly in the context of severe illness.
The rare occurrence of T-cell infection with EBV suggests that the necessary conditions are infrequently met. Susceptible T-cell targets could be rare or represent an abnormal T-cell population, and conditions permitting T-cell infection could be stringent and peculiar. Rare viral mutants may be required for T-cell infection. Previously, genetic studies of EBV have documented polymorphism in the EBNA2, EBNA3, LMP-1 genes, and BamHI F region of the genome, but EBV isolates from normal individuals and EBV–positive tumors in the same geographic area generally exhibited similar gene polymorphisms.27 We found that the EBNA-1 variant detected here in both T cell lymphomas and in the peripheral blood T cells of one of the patients was not previously identified in peripheral blood of normal individuals,28 including ten normal Japanese (data not shown), suggesting that further studies are necessary to study the potential role of EBNA-1 mutations to EBV–associated diseases. The severity of disease associated with T-cell infection with EBV may be due, in part, to infection and secondary functional impairment of the same immune T cells that are primarily involved in host immunity against the virus.40 In addition, although EBV has evolved an effective strategy for latent survival in B cells, such strategy may not apply to the establishment of latency in T cells.
Severe, chronic active EBV infection is a life-threatening illness leading to the development of lymphoma, myelodysplastic syndrome, opportunistic infection, or multiple organ failure over a period of months to several years. High titer antibodies to EBV and the abundance of EBV DNA or antigens in the tissues have suggested a pathogenetic role for the virus that remains undefined. The present study, in conjunction with previous observations of EBV–infecting T cells, raises the possibility that T-cell infection with EBV represents a key step in the pathogenesis of the disease and its complications.
Address correspondence to Dr G. Tosato, Center for Biologics Evaluation and Research, Building 29A, Room 2D16, HFM-535, 1401 Rockville Pike, Rockville, Maryland 20852.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
This is a US government work. There are no restrictions on its use.

![Fig. 4. EBV gene expression in lymphoma tissues assessed by RT-PCR analysis. Total RNA extracted from lymphoma tissues of case 1 (lymph node) and case 2 (spleen), and from control EBV–negative BJAB and EBV–positive B95-8 cell lines, was reverse transcribed and subjected to PCR amplification with specific primers. The amplified products containing α-[32P]dCTP were electrophoresed through 6% acrylamide Tris-borated EDTA gels, followed by autoradiography. For GAPDH control, the amplified products were electrophoresed through a 1.5% agarose gel prestained by ethidium bromide.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/6/10.1182_blood.v91.6.2085/2/m_blod4063104.jpeg?Expires=1769844834&Signature=issDBKySJ2yBcR81hu0d2n5nz7uViFpVuAfoh3CNhJiWc769aV8vaCLmUEBbJKcyAo77ZC5tkZ~cBQp~9rnKyh-RbQ1XnYpQS5lpN~iOyxAeYfWwxD6q~bTVhbQpD3GICNVS4ymSme1e1BlcNQfMhyOT-EbyGSAhzVXy0y2aTgtcauYvmWbdZYI2uGeDPJggfUHp3n44CBRMllUSr7ioM422jN5Ahz7wkrZbAxU2pIvWUKs4vqrSXgaaTsnFwPQBEgciIepmv2PKvia56elCl3WA3N~KS1uhOTz4eSr~fTCG837X~KXR2zt6Xu7udhflF5V4GO1S23~S8niB2~Cdeg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal