Abstract
A region of chromosome 13q14.3, telomeric to the Retinoblastoma gene RB-1 is frequently deleted in patients with B-cell chronic lymphocytic leukemia (B-CLL). A cosmid and P1-derived artificial chromosome (PAC) contig spanning over 600 kb has been constructed, which encompasses this locus. The contig clones have been used to order a number of markers along the minimally deleted region and to localize a series of CpG islands corresponding to possible candidate genes. A novel polymorphic dinucleotide repeat, 6E3.2, present in one of the ordered cosmid clones has been isolated for use in deletion mapping studies of patient DNA. Leukemic samples from 229 CLL patients have been screened for loss of heterozygosity using microsatellite markers and analyzed for hemizygous and homozygous deletions by Southern blot techniques using genomic probes selected from cosmids across the region. Hemizygous deletions were found in 31% of cases with an additional 10% showing homozygous loss. The use of these probes has defined the commonly deleted area to less than 130 kb, centromeric to the locus D13S272.
B-CELL CHRONIC LYMPHOCYTIC leukemia (B-CLL) is the most frequently found leukemia among adults in Western Europe and North America.1,2 However, little is known about its etiology and pathogenesis. Cytogenetic analysis has shown recurring abnormalities of which the most common are trisomy 12 and structural abnormalities of chromosome 13q.3,4 Deletions or translocations of chromosome 13q14 are found in approximately 20% of cases5 and are often the sole abnormality in early stage CLL, suggesting that the region contains one or more genes of importance in the pathogenesis of B-CLL.6-8 The locus D13S259 located 1.6 centimorgans telomeric to the retinoblastoma gene has been found to be deleted homozygously in B-CLL cases, indicating the likely presence of a tumor suppressor gene in its vicinity.10-18 More recently we have shown a higher incidence of homozygous loss at another locus, D13S319, located between D13S25 and RB-1.19 With the availability of a series of YAC contigs, cosmid clones, and microsatellite markers,20-23subsequent studies have sought to further define the minimum critical region of genetic loss.
We have produced a library of cosmids covering the region between D13S273 and D13S31 including a contig covering over 600 kb that contains the markers D13S319, D13S273, 206XF12, D13S25, and D13S262. A series of nonrepetitive genomic probes were isolated from these cosmids and used in the isolation of a series of P1-derived artificial chromosome (PAC) clones that were integrated into the genomic map of the region. Previously identified microsatellite markers20 22 have been accurately ordered with respect to each other and a novel polymorphic microsatellite has been isolated for use in deletional mapping of B-CLL patient samples. A map of the region has been created by integrating the cosmid and PAC data with restriction maps of the yeast artificial chromosome (YAC) clones used in the study. This has enabled the minimal region of loss in patient samples to be defined within the contig borders and narrowed to less than 130 kb.
MATERIALS AND METHODS
Cosmid Subcloning
A series of three YACs, YAC ICRF C161 (340 kb), YAC ICRF A9126 (540 kb), and YAC CEPH 922A8 (1800 kb) isolated from the Institute of Cancer Research Fund (ICRF) human YAC library and the Centre d'Etudes du Polymorphisme Humain (CEPH) 2 human YAC library, respectively, have previously been shown to encompass the critical region.23 They appeared nonchimeric by fluorescent in situ hybridization (FISH) studies and were chosen for a cosmid subcloning procedure.24 Total yeast and YAC DNA were partially digested with the restriction endonuclease Mbo I, alkaline phosphatase treated, ligated to the prepared vector Supercos1, packaged (Stratagene Gigapack II XL; Stratagene, La Jolla, CA), and used to introduce the cosmids to a suitable host strain XL1 Blue MR′. The bacteria were plated out on filters on selective media and human sequence-specific cosmids were isolated after hybridization of replica filters with a repetitive human probe (Cot1 DNA; GIBCO BRL, Gaithersburg, MD) and a total yeast genomic probe.
The cosmid clones positive for the human probe alone were purified and allocated to a specific address in a gridded library for each YAC subcloned. At least a fourfold degree of coverage of YAC insert area was aimed for with 36 cosmids from YAC C161, 60 cosmids from YAC A9126, and 200 cosmids from YAC 922A8.
Contig Formation
Cosmids in each of the three libraries were screened by polymerase chain reaction (PCR) or hybridization for the following markers: D13S273, D13S272, D13S319, 206XF12, D13S25, D13S262, D13S284, and D13S31. Cosmids positive for one or more of these markers were used as the basis of contig formation. Cosmids from each library were fingerprinted using EcoRI digestion, agarose gel electrophoresis, Southern blotting, and hybridization to a repetitive human genomic probe (Cot1; GIBCO BRL). A series of single copy genomic subclones from various cosmids in the libraries were isolated and used to identify and link cosmids lacking the previously screened markers. Nonrepetitive fingerprinting was performed using whole cosmid hybridization to EcoRI-digested cosmid blots to define minimally overlapping cosmid clones within the contig25 in addition to Alu-PCR fingerprinting using the primers Alu517 and BK33 followed by agarose gel electrophoresis.26 27
PAC Library Screening
A human genomic PAC library in the vector pCYPAC2N produced by Pieter de Jong's group at the Roswell Park Cancer Institute, Buffalo, NY, and obtained from the United Kingdom Human Genome Mapping Resource Centre, Cambridge, UK, was screened with a series of single copy probes isolated from four cosmids in separate locations within the deletion region. A series of nine PACs were isolated and mapped back to the chromosome 13q14.3 region by means of FISH and integrated into the cosmid and YAC map of the region.
Identification of Potential Microsatellite Polymorphisms
Cosmids from the contig were hybridized with a CA repeat probe (Pharmacia, Uppsala, Sweden) to indicate the presence of a number of potentially polymorphic repeat containing areas within the region. From one of these cosmids (cosmid C6a) the regions surrounding the dinucleotide repeat sequence (CA)n were sequenced by dideoxy chain termination sequencing and used in the design of primers to produce an informative polymorphic PCR probe for use in deletion studies.
Pulse Field Gel Electrophoresis
Total yeast high molecular weight DNA from the YACs C161, 9126, and 922A8 was prepared in 1% low melting temperature agarose. The YACs C161, 9126, and 922A8 were used in partial digestion mapping studies to identify distances between clusters of cutting sites for infrequently cutting enzymes using probes that recognize the right (URA3) or left (ARS 1 Trp 1) YAC vector arms.28
Patient Samples
Blood samples were obtained with informed consent from patients with B-CLL, whose diagnosis was based on lymphocyte morphology, CD19, CD23, CD5, and the weak expression of monotypic immunoglobulin. Peripheral blood samples were subjected to ficoll gradient centrifugation. In patients whose lymphocyte count was below 30 × 109cells/L the mononuclear cells were either T-cell depleted using sheep red cell rosetting or positively selected for B cells using CD19-positive magnetic beads (Dynal, Oslo, Norway). The resulting mononuclear cells consisted of >95% leukemic cells as determined by immunophenotyping studies. Polymorphonuclear cells or T lymphocytes obtained with CD3-positive magnetic beads were used as control fractions. DNA was extracted from the separated leukemic and control cell fractions using a standard proteinase K phenol-chloroform extraction.29
DNA Probes
Probe p68RS2.0 recognizes a variable number tandem repeat (VNTR) polymorphism within the retinoblastoma gene30; ph2-42 recognizes the D13S25 locus; MGG15 recognizes a 5-kb EcoRI fragment from cosmid C9a, which contains the markers D13S319 and D13S272 within it; p9E4.3 is a 4.3-kbEcoRI fragment, which hybridizes to a CpG island <30 kb centromeric to D13S319; p6E4.5 recognizes a CpG island <200 kb centromeric to D13S319, p31a is located within 40 kb centromeric to p6E4.5; and p68c is located approximately 200 kb telomeric to D13S319. Other genomic probes used were p6E3.2, p9a, p9.19, p29a, p18a, p92c, and p25b, isolated from cosmids C9a, C29a, C18a, C92c, and C25b, respectively (Fig 1). Control probes used to help in the quantification of loss were a genomic clone from the renin gene located on chromosome 1, β-interferon on chromosome 9, pb16 (bcl-2 cDNA clone corresponding to the 1.6-kb 3′ part of the first exon) on chromosome 18,31 p105-153A (5q11.2-5q13.3; D5S39),32 and pc117.3, a genomic fragment from the bcl-1 gene on chromosome 11.
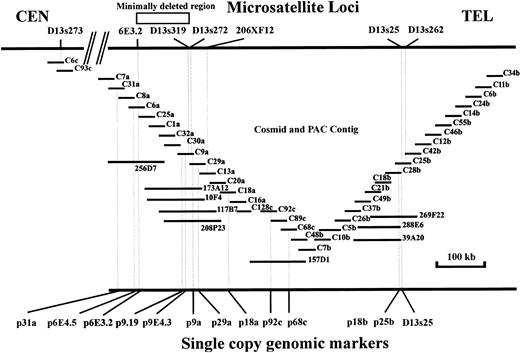
Cosmid and PAC map of region contained within contig. Locations of microsatellites are shown on upper bar with genomic single copy probes shown on the lower bar.
Cosmid and PAC map of region contained within contig. Locations of microsatellites are shown on upper bar with genomic single copy probes shown on the lower bar.
Southern Analysis
Southern hybridization analysis was performed as described previously.15 19 Eight micrograms of leukemic DNA was digested with the restriction endonucleases EcoRI, SacI, or HindIII, separated by 1% agarose gel electrophoresis and transferred to nylon filter (Hybond N, Amersham, Little Chalfont, UK). The samples were probed with a series of single copy probes from the region. The ratios between the signals obtained using 13q14.3 probes and signals obtained using the control probes was determined for each of the samples either by visual inspection or using a laser scanning densitometer (Pharmacia). Control cell DNA was included on all filters. Samples were determined to show hemizygous loss if the tumor signal was between 40% to 50% less intense compared with the control signal and to show homozygous loss if the tumor signal was below 10% of the control signal.
Microsatellite Studies
Tumor and normal DNA samples were PCR amplified to test for loss of heterozygosity at the following microsatellite loci: D13S273, D13S272, D13S319, and 6E3.2, in addition to PCR restriction fragment length polymorphisms (RFLP) present in the RB-1 gene33 and the polymorphic dinucleotide repeat and Ssp I site at the D13S25 locus.18 PCR amplification was performed using 100 ng of template DNA, 50 μmol/L of each of dCTP, dATP, dTTP, and dGTP and 2 μCi of 32P dCTP using 1 U Thermoprime polymerase (Applied Biotechnologies, Leatherhead, UK) in a total volume of 25 μL. The cycle conditions used for the primers were as follows: 94°C for 30 seconds, 58°C for 30 seconds, and 72°C for 30 seconds for the loci D13S319, D13S272, and 6E3.2; 94°C for 30 seconds, 57°C for 30 seconds, and 72°C for 30 seconds for D13S25; and 94°C for 30 seconds, 53°C for 30 seconds, and 72°C for 30 seconds for D13S273. The primer sequences used for the 6E3.2 polymorphism were 6E3.2FOR 5′-GCAAGTGGATTGGTTTGGTTG-3′ and 6E3.2REV 5′-ACATAGCAAGACCCAGTCTC-3′. All other microsatellite primer sequences were obtained from previous studies.19 20 All samples were amplified on two separate occasions for 24 cycles to verify loss of heterozygosity (LOH) results. Microsatellite products were separated on a 6% polyacrylamide gel for between 2 and 5 hours depending on product size and exposed overnight to autoradiography film (Hyperfilm MP; Amersham). LOH results for each tumor/normal pair were determined by visual inspection of the bands resulting from the PCR. LOH was scored positive if one of the polymorphic band's intensity was reduced by >75% between normal and tumor samples. Homozygous loss was not determined by microsatellite analysis.
FISH Analysis
PAC clone DNA (1 μg) was labeled with digoxigenin 11-dUPT by nick translation and hybridized at a concentration of 10 ng/μL with an excess of blocking DNA (Cot1; GIBCO BRL) in a buffer containing 2 × SSC (1 × SSC = 0.15 mol/L NaCl/0.015 mol/L sodium citrate), 10% dextran sulphate and 1% sodium dodecyl sulfate (SDS) to metaphase spreads of control samples. A chromosome 13/21 centromeric probe (Alpha laboratories, Eastleigh, UK) labeled with biotin was used as a control for hybridization and chromosomal localization.
RESULTS
Cosmid and PAC Contig
A total of 296 human specific cosmids were isolated from the subcloning of three nonchimeric YACs encompassing the minimally deleted region of 13q14.3 to create a highly enriched library of cosmids from this region. These cosmids were used to create several contigs including an overlapping group of 38 clones extending from approximately 200 kb centromeric of the marker D13S319 to over 100 kb telomeric to the marker D13S25. Nine PAC clones isolated from the Human Genome Mapping Project (HGMP) human PAC library were integrated into this detailed map (Fig 1). The cosmid clones, with an average insert size of 38 kb, have been used as a source of single copy markers from across the region and as the basis for the isolation of novel polymorphic microsatellites. The contig has been used in the ordering of a series of previously identified loci. The order centromeric to telomeric of these loci is D13S273, D13S319, D13S272, 206XF12, D13S25, and D13S262.
The maximum distance between the ordered markers can now be estimated from the overlapping cosmid insert size determination. The marker 6E3.2 lies within 3 kb of the 230-kb Mlu I site found in the restriction analysis of YAC C161 (Fig 2). The markers D13S319 and D13S272 are situated within 30 kb telomeric to theMlu I site present in the probe p9E4.3 and 130 kb from the left vector arm of this YAC, thus placing 6E3.2 within 100 kb centromeric of p9E4.3. Cosmid C9a contains the microsatellite markers D13S319 and D13S272, which are both located within a 5-kb EcoRI fragment. Cosmid C29a, which also contains the marker D13S272, overlaps with cosmid C13a, which contains the more telomeric marker 206XF12, thus ordering D13S272 more telomeric to D13S319 and indicating the distance between D13S272 and 206XF12 to be less than the combination of the two cosmid inserts, a total of 73 kb. The distance between the marker p92c and the marker 206XF12 can be estimated from cosmid and PAC insert sizes spanning the intervening distance, which by our estimate should not be greater than 180 kb. Likewise the distance between 206XF12 and D13S25 can be estimated by the addition of the intervening cosmid and PAC genomic insert sizes and should be no greater than 400 kb. Cosmid C25b, which has an insert size of 36 kb, contains the marker D13S25 and the microsatellite marker D13S262. A nonrepetitive genomic subclone p25b was isolated from this cosmid and used to link it to an overlapping cosmid C28b, which contains D13S25 but not D13S262. Cosmid C28b in turn was linked to a series of centromeric cosmid and PAC clones, which eventually overlap with clones in the 206XF12 region. This positions D13S25 within 36 kb and centromeric to D13S262.
Partial digestion mapping of YAC C161. YAC C161 was partially digested, as specified in the Materials and Methods, and hybridized with a probe recognizing the left (ARS 1 Trp 1) vector arm. Lanes numbered 1 to 8, respectively, are undigested Mlu I cut,BssHII cut, Sfi I cut, Not I cut, Sal I cut, Nru I cut, and Sac II cut. This result indicates a large cluster of rare cutting sites located 130 kb from the left YAC vector arm. Other clusters of rare cutting sites are located 170 kb and 230 kb from this vector arm.
Partial digestion mapping of YAC C161. YAC C161 was partially digested, as specified in the Materials and Methods, and hybridized with a probe recognizing the left (ARS 1 Trp 1) vector arm. Lanes numbered 1 to 8, respectively, are undigested Mlu I cut,BssHII cut, Sfi I cut, Not I cut, Sal I cut, Nru I cut, and Sac II cut. This result indicates a large cluster of rare cutting sites located 130 kb from the left YAC vector arm. Other clusters of rare cutting sites are located 170 kb and 230 kb from this vector arm.
Pulse Field Gel Electrophoresis and CpG Island Screening
Partial digestion mapping studies of the YACs 922A8, A9126, C161, and 750 were used to define the distances between clusters of rare cutting restriction enzyme sites within the region (Fig 2). Three Not I sites were located within 60 kb centromeric to D13S272, two of these being present within the genomic probe p9E4.3a, which also contains sites for Mlu I, BssHII, Sal I, and SacII. Another Not I site was located approximately 300 kb in a centromeric direction of this marker in the restriction mapping of the YAC 922A8 (data not shown). In addition clusters of rare cutting restriction endonuclease sites were identified in cosmids C7a, C6a, C30a, and C18b through rare cutting enzyme screening of cosmid inserts.
Deletion Mapping
Southern blot analysis.
A series of 229 leukemic DNA samples, in the majority of cases paired with control DNA from the same individuals, has been screened by hybridization with single copy probes including Rb1, Mgg15, and D13S25 and the novel markers p6E4.5a, p31a, p6E3.2, p9E4.3a, p9a, p25b, and p92c (Figs 3 and4 ). The frequency of hemizygous loss varied between 20% (16 of 80) at RB-1, 30% (46 of 152) at D13S25, and 31% (71 of 229) at p9E4.3, which is within 30 kb centromeric of the D13S319 locus. The greatest incidence of homozygous loss was observed using this probe, with a total of 10% (22 of 229) of cases (Table 1).
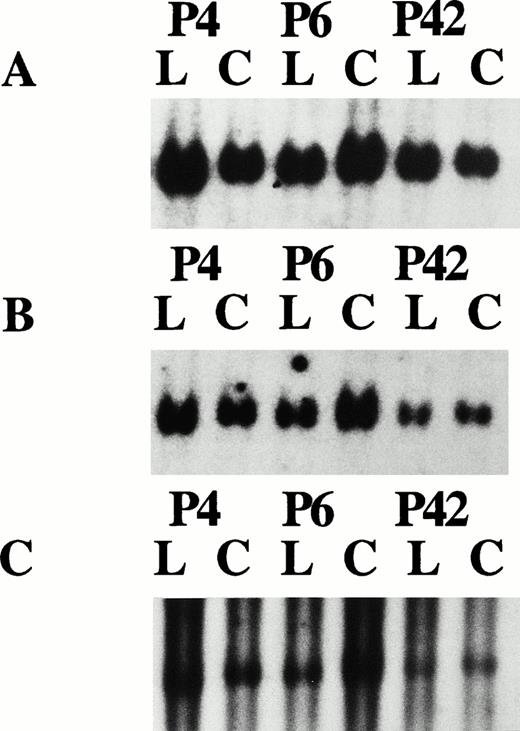
Autoradiograph results for patients P4, P6, and P42. The leukemic (L) and control (C) DNA was EcoR1 digested, size fractionated on 1% agarose gel, and hybridized with the following probes: (A) Control probe β-interferon; (B) p92c, with densitometry results of 0.94, 0.92, and 0.52, respectively, compared with the control signals showing homozygous retention, homozygous retention, and hemizygous loss, respectively; and (C) p25b, with densitometry results of 1.12, 0.92, and 0.57, respectively, compared with the control showing homozygous retention, homozygous retention, and hemizygous loss, respectively.
Autoradiograph results for patients P4, P6, and P42. The leukemic (L) and control (C) DNA was EcoR1 digested, size fractionated on 1% agarose gel, and hybridized with the following probes: (A) Control probe β-interferon; (B) p92c, with densitometry results of 0.94, 0.92, and 0.52, respectively, compared with the control signals showing homozygous retention, homozygous retention, and hemizygous loss, respectively; and (C) p25b, with densitometry results of 1.12, 0.92, and 0.57, respectively, compared with the control showing homozygous retention, homozygous retention, and hemizygous loss, respectively.
Autoradiographs of leukemic DNA samples from patients P15, P4, P7, and P13 and control (C) and leukemic (L) samples were digested with the restriction enzyme EcoRI, size fractionated through 1% agarose gel, and hybridized with the following probes: (A) control probe renin; (B) p9E4.3, with densitometry results compared with the control signals of 0.05, 0.04, 0.40, and 0.53, showing homozygous loss for P15 and P4 and hemizygous loss for samples P7 and P13, respectively; and (C) showing the same samples hybridized with the probes (I, upper band) p6E3.2, a 800-bp EcoRI MluI probe isolated from the same genomic 3-kb EcoRI fragment as the polymorphic 6E3.2 marker, with densitometry results compared with the control signals of 0.08, 0.46, 0.41, and 0.47, respectively, showing homozygous loss, hemizygous loss, hemizygous loss, and hemizygous loss, respectively and (II, lower band) p9a, a 2-kb EcoRI fragment isolated in the overlapping region between cosmids C9a and C29a, with densitometry results compared with the control signals of 0.03, 0.48, 0.42, and 0.49, respectively, showing homozygous loss, hemizygous loss, hemizygous loss, and hemizygous loss, respectively.
Autoradiographs of leukemic DNA samples from patients P15, P4, P7, and P13 and control (C) and leukemic (L) samples were digested with the restriction enzyme EcoRI, size fractionated through 1% agarose gel, and hybridized with the following probes: (A) control probe renin; (B) p9E4.3, with densitometry results compared with the control signals of 0.05, 0.04, 0.40, and 0.53, showing homozygous loss for P15 and P4 and hemizygous loss for samples P7 and P13, respectively; and (C) showing the same samples hybridized with the probes (I, upper band) p6E3.2, a 800-bp EcoRI MluI probe isolated from the same genomic 3-kb EcoRI fragment as the polymorphic 6E3.2 marker, with densitometry results compared with the control signals of 0.08, 0.46, 0.41, and 0.47, respectively, showing homozygous loss, hemizygous loss, hemizygous loss, and hemizygous loss, respectively and (II, lower band) p9a, a 2-kb EcoRI fragment isolated in the overlapping region between cosmids C9a and C29a, with densitometry results compared with the control signals of 0.03, 0.48, 0.42, and 0.49, respectively, showing homozygous loss, hemizygous loss, hemizygous loss, and hemizygous loss, respectively.
Table of Percentage Loss at Loci Tested
| . | RB1 . | 6E4.5 . | 9E4.3/D13S319 . | p92c . | D13S25 . |
|---|---|---|---|---|---|
| Hemizygous loss/total | 16/80 | 44/172 | 71/229 | 28/101 | 46/152 |
| no. analyzed | 20% | (25%) | (31%) | (28%) | (30%) |
| Homozygous loss/total | 0/80 | 17/172 | 22/229 | 8/101 | 8/152 |
| no. analyzed | 0% | (10%) | (10%) | (8%) | (5%) |
| Total loss/total | 16/80 | 61/172 | 93/229 | 36/101 | 51/152 |
| no. analyzed | 20% | (35%) | (41%) | (36%) | (35%) |
| . | RB1 . | 6E4.5 . | 9E4.3/D13S319 . | p92c . | D13S25 . |
|---|---|---|---|---|---|
| Hemizygous loss/total | 16/80 | 44/172 | 71/229 | 28/101 | 46/152 |
| no. analyzed | 20% | (25%) | (31%) | (28%) | (30%) |
| Homozygous loss/total | 0/80 | 17/172 | 22/229 | 8/101 | 8/152 |
| no. analyzed | 0% | (10%) | (10%) | (8%) | (5%) |
| Total loss/total | 16/80 | 61/172 | 93/229 | 36/101 | 51/152 |
| no. analyzed | 20% | (35%) | (41%) | (36%) | (35%) |
Microsatellite studies.
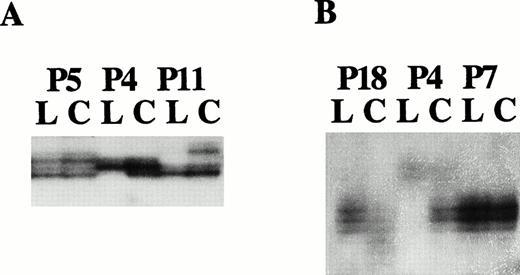
Fifty-eight patient samples were screened across the following microsatellite/RFLP polymorphism loci: RB1, D13S273, D13S319, D13S272, and D13S25. In most of the cases, large deletions encompassing several of these markers were observed. The results obtained from a select number of key patients are shown in Fig5. The novel microsatellite 6E3.2 was found to be approximately 60% informative in the patient population screened. Hemizygous loss of the markers D13S272 and 6E3.2 is shown in Fig 5A and B in a patient (P4) who showed homozygous loss by hybridizational screening with an intervening marker.
Examples of microsatellite analysis with markers (A) D13S272 and (B) 6E3.2. (A) The lanes corresponding to the leukemic (L) and control (C) cell populations of patients P5, P4, and P11 show biallelic retention, hemizygous loss, and hemizygous loss, respectively. (B) The lanes corresponding to the leukemic (L) and control (C) cell populations of patients P18, P4, and P7 show hemizygous loss, hemizygous loss, and noninformativity, respectively.
Examples of microsatellite analysis with markers (A) D13S272 and (B) 6E3.2. (A) The lanes corresponding to the leukemic (L) and control (C) cell populations of patients P5, P4, and P11 show biallelic retention, hemizygous loss, and hemizygous loss, respectively. (B) The lanes corresponding to the leukemic (L) and control (C) cell populations of patients P18, P4, and P7 show hemizygous loss, hemizygous loss, and noninformativity, respectively.
Minimal region of deletion.
Although the majority of cases showing loss had large deletions across the region, data from a few patients enabled us to delineate a minimal region of deletion within our contig. These cases are discussed next and represented diagrammatically in Fig 6.
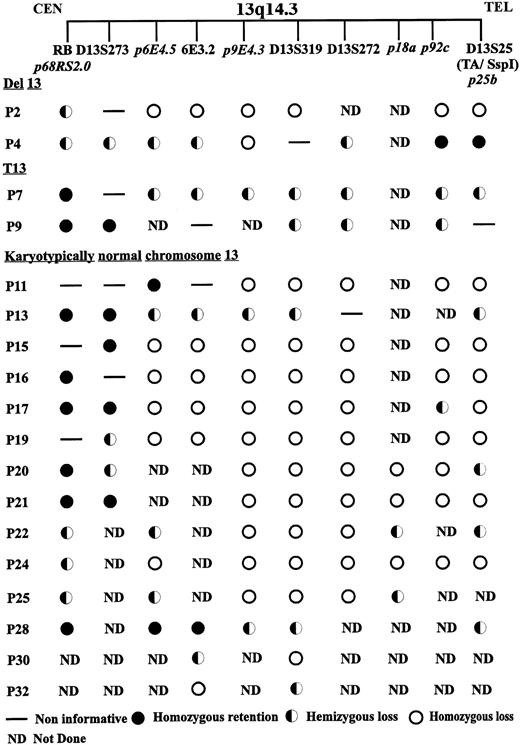
Deletion summary. Examples of microsatellite (D13S273, 6E3.2, D13S319, D13S272, and D13S25) and Southern (Rb1, p6E4.5, p9E4.3, p25b, and D13S25) analysis on key patients showing deletions in the 13q14.3 region. The table is divided into three sections with patients with karyotypic 13q deletions (del 13), karyotypic 13q translocations (t13), and patients not showing chromosome 13 rearrangement by cytogenetic analysis (karyotypically normal chromosome 13).
Deletion summary. Examples of microsatellite (D13S273, 6E3.2, D13S319, D13S272, and D13S25) and Southern (Rb1, p6E4.5, p9E4.3, p25b, and D13S25) analysis on key patients showing deletions in the 13q14.3 region. The table is divided into three sections with patients with karyotypic 13q deletions (del 13), karyotypic 13q translocations (t13), and patients not showing chromosome 13 rearrangement by cytogenetic analysis (karyotypically normal chromosome 13).
Patient no. 4, with hemizygous loss of the microsatellite markers D13S273, 6E3.2, and D13S272, and the single copy genomic probes p6E4.5, p6E3.2, and p9a (the latter two shown in Fig 4C [I] and [II]) was found to have homozygous loss of the single copy probe p9E4.3 by Southern hybridization (Fig 4B) and hemizygous retention of the MGG15 probe located at D13S319 (data not shown). Homozygous retention was observed for the probes p92a, p25b (Fig 3B and C, respectively), and the D13S25 probe. Patient no. 30, with monoallelic retention of the centromeric probe p6E3.2, showed homozygous loss of the probe p9.19, located on cosmid C9a centromeric to the marker D13S272 (Fig 7B and C, respectively). Also for the centromeric limit of deletion, patient no. P28 showed biallelic retention of probe p6E4.5 with hemizygous loss at the telomeric marker D13S25. This patient also showed hemizygous loss of the probe p9E4.3 (Fig 8B). In addition to patient P4 in determining the telomeric border to the critical region of loss, patient sample P32 showed hemizygous retention of the probe p9E4.3 and homozygous deletion of the more centromeric probe p6E3.2 (Fig 8B and C, respectively). P22 and P25 showed retention of probe p18a, hemizygously, with concurrent homozygous loss of the MGG15 probe signal at D13S319. Patient samples P26 and P27 also showed biallelic retention of the p18a probe with the retention of both p29a and the Mgg15 probes monoallelically. Two further patients, P20 and P21, also showed a similar common region of deletion. Patient no. 20 showed hemizygous loss for the p68c single copy probe, with homozygous loss of p9E4.3, and patient no. 21 had biallelic retention of the single copy probe p31a while showing homozygous loss of the MGG15 single copy probe located at the D13S319 locus. Patient no. 18 showed biallelic retention of D13S272 and hemizygous loss of D13S273 and 6E3.2 by microsatellite analysis, although Southern analysis at the D13S319 locus was not performed on this patient because of insufficient DNA. However, homozygous loss with apparent biallelic microsatellite results caused by contamination from normal cells cannot be ruled out for this patient sample at this locus, because monoallelic results were obtained at the D13S25 locus (see Fig 6). Patient sample P17 showed homozygous deletion at the markers p6E4.5, p9E4.3, p9a, and at the D13S25 locus, although uniquely this patient showed monoallelic retention of the probe p92c. These results allow us to narrow down the critical region of homozygous deletion to approximately 130 kb, between the novel marker 6E3.2 situated on cosmid C6a and D13S272 located within 5 kb telomeric to the D13S319 marker on cosmid C9a.
Leukemic samples from patients P30 and P52 wereEcoRI digested, size fractionated on 1% agarose gel, and hybridized with the following probes: (A) control probe bcl-1; (B) probe p6E3.2, with densitometry results of 0.52 in patient sample P30 compared with the P52, which had no loss detected with any 13q14 probe, showing hemizygous loss for P30; and (C) probe P9.19, which recognizes a 19 EcoRI band present in cosmid C9a, with densitometry results of 0.02 for the P30 sample compared with the P52 sample, showing homozygous loss for P30.
Leukemic samples from patients P30 and P52 wereEcoRI digested, size fractionated on 1% agarose gel, and hybridized with the following probes: (A) control probe bcl-1; (B) probe p6E3.2, with densitometry results of 0.52 in patient sample P30 compared with the P52, which had no loss detected with any 13q14 probe, showing hemizygous loss for P30; and (C) probe P9.19, which recognizes a 19 EcoRI band present in cosmid C9a, with densitometry results of 0.02 for the P30 sample compared with the P52 sample, showing homozygous loss for P30.
Control (C) and leukemic (L) samples from patient P32, used to create the cell lines 183-Ccl and 183-E95, respectively, and leukemic sample from patient 28 were HindIII digested, fractionated on 1% agarose, and hybridized with the following probes: (A) control probe bcl-1; (B) p9E4.3, with densitometry results compared with control signals of 0.45 and 0.45, respectively, showing hemizygous loss and hemizygous loss, respectively; and (C) probe p6E.32, with densitometry results compared with control signals of 0.02 and 0.79 showing homozygous loss and biallelic retention, respectively.
Control (C) and leukemic (L) samples from patient P32, used to create the cell lines 183-Ccl and 183-E95, respectively, and leukemic sample from patient 28 were HindIII digested, fractionated on 1% agarose, and hybridized with the following probes: (A) control probe bcl-1; (B) p9E4.3, with densitometry results compared with control signals of 0.45 and 0.45, respectively, showing hemizygous loss and hemizygous loss, respectively; and (C) probe p6E.32, with densitometry results compared with control signals of 0.02 and 0.79 showing homozygous loss and biallelic retention, respectively.
DISCUSSION
Recessive genes involved in the prevention of oncogenesis are known as tumor suppressor genes, and biallelic loss of function is required for phenotypic expression of neoplasia.34 With the aim of delineating the critical region of homozygous 13q14.3 loss in patients with B-CLL we have constructed a detailed cosmid and PAC contig across the region. High resolution mapping studies were focussed on the region centromeric to the marker D13S25 and telomeric to the Rb1 locus.15,19 The contig produced has enabled not only the ordering of closely spaced markers, but also the isolation of novel polymorphic markers and the definition of the relevant area to a 130-kb region. A number of CpG island probes have been identified and isolated. These have proven useful for deletion studies performed on patient samples, as landmark probes in the physical map of the region, for pulse field gel studies, and as markers for the probable location of genes in the region. In the majority of cases in which loss was observed, the extent of the hemizygous or homozygous deletion detected by the methods used in this study, ie, Southern hybridization and microsatellite analysis, extended beyond the centromeric or telomeric limits of the contig. However, in a select group of patients the boundaries of deletion could be delineated within the contig borders. Combining the results from 12 key patients defines the critical region to between the markers 6E3.2 and D13S272, a distance of less than 130 kb, which is covered by a series of overlapping cosmid and PAC clones in our map (Fig 1). The critical region defined in this study is centromeric to a recently mapped minimal area of deletion by Bullrich et al35 in a study of sixty cases. Stilgenbauer et al,36 who screened 131 cases by FISH alone, determined a similar centromeric pattern of deletion to the present investigation, defining the critical area between D13S273 and D13S272, a distance of 210 kb to 600 kb according to our map.
Although 229 patient samples were screened in this study, only 12 cases significantly contributed to the resolution of the region of critical loss. This suggests that a large patient sample may be necessary for accurate analysis of the region. Nevertheless, Kalachikov et al37 defined a minimally deleted region of 300 kb based on the analysis of 58 patients, which encompassed the region defined in this study and partially overlapped the centromeric limit of the region defined by Bullrich et al.35 The markers telomeric to D13S319 have been examined in some detail in these studies. The data presented here resolved more precisely the region between 6E3.2 and D13S272 and suggested that telomeric loci lie outside the region of loss seen in the majority of 13q14.3 deletions.
The possibility of several distinct regions of chromosome 13 deletion has been recently proposed. Garcia-Marco et al38 show 13q12 deletions in B-CLL cases, which also show loss of the D13S25 locus despite the retention of an intervening locus, D13S171. Bullrich et al35 also found evidence of a number of separate regions of deletion telomeric to the locus D13S272. One patient sample in our study, P17, does indeed show evidence of two separate regions of biallelic deletion, one encompassing the D13S272 region and another at the D13S25 locus with an intervening region of hemizygous loss. In a recent study of chromosome 11q23 deletion in patients with lymphoproliferative disorders several different regions of deletion were described.39 This study concluded that one major region was found deleted in the majority, although by no means not all of the cases showing loss by FISH. Our results indicated, likewise, that a single region of deletion centromeric to D13S272 can be observed in the majority of 13q14.3 loss cases. This area of deletion contains at least three CpG islands present in cosmids C6a, C30a, and C9a and includes three Not I sites. As yet no genes proven to be associated with hematologic conditions have been located within this area.40 The recent identification of multiple putative expressed transcripts located between the markers D13S273 and D13S25,37,41 in addition to the clustering of CpG islands in the region centromeric to D13S319 reported here, suggests that the area is relatively gene rich.42 The detailed map produced in this study facilitates the further delineation of the minimal region of loss found at 13q14.3 in many B-CLL patients and the subsequent isolation of the putative tumor suppressor gene. This work is now in progress.
M.M.C., O.R., D.G.O., and S.E. contributed equally to this work.
Supported by grants from the Kay Kendall Leukaemia Research Fund, the King Gustav V Jubilee Fund, the Cancer Society of Sweden, the Felix Mindus Fund for Leukemia Research, the Russian HUGO program, and the US DOE Human Genome Program Grant No. OR00033-93CIS016.
Address reprint requests to David G. Oscier, FRCP, Molecular Biology Laboratory, Royal Bournemouth General Hospital, Bournemouth BH7 7DW, UK.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal