Abstract
The fact that the p16/INK4a and p15/INK4b genes are frequently inactivated in human malignancies and that p16/INK4a null mice spontaneously develop B-cell lymphomas prompted us to examine the status of both genes in Burkitt's Lymphoma (BL). We found a low frequency of p16/INK4a and p15/INK4b deletions and mutations in BL cell lines and biopsies. However, p16/INK4a exon 1 was methylated in 17 out of 19 BL lines (89.5%) and in 8 out of 19 BL biopsies (42%) analyzed. p15/INK4b Exon 1 was also methylated, although at a lower frequency. p16/INK4a mRNA was readily detected in BL lines carrying unmethylated p16/INK4a, but not in those carrying methylated p16/INK4a. No p16/INK4a protein was detected in any of the BL lines and biopsies examined. In contrast, only one out of seven lymphoblastoid cell lines (LCLs) examined was methylated in p16/INK4a exon 1, and three out of the six LCLs with unmethylated p16/INK4a expressed detectable levels of p16/INK4a protein. Thus, the frequent p16/INK4a methylation in BL lines correlates with downregulation of p16/INK4a expression, suggesting that exon 1 methylation is responsible for silencing the p16/INK4a gene in BL.
CELL CYCLE progression is regulated by complexes formed between cyclins and cyclin-dependent kinases (CDKs). The kinase activity of CDK4 and CDK6 is activated on association with D-type cyclins in the G1 phase of the cell cycle.1 The cyclin D-CDK 4/6 complexes can phosphorylate the retinoblastoma (RB) protein in vitro, suggesting that cyclin D-CDK4/6 complexes positively regulate progression through the G1 phase of the cell cycle by phosphorylating RB, thereby eliminating the cell cycle block imposed by RB.2 The activity of cyclin D-CDK4/6 complexes is blocked by cyclin-dependent kinase inhibitors (CDIs).3 The p16/INK4a and p15/INK4b proteins, both members of the INK4 family of CDIs, bind CDK4/6 and block their complexing with D type cyclins.4-6 This prevents RB phosphorylation and progression into S phase, thus arresting the cell in the G1 phase.
Homozygous deletion of the p16/INK4a gene and the p15/INK4b gene that maps in tandem with p16/INK4a on chromosome 9p21 was first observed in many human tumor cell lines7,8 and has subsequently been found in a wide variety of human primary tumors, for instance gliomas, acute lymphoblastic leukemias, and prostate and bladder carcinomas.1 In addition, point mutations and small deletions in the p16/INK4a gene occur in certain tumor types, including pancreatic adenocarcinomas and esophageal and biliary tract carcinomas, and are associated with familial melanoma.1 Methylation of the CpG islands within the p16/INK4a and p15/INK4b genes, detected in breast and colon carcinomas, is yet another mechanism for silencing these genes.9-13 All these findings indicate that at least p16/INK4a is an important tumor suppressor gene.
A second p16/INK4a transcript that initiates from a more 5′ promoter encodes an entirely different protein, p16β or p19ARF, caused by splicing of the alternative exon 1β to the common exon 2, initiation of translation from an AUG within exon 1β, and usage of an alternative reading frame in exon 2.14-17 The p16β/p19ARF protein has been shown to induce both G1 and G2 arrest in NIH3T3 cells.17 Although at least some of the point mutations in p16/INK4a found in human tumors affect both the regular p16 protein and p16β/p19ARF, the role of the latter in tumorigenesis remains unclear.
Because inactivation of p16/INK4a and possibly p15/INK4b is a common mechanism for disruption of cell cycle control in human tumors, and because p16/INK4a null mice develop B-cell lymphomas with a high incidence,18 we decided to study the involvement of these genes in Burkitt's lymphoma (BL). Multiple factors contribute to the origin of this tumor. All BL carry chromosomal translocations that activate the c-myc proto-oncogene by juxtaposition to one of the Ig heavy and light chain loci. Most endemic BL and some sporadic BL carry Epstein-Barr virus (EBV).19 In addition, the p53 gene is mutated in a large fraction of BL biopsies and lines.20-22Non-neoplastic EBV-immortalized lymphoid cell lines (LCLs), on the other hand, lack Ig-myc translocations and express wild type p53. Here we show that the p16/INK4a and p15/INK4b loci are frequently methylated in BL but not in LCL, and that p16/INK4a methylation correlates with silencing of the p16/INK4a gene in BL lines.
MATERIALS AND METHODS
Tumor biopsies and cell lines.
Twenty-one primary BL biopsies from endemic areas in Africa were provided by George Klein, Microbiology & Tumor Biology Center, Karolinska Institute. More than 90% of the cells were tumor cells in these biopsies. Forty-three BL cell lines and 12 LCLs were also included (Table 1). Cells were grown in RPMI 1640 or Iscove's modified Dulbecco's medium (IMDM) supplemented with antibiotics and 10% fetal bovine serum (FBS). Normal human B cells were obtained from healthy donors using CD19-loaded magnetic beads (Dynal, Oslo, Norway).
p16/INK4a and p15/INK4b Status of BL and LCLs
| . | Gene Status . | Exon 1 Methylation . | RT-PCR . | Northern . | Western . | |||
|---|---|---|---|---|---|---|---|---|
| p16 . | p15 . | p16 . | p15 . | p16 . | p15 . | p16 . | p16 . | |
| BL lines | ||||||||
| Akata | + | + | + | − | − | + | − | ND |
| BL2 | − | − | NA | NA | − | − | − | − |
| BL18 | wt | + | − | + | + | − | − | − |
| BL28 | − | − | NA | NA | − | − | − | − |
| BL36 | + | + | + | − | − | + | − | − |
| BL41 | + | + | + | − | − | + | − | − |
| BL60 | + | + | + | − | (+) | + | − | − |
| CA46 | + | + | + | − | (+) | + | − | − |
| Daudi | + | + | + | + | − | + | − | − |
| DG75 | + | + | + | − | − | + | − | − |
| Ew36 | − | + | NA | + | − | + | − | − |
| Jijoye | + | + | + | + | − | + | ND | − |
| Naliaka | + | + | + | + | − | + | − | − |
| Namalwa | + | + | + | + | − | + | − | − |
| P3H3 | + | + | + | + | − | + | − | − |
| Rael | + | + | + | + | − | + | − | − |
| Raji | + | + | + | + | − | + | ND | − |
| Ramos | + | + | + | − | − | + | − | − |
| Seraphine | mut | + | − | + | + | + | (+) | − |
| WW2-BL | + | + | + | + | − | + | − | − |
| Akuba | wt | + | + | + | − | + | − | − |
| BL72 | + | + | + | − | − | ND | − | − |
| BL biopsies | ||||||||
| JN | + | + | + | + | − | |||
| JO | wt | wt | − | − | − | |||
| MM | wt | wt | + | − | − | |||
| MY | + | + | + | − | − | |||
| AM | + | + | + | − | ||||
| CC | wt | wt | + | + | ||||
| DBV | + | + | − | − | ||||
| HA | + | + | + | − | ||||
| HK | + | + | − | − | ||||
| JK | + | + | + | − | ||||
| KO | + | + | − | − | ||||
| MA | + | + | − | − | ||||
| MO | + | + | − | + | ||||
| NW | wt | wt | − | − | ||||
| ND | wt | wt | − | − | ||||
| PK | wt | wt | + | + | ||||
| RM | wt | wt | − | + | ||||
| SO | + | + | − | − | ||||
| WM | + | + | − | − | ||||
| LCLs | ||||||||
| Cherry | + | + | − | − | + | + | − | − |
| larc139 | + | + | − | − | + | + | + | + |
| larc171 | + | + | + | + | − | + | − | − |
| larc174 | + | + | − | − | + | + | (+) | (+) |
| larc307 | + | + | − | − | − | + | − | − |
| Nad20 | + | + | − | − | + | + | ND | (+) |
| WW2-LCL | + | + | − | − | − | + | − | − |
| Normal B cells | + | + | − | − | − | + | − | − |
| . | Gene Status . | Exon 1 Methylation . | RT-PCR . | Northern . | Western . | |||
|---|---|---|---|---|---|---|---|---|
| p16 . | p15 . | p16 . | p15 . | p16 . | p15 . | p16 . | p16 . | |
| BL lines | ||||||||
| Akata | + | + | + | − | − | + | − | ND |
| BL2 | − | − | NA | NA | − | − | − | − |
| BL18 | wt | + | − | + | + | − | − | − |
| BL28 | − | − | NA | NA | − | − | − | − |
| BL36 | + | + | + | − | − | + | − | − |
| BL41 | + | + | + | − | − | + | − | − |
| BL60 | + | + | + | − | (+) | + | − | − |
| CA46 | + | + | + | − | (+) | + | − | − |
| Daudi | + | + | + | + | − | + | − | − |
| DG75 | + | + | + | − | − | + | − | − |
| Ew36 | − | + | NA | + | − | + | − | − |
| Jijoye | + | + | + | + | − | + | ND | − |
| Naliaka | + | + | + | + | − | + | − | − |
| Namalwa | + | + | + | + | − | + | − | − |
| P3H3 | + | + | + | + | − | + | − | − |
| Rael | + | + | + | + | − | + | − | − |
| Raji | + | + | + | + | − | + | ND | − |
| Ramos | + | + | + | − | − | + | − | − |
| Seraphine | mut | + | − | + | + | + | (+) | − |
| WW2-BL | + | + | + | + | − | + | − | − |
| Akuba | wt | + | + | + | − | + | − | − |
| BL72 | + | + | + | − | − | ND | − | − |
| BL biopsies | ||||||||
| JN | + | + | + | + | − | |||
| JO | wt | wt | − | − | − | |||
| MM | wt | wt | + | − | − | |||
| MY | + | + | + | − | − | |||
| AM | + | + | + | − | ||||
| CC | wt | wt | + | + | ||||
| DBV | + | + | − | − | ||||
| HA | + | + | + | − | ||||
| HK | + | + | − | − | ||||
| JK | + | + | + | − | ||||
| KO | + | + | − | − | ||||
| MA | + | + | − | − | ||||
| MO | + | + | − | + | ||||
| NW | wt | wt | − | − | ||||
| ND | wt | wt | − | − | ||||
| PK | wt | wt | + | + | ||||
| RM | wt | wt | − | + | ||||
| SO | + | + | − | − | ||||
| WM | + | + | − | − | ||||
| LCLs | ||||||||
| Cherry | + | + | − | − | + | + | − | − |
| larc139 | + | + | − | − | + | + | + | + |
| larc171 | + | + | + | + | − | + | − | − |
| larc174 | + | + | − | − | + | + | (+) | (+) |
| larc307 | + | + | − | − | − | + | − | − |
| Nad20 | + | + | − | − | + | + | ND | (+) |
| WW2-LCL | + | + | − | − | − | + | − | − |
| Normal B cells | + | + | − | − | − | + | − | − |
Another 21 BL lines (AG876, BL10, BL16, BL29, BL37, BL49, BL57, BL67, JBL2, JD38, JI, KK125, Ly67, MC116, Mutu, Odour, PA682, PP984, Silfere, ST486, and WW1-BL), 5 LCLs (CBN1 12, LCL2, LCL6, Nad118, and WW1-LCL), and 2 BL biopsies (MK and TO), were screened for homozygous deletion of p16/INK4a and p15/INK4b and were found to retain both genes. Six of these BL lines (BL37, BL49, BL67, JI, Mutu, PA682, ST486, and Silfere) did not express any p16 protein. BL biopsies were not examined by Northern blotting and RT-PCR. Four BL biopsies were analyzed by Western blotting.
Abbreviations: NA, not applicable; ND, not determined; +, present; −, absent; wt, wildtype; mut, mutant; (+), only detected after long exposure.
Preparation of genomic DNA and polymerase chain reaction (PCR) amplification.
Genomic DNA was prepared as described.23 PCR was performed using 10 pmol of each primer, 250 μmol/L of dNTP mix, 5% dimethyl sulfoxide (DMSO), 1 U Taq Polymerase (Pharmacia, Uppsala, Sweden, or Perkin Elmer, Norwalk, CT), standard Taq buffer, and 50 to 100 ng of genomic DNA. p16/INK4a Exon 2 was amplified using the 42F and 551R primers, and p15/INK4b exon 2 was amplified using the 89F and 50R primers.7 PCR was performed for a total of 35 cycles using an annealing temperature of 58°C (p16/INK4a) or 56.5°C (p15/INK4b). PCR products were analyzed on EtBr-stained 1.2% agarose gels. The quality of the genomic DNA was confirmed by parallel PCR amplification of a GAPDH gene exon 8 fragment, using the same conditions as for PCR amplification of p15/INK4b, and the primers 5′-CCCTCCGGGAAACTGTGGCGT-3′ (GAPDHE8F) and 5′-ATGCCAGCCCCAGCGTCAAAG-3′ (GAPDHE8R). All PCR reactions were repeated at least twice.
Southern blot analysis.
Southern blotting was performed using standard procedures.23 A 126-bp fragment corresponding to nt 107-233 of p16/INK4a exon 2 was used as a probe to detect both p16/INK4a and p15/INK4b.7 The probe was radioactively labeled using Megaprime DNA labeling system (Amersham, Buckinghamshire, UK) following the manufacturer's instructions. Ten micrograms ofEcoRI-digested genomic DNA from each sample was separated on 1% agarose gels and transferred onto Hybond N+ filters (Amersham). Hybridizing bands were visualized by autoradiography or Phospholmager.
DNA sequencing.
Sequencing reactions were performed using the Taq DyeDeoxy Terminator Cycle Sequencing Kit (Applied Biosystems, CT) according to the manufacturer's instructions. All sequencing reactions were performed on pooled PCR samples from three different reactions. The primers used for sequencing of p16/INK4a exon 1 were 2F and 1108R.7 The primers used for sequencing of exon 2 of p16/INK4a and p15/INK4b were 42F and 551R, and 89F and 50R, respectively.
Reverse transcription (RT)-PCR.
cDNA was synthesized from total RNA using M-MLV Reverse Transcriptase (GIBCO-BRL, Gaithersburg MD) according to the manufacturer's instructions. RT-PCR was performed using 10 pmole of each primer, 250 μmol/L of dNTP mix, 5% (p16 and p16β) or 3% (p15) DMSO, and 1 U of Taq Polymerase (Pharmacia) and standard Taq buffer. PCR was performed for a total of 35 cycles using an annealing temperature of 61°C and the primers 5′-ACTAGATCTTCGCACGAGGCAGCATGG-3′ (p16E1F) and 5′-GTTGTGGCGGGGGCAGTTGT-3′ (p16E3R) for p16 cDNA; 5′-AAGGATCCATGGTGCGCAGGTTCTTGG-3′ (p16BF) and p16E3R (see above) for p16β cDNA; and 5′-GTTTACGGCCAACGGTGGA-3′ (p15E1F) and 5′-GCAGAATTCATCGAATTAGGTGGGTGG-3′ (p16E2R) for p15 cDNA. Ten microliters from each reaction was run on 1% agarose gels and then transferred onto Hybond N+ membranes (Amersham). Membranes were probed with 32P-dCTP–labeled probes corresponding to exon 1 of the respective genes. As control, GAPDH cDNA was amplified by RT-PCR using the primers 5′-TGCCTCCTGCACCACCAACTG-3′ (GAPDHE7F) and GAPDHE8R (see above) for each sample tested. Hybridizing bands were visualized by PhosphoImager.
Northern blot analysis.
Northern blotting was performed as described.23 A 175-bpBamHI-BgII p16/INK4a cDNA fragment corresponding to the entire coding region of exon 1 and the first 20 bp of the coding region of exon 2 was used as probe. Twenty micrograms of total RNA was loaded in each lane. Hybridizing bands were visualized by PhosphoImager.
Western blot analysis.
Detection of p16 protein by immunoblotting was performed as outlined.24 Cell extracts were prepared from subconfluent cultures by resuspending 1 × 107 cells in 100 μL of Laemmli sample buffer. Total protein from each sample (50 to 150 μg) was separated by sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis and transferred onto nitrocellulose. p16 was detected with the rabbit polyclonal antiserum 15126E and/or the mouse monoclonal antibody 13251A (Pharmingen, CA), followed by ECL (Amersham) according to the manufacturer's instructions. Actin was detected with the mouse antibody N350 (Amersham).
Methylation assay.
Genomic DNA from primary tumors and cell lines was digested overnight with SacII (Pharmacia), HpaII (MBI Fermentas, Vilnius, Lithuania; GIBCO BRL; and Pharmacia) or MspI (New England Biolabs, Beverly, MA), using 20 U enzyme/μg DNA. PCR was performed using 10 pmol of each primer, 250 μmol/L of dNTP mix, 5% DMSO, 1 U Taq Polymerase (Pharmacia) standard Taq buffer, and 125 ng of genomic DNA. PCR was performed for a total of 21 cycles using an annealing temperature of 59°C and the primers 5′-ATGGAGCCTTCGGCTGAC-3′ (p16metF), 2F and 1108R for p16/INK4a exon 1, and 5′-TTTCCCAGAAGCAATCCA-3′ (p15metF) and 5′-TGTCGCACCTTCTCCACT-3′ (p15 metR) for p15/INK4b exon 1. Ten microliters from each reaction was run on 1% agarose gels and transferred onto nylon membranes (Hybond N+; Amersham). Membranes were probed with 32P-dCTP–labeled probes corresponding to part of exon 1 of the respective genes (Fig1). Hybridizing bands were visualized by PhosphoImager. All restriction digestions, PCR reactions, and transfers were repeated at least twice.
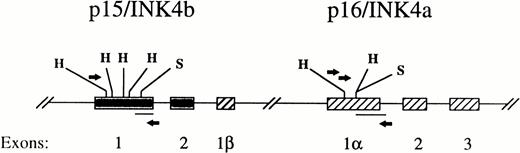
Strategy for detection of gene methylation using PCR followed by hybridization with an internal probe. Primers used are indicated by the arrows. Filled lines represents probes. Cleavage sites for HpaII/Msp I (H) and SacII (S) are indicated. The gene map is not drawn to scale.
Strategy for detection of gene methylation using PCR followed by hybridization with an internal probe. Primers used are indicated by the arrows. Filled lines represents probes. Cleavage sites for HpaII/Msp I (H) and SacII (S) are indicated. The gene map is not drawn to scale.
RESULTS
Analysis of p16/INK4a and p15/INK4b by PCR and Southern blotting.
Forty-three BL lines and 12 LCLs were screened for homozygous deletion of p16/INK4a and p15/INK4b by PCR, using primers surrounding exon 2.7 All 12 LCLs and 40 out of 43 BL lines retained exon 2 of both p16/INK4a and p15/INK4b. Three BL lines (BL2, BL28, and Ew36; 7%) showed homozygous deletion of exon 2 of p16/INK4a, and two lines (BL2 and BL28; 4.6%) also had homozygous deletion of exon 2 of p15/INK4b (Fig 2, lanes 3, 5, and 7; Table1). These results were confirmed by Southern blotting (data not shown). Iarc139, an LCL derived from the same patient as BL28, retained both p16/INK4a and p15/INK4b, showing that homozygous deletion of p16/INK4a and p15/INK4b is a somatic rather than a constitutional change.
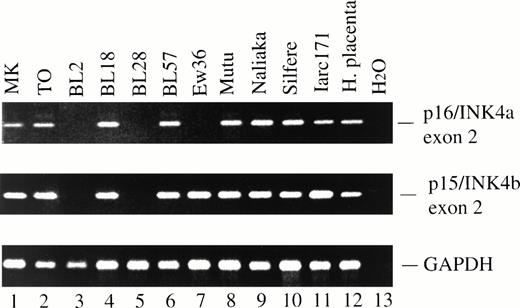
PCR amplification of exon 2 of p16/INK4a (top) and p15/INK4b (middle). BL2 (lane 3) and BL28 (lane 5) had homozygous deletion of exon 2 of both p16/INK4a and p15/INK4b, whereas Ew36 (lane 7) had homozygous deletion of p16/INK4a exon 2 but retained p15/INK4b exon 2. The expected p16/INK4a and p15/INK4b PCR products were obtained from genomic DNA from all primary BL biopsies indicated in Table 1. Two examples, MK and TO, are shown (lanes 1 and 2). Lanes 3 to 10, BL lines; lane 11, LCL. DNA integrity was confirmed using primers specific for GAPDH exon 8 (bottom).
PCR amplification of exon 2 of p16/INK4a (top) and p15/INK4b (middle). BL2 (lane 3) and BL28 (lane 5) had homozygous deletion of exon 2 of both p16/INK4a and p15/INK4b, whereas Ew36 (lane 7) had homozygous deletion of p16/INK4a exon 2 but retained p15/INK4b exon 2. The expected p16/INK4a and p15/INK4b PCR products were obtained from genomic DNA from all primary BL biopsies indicated in Table 1. Two examples, MK and TO, are shown (lanes 1 and 2). Lanes 3 to 10, BL lines; lane 11, LCL. DNA integrity was confirmed using primers specific for GAPDH exon 8 (bottom).
The status of the p16/INK4a and p15/INK4b genes was also examined in 21 primary BL biopsies. PCR analysis showed that all BL biopsies retained exon 2 of both genes. Representative examples of this analysis are shown in Fig 2 (lanes 1 and 2). These results were confirmed by Southern blotting (data not shown).
Analysis of p16/INK4a and p15/INK4b mRNA and protein expression.
p16/INK4a mRNA levels in the BL lines were examined by Northern blotting and RT-PCR. As expected, no p16/INK4a mRNA was detected in the BL lines that carried homozygous p16/INK4a deletion. Surprisingly, however, Northern blot analysis did not reveal any p16/INK4a mRNA in a majority of the BL lines that had retained the p16/INK4a gene. Only 1 out of 20 BL lines (Seraphine) and 2 out of 6 LCLs analyzed (Iarc139 and Iarc174) expressed levels of p16/INK4a mRNA detectable by Northern blotting (data not shown). Using the more sensitive RT-PCR technique followed by hybridization with an internal probe, p16/INK4a mRNA was detected in Seraphine and BL18, and in the LCLs Cherry, Iarc139, Iarc174, and Nad20 (Fig 3; Table 1). After long exposure, low levels of p16/INK4a mRNA were also found in the BL lines BL60 and CA46 (not shown). The alternative p16/INK4a transcript, p16β, was expressed in all BL lines with intact p16/INK4a gene (Fig3). Using RT-PCR, low levels of p15/INK4b mRNA were detected in all BL lines that carried an intact p15/INK4b gene, except BL18. p15/INK4b mRNA was also detected in all six LCLs tested, and in normal B cells (Fig 3; Table 1).
Detection of p16/INK4a, p16β/INK4a, and p15/INK4b mRNA by RT-PCR and hybridization with an internal probe. After electrophoresis, PCR products were transferred to nylon filters and hybridized with probes corresponding to exon 1 of p16/INK4a, p16β/INK4a, p15/INK4b, and rat GAPDH cDNA. Hybridizing bands were visualized by Phospholmager.
Detection of p16/INK4a, p16β/INK4a, and p15/INK4b mRNA by RT-PCR and hybridization with an internal probe. After electrophoresis, PCR products were transferred to nylon filters and hybridized with probes corresponding to exon 1 of p16/INK4a, p16β/INK4a, p15/INK4b, and rat GAPDH cDNA. Hybridizing bands were visualized by Phospholmager.
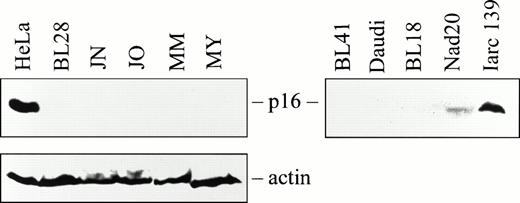
Western blotting revealed high levels of p16 protein in the Saos-2 and HeLa cell lines used as positive controls. However, no p16 protein was observed in any of the 4 BL biopsies and 27 BL lines examined (Fig4; Table 1). Among the LCLs, Iarc139 expressed comparatively high levels of p16 protein (Fig 4), and Nad20 and Iarc174 expressed low levels detectable after long exposure (Fig 4and data not shown). The remaining LCLs and normal human B cells did not express detectable levels of p16 protein (Table 1).
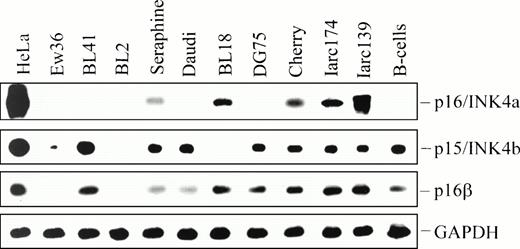
Western blot analysis of p16 expression in BL biopsies (left panel), cell lines, and LCLs (right panel). HeLa cells were used as positive control. An actin antibody was used as an internal control to confirm protein integrity.
Western blot analysis of p16 expression in BL biopsies (left panel), cell lines, and LCLs (right panel). HeLa cells were used as positive control. An actin antibody was used as an internal control to confirm protein integrity.
Methylation of the p16/INK4a and p15/INK4b genes.
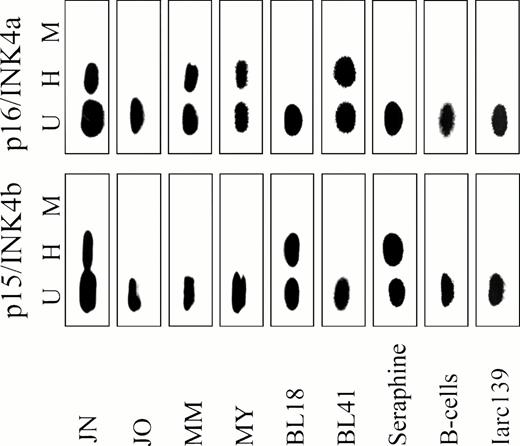
To determine whether the lack of p16/INK4a expression in the BL lines was caused by methylation of the CpG island in p16/INK4a exon 1, we used a previously described PCR strategy9 (Fig 1). Genomic DNA from primary BL biopsies, BL lines, LCLs, and normal human B cells were digested with the methylation-sensitive restriction enzymesSacII or HpaII, or the methylation-insensitive enzymeMsp I, an isoschizomer of HpaII. Digested and intact DNA were subsequently analyzed by PCR followed by hybridization with an internal probe (Fig 1). Representative examples of this analysis using the 3′ of the two forward primers are shown in Fig5. Eight out of 19 primary BL biopsies (42%) examined were methylated at the 3′ HpaII site. Methylation of both HpaII sites, as shown by the appearance of a PCR product following HpaII cleavage when the 5′ of the two forward primers was used, was not observed in any of the primary BL. Seventeen out of 19 BL lines (89.5%) were methylated at the 3′HpaII site. The 5′ HpaII site and the SacII site were less frequently methylated than the 3′ HpaII site in the BL lines (not shown). Only two BL lines, Seraphine and BL18, both of which expressed p16/INK4a mRNA, were unmethylated in the region examined (Fig 5; Table 1). Thus, p16/INK4a exon 1 methylation occurs with high frequency in BL. In contrast, six out of seven LCLs and normal human B cells were unmethylated in this region (Fig 5; Table 1).
Methylation of exon 1 of p15/INK4b (bottom panel) and p16/INK4a (top panel) in representative BL biopsies, BL lines, LCLs, and normal B cells. Genomic DNA was digested with HpaII (H),Msp I (M), or left undigested (U), and analyzed by PCR followed by hybridization with an internal probe, as shown in Fig 1.
Methylation of exon 1 of p15/INK4b (bottom panel) and p16/INK4a (top panel) in representative BL biopsies, BL lines, LCLs, and normal B cells. Genomic DNA was digested with HpaII (H),Msp I (M), or left undigested (U), and analyzed by PCR followed by hybridization with an internal probe, as shown in Fig 1.
Methylation of the p15/INK4b gene was studied using the same approach, except that only one forward primer was used (Fig 1). Twelve out of 20 BL lines (60%) and 5 out of 19 BL biopsies (26%) were methylated at all four HpaII sites in p15/INK4b exon 1. p15/INK4b Methylation was not found in normal B cells nor in six out of seven LCLs investigated (Fig 5; Table 1).
DNA sequence analysis of p16/INK4a and p15/INK4b.
DNA sequencing of exon 2 of p16/INK4a and p15/INK4b did not reveal any mutations in the 7 BL biopsies analyzed (Table 1). In addition, exons 1 and 2 of p16/INK4a in the BL lines Seraphine, BL18 (both carrying unmethylated p16/INK4a) and Akuba (carrying methylated p16/INK4a) were sequenced. This analysis showed a G to T substitution at nucleotide 298 in exon 2 of the p16/INK4a gene in Seraphine, resulting in an Ala to Ser substitution at codon 100. No mutations were found in exons 1 and 2 of p16/INK4a in BL18 and Akuba.
DISCUSSION
We found that only a minor fraction of the BL lines had homozygous deletion of the p16/INK4a and p15/INK4b genes, confirming studies by Stranks et al25 and Herman et al.26 Thus, BL differs in this respect from for instance acute lymphoblastic leukemias, which often carry homozygous deletion of p16/INK4a.27 Moreover, DNA sequencing did not reveal any small deletions or point mutations in exon 2 of p16/INK4a and p15/INK4b in the BL biopsies examined, nor in p16/INK4a exon 1 and 2 in the BL lines BL18 and Akuba. These results thus indicate that inactivation of the p16/INK4a and p15/INK4b genes by structural alterations is infrequent in BL.
Although most BL lines had retained the p16/INK4a gene, only 4 BL lines expressed detectable levels of p16/INK4a mRNA as determined by RT-PCR followed by hybridization with an internal probe. Because methylation of the p16/INK4a and p15/INK4b genes has been associated with downregulation of their expression in several tumor types,9-13 we asked whether p16/INK4a and p15/INK4b gene methylation occurred in our BL lines and biopsies. We found methylation of exon 1 of the p16/INK4a gene in a majority (89.5%) of the BL lines and in a large fraction (42%) of the primary BL biopsies. A similar frequency was reported in a study of eight primary BL tumors.26 We did not observe p16/INK4a gene methylation in any of the LCLs examined except Iarc171, nor in normal human B cells, consistent with the possibility that methylation is a BL-associated phenomenon responsible for silencing the p16/INK4a gene. In support of this idea, methylation of p16/INK4a exon 1 showed an inverse correlation with p16 mRNA expression in the BL lines and LCLs. The only two BL lines that were not methylated in p16/INK4a exon 1, BL18 and Seraphine, expressed p16/INK4a mRNA levels detectable by RT-PCR (Fig3). Two other BL, CA46 and BL60, both carrying a methylated p16/INK4a gene, also expressed p16/INK4a mRNA. However, the levels of p16/INK4a mRNA expressed by CA46 and BL60 were very low and could only be detected after long exposure of the RT-PCR blots. It is possible that complete silencing of the p16/INK4a gene requires methylation of additional sites outside the region studied here, and that these sites are not methylated in CA46 and BL60. At least four of the LCLs that carried unmethylated p16/INK4a expressed p16/INK4a mRNA as determined by Northern analysis and/or RT-PCR; the only LCL that carried methylated p16/INK4a (Iarc171) did not express p16/INK4a mRNA and protein. Thus, p16/INK4a methylation occurred with high frequency in the BL biopsies and lines but not in the LCLs, and was associated with silencing of this gene in the BL lines, suggesting the possibility that p16/INK4a inactivation through this mechanism is involved in the development of BL. The observation that the frequency of p16/INK4a gene methylation is higher in BL lines than primary tumors indicates a selection for p16/INK4a gene inactivation also during in vitro culture. A corresponding discrepancy between the mutation rate in primary BL biopsies and BL lines exists for the p53 gene.21 28
Exon 1 of p15/INK4b was also methylated in the BL lines and primary tumors, although at a lower frequency than p16/INK4a. Many BL lines with methylated p15/INK4b expressed p15/INK4b mRNA, as shown by RT-PCR followed by hybridization with an internal probe (Table 1). This is consistent with the observation that p15/INK4b gene methylation does not necessarily lead to complete silencing of the gene.9However, we cannot exclude the possibility that a fraction of each sample contains cells with unmethylated p15/INK4b that account for the p15/INK4b mRNA detected.
DNA sequencing revealed a point mutation in the coding sequence of p16/INK4a in the BL line Seraphine. This gives rise to an Ala to Ser substitution at residue 100 in the p16 protein. We have no experimental data regarding the consequences of this mutation for p16 function, but the fact that it occurred in one of the two BL lines carrying an unmethylated p16/INK4a gene is consistent with the idea that point mutation represents an alternative, uncommon mechanism for p16/INK4a inactivation in BL.
The cytogenetic hallmark of BL is the chromosomal translocations that activate the c-myc proto-oncogene. The c-myc protein has been shown to activate cdc25A, a gene encoding a CDK-activating phosphatase expressed in the early G1 phase of the cell cycle.29 CDK4 and CDK6, two kinases that form complexes with D cyclins in the G1 phase and phosphorylate the RB protein, are putative substrates of cdc25A. Thus, one consequence of constitutive c-myc activation in BL may be disruption of normal G1 cell cycle control through increased RB phosphorylation. Nevertheless, our observation that p16/INK4a is often methylated in BL indicates that p16/INK4a inactivation may provide a selective growth advantage even in the presence of constitutively active c-myc. The reason may be that activation of the cdc25A phosphatase would not result in CDK4 or CDK6 activation if p16/INK4a is expressed. p16/INK4a could thus be considered dominant over cdc25A in this respect. This notion is supported by the observation that p16/INK4a can block transformation of primary rat embryo fibroblasts by c-myc and mutant ras.30
A large fraction of BL also carry p53 mutations. Because c-myc is known to induce apoptosis in a p53-dependent manner,31 it is possible that constitutive c-myc expression would lead to a selection for p53 mutations that would prevent p53-induced apoptosis. Moreover, functional RB has been shown to inhibit p53-dependent apoptosis.32 Loss of p16/INK4a, resulting in functional RB inactivation through CDK4/6-mediated phosphorylation, may therefore promote p53-dependent apoptosis as well, and further increase the selection for p53 mutation.
EBV is another factor involved in the genesis of BL. All BL lines that had homozygous deletion of p15/INK4b and/or p16/INK4a, namely BL2, BL28, and Ew36, are EBV negative. However, the BL lines and biopsies that carried p16/INK4a methylation included both EBV positive and EBV negative BL (for example the EBV positive Daudi, Namalwa, and WW2-BL, and the EBV negative BL41, DG75, and Ramos).22 This suggests that EBV does not reduce the selection for p16/INK4a gene inactivation during BL development. The situation may be different in LCLs. In contrast to primary BL and phenotypically representative BL lines, LCLs express the EBV-encoded nuclear antigens EBNA-2-6 and the membrane proteins LMP-1, -2A, and -2B, several of which are important for transformation of B cells.19 Conceivably, these transformation-associated EBV-encoded proteins may allow unlimited growth despite the expression of p16/INK4a in LCLs.
We detected the alternative p16/INK4a transcript, p16β, by RT-PCR and/or Northern blotting in all BL lines carrying an intact p16/INK4a gene (Fig 3; data not shown). This is in agreement with the notion that methylation of p16/INK4a exon 1 does not silence p16β transcription, and that p16β is usually not targeted during BL development. The BL with homozygous deletion of p16/INK4a, which knocks out both the regular and the p16β transcript, are obvious exceptions. Moreover, the G to T base substitution in exon 2 of p16/INK4a in Seraphine that replaces Ala with Ser in the regular p16 protein also affects the p16β protein, causing a Gly to Val change at codon 114. We cannot exclude that p16/INK4a exon 1β or other regions of the p16/INK4a gene that were not analyzed have suffered point mutations or minor deletions that inactivate p16β in other BL lines studied here. Nevertheless, the frequent p16/INK4a methylation and the relatively infrequent homozygous deletion of this gene as shown in this study indicate a selection for inactivation of the regular p16INK4a protein but not the p16β protein in BL.
ACKNOWLEDGMENT
We thank George Klein, Karolinska Institute, for providing BL lines and biopsies and LCLs, and Alexander Kamb, Myriad Genetics, for the p16/INK4a cDNA.
Supported by grants from the Gustaf V Jubilee Fund and the Swedish Cancer Society (Cancerfonden). U.K. was supported by Pharmacia & Upjohn.
Address reprint requests to Klas G. Wiman, Microbiology & Tumor Biology Center, Karolinska Institute, Doktorsringen 13, S-171 77 Stockholm, Sweden.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal